ABSTRACT
The full-length cDNA sequence of a porcine gene, MOSPD2, was amplified using the rapid amplification of cDNA ends method based on a pig expressed sequence tag sequence which was highly homologous to the coding sequence of the human MOSPD2 gene. Sequence prediction analysis revealed that the open reading frame of this gene encodes a protein of 491 amino acids that has high homology with the motile sperm domain-containing protein 2 (MOSPD2) of five species: horse (89%), human (90%), chimpanzee (89%), rhesus monkey (89%) and mouse (85%); thus, it could be defined as a porcine MOSPD2 gene. This novel porcine gene was assigned GeneID: 100153601. This gene is structured in 15 exons and 14 introns as revealed by computer-assisted analysis. The phylogenetic analysis revealed that the porcine MOSPD2 gene has a closer genetic relationship with the MOSPD2 gene of horse. Tissue expression analysis indicated that the porcine MOSPD2 gene is generally and differentially expressed in the spleen, muscle, skin, kidney, lung, liver, fat and heart. Our experiment is the first to establish the primary foundation for further research on the porcine MOSPD2 gene.
Introduction
Motile sperm domain-containing protein 2 (MOSPD2) is one of the major sperm proteins which are involved in sperm motility. These proteins, including MOSPD1 and MOSPD3, oligomerize to form filaments and are defined by the presence of the major sperm protein domain and two transmembrane domains. These genes are generally strongly expressed in mesenchymal tissues and are up-regulated during differentiation in osteoblastic, myoblastic and adipocytic cell lines. It has been suggested that MOSPD1 plays a pivotal role in the developmental regulation at the switch between mesenchymal and epithelial cells [Citation1–3]. Gene-environment analysis reported that MOSPD2 may be related to the obesity traits among postmenopausal African-American and Hispanic women [Citation4]. This gene is also involved in the dynamic protein interaction of the human centrosome--cilium interface [Citation5].
As mentioned earlier, the MOSPD2 gene is an important gene which has many biological functions. Until today, the MOSPD2 gene has been reported in horse, human, chimpanzee, rhesus monkey, mouse and other animals. To the best of our knowledge, the pig MOSPD2 gene has not been reported yet.
The aim of the present study was to clone the full-length cDNA sequence of the porcine MOSPD2 gene, and further do necessary sequence analysis and tissue expression analysis. These will establish the primary foundation of understanding this porcine gene.
Materials and methods
Animals and sample preparation
Five adult Yunnan local pigs were slaughtered. Spleen, muscle, skin, kidney, lung, liver, fat and heart samples were collected, frozen in liquid nitrogen and then stored at −80 °C. The total RNA was extracted using the Total RNA Extraction Kit (Invitrogen, Carlsbad, CA, United States) [Citation6–8]. These RNA samples were used to perform rapid amplification of cDNA ends (RACE) polymerase chain reaction (PCR) and tissue expression profile analysis.
5'- and 3'-RACE
5'- and 3'-RACE were performed to isolate the full-length cDNA of the porcine MOSPD2 gene, according to the instructions of the SMART™ RACE cDNA Amplification Kit (Clontech, Palo Alto, CA, United States). For the porcine MOSPD2 gene, the gene-specific primers (GSPs) were designed based on one pig expressed sequence tag (EST) sequence whose sequence is highly homologous to the coding sequence of the human MOSPD2 gene: EW335266. The GSPs were: 5′-RACE GSP: 5′-CCCCCGTGGGGAGACACAACTATAT-3′, 3′-RACE GSP: 5′-GGGCCCTTATTACACATCAGCCCAG-3′. RACE touchdown PCRs were carried out with 5 cycles of 94 °C/30 s and 72 °C/3 min, followed by 5 cycles of 94 °C/30 s, 68 °C/30 s and 72 °C/3 min, finally with 30 cycles of 94 °C/30 s, 67 °C/30 s and 72 °C/3 min to terminate the reaction. The RACE PCR products were then cloned into a pMD18-T vector (TaKaRa, Dalian, China) and sequenced bidirectionally with the commercial fluorometric method (SHENGGONG, Shanghai, China). At least five independent clones were sequenced for each PCR product.
Quantitative real-time PCR (qRT-PCR) for tissue expression profile analysis
qRT-PCR for evaluating the level of mRNA for the porcine MOSPD2 gene was performed on the ABI Prism 7300 Sequence Detection Systems (Applied Biosystems, Foster City, CA, United States). A 25 µL PCR reaction volume contained 1 µL of SYBR Green real-time PCR Master Mix, 100 ng of cDNA template and 200 nmol/L of each primer. The housekeeping gene beta-actin (DQ845171) was used as the internal control. The control gene primers used were: 5′-CGGGACATCAAGGAGAAGC-3′ (forward primer 1) and 5′-TACTTGCGCTCTGGAGGC-3′ (reverse primer 1). The PCR product is 383 bp in length. The MOSPD2 GSPs were: 5′-CCCATTGCCAGCGAGGAT-3′ (forward primer 2) and 5′-CACCAGGATCACAGCTGC-3′ (reverse primer 2). The PCR product is 382 bp in length. The conditions for real-time PCR were: an initial denaturation at 95 °C for 3 min, 40 cycles of 95 °C for 15 s, 58 °C for 15 s and 72 °C for 20 s. For each sample, reactions were set up in triplicate to ensure the reproducibility of the results. The gene relative expression levels were quantified relative to the expression of the reference gene, beta actin, by employing the 2−ΔΔCt value mode [Citation9].
Sequence analysis
The gene sequence prediction was done using GenScan software (http://genes.mit.edu/GENSCAN.html). The protein analysis was performed using the Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information (NCBI) server (http://www.ncbi.nlm.nih.gov/BLAST) and the ClustalW software (http://www.ebi.ac.uk/clustalw). The pig genome database is publicly available at the NCBI Pig Genome Resources (http://www.ncbi.nlm.nih.gov/projects/genome/guide/pig/).
Results and discussion
RACE results for the porcine MOSPD2 gene
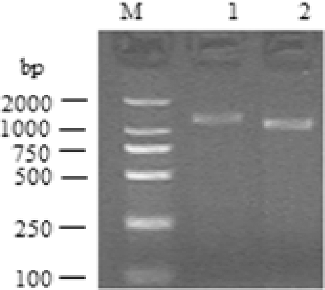
In the present study, we first obtained the full length of the porcine MOSPD2 gene cDNA by using 5′- and 3′-RACE. For the porcine MOSPD2 gene, through 5′-RACE, one PCR product of 1301 bp was obtained. The 3′-RACE product was 1062 bp. These products were then cloned into a T-vector and sequenced. Taken together, a 2148 bp cDNA complete sequence was finally obtained ().
Sequence analysis
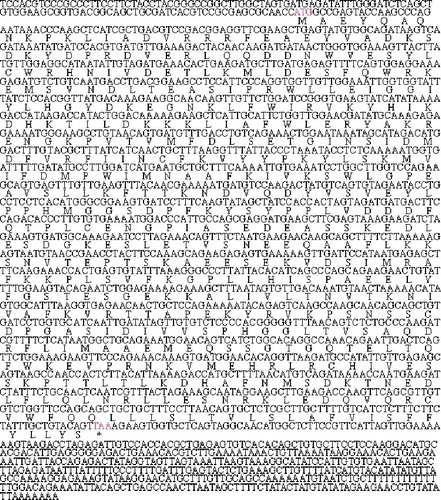
The nucleotide sequence analysis using the BLAST software at the NCBI server revealed that this gene was not homologous to any of the known porcine genes and it was then deposited into the GenBank database (accession number: GU373667). The sequence prediction carried out using the GenScan software revealed an open reading frame (ORF) encoding 416 amino acids in the 2148 bp cDNA sequence. The complete cDNA sequence of this gene and the encoded amino acids are shown in
Figure 2. Complete cDNA sequence and encoded amino acids of the MOSPD2 gene (GenBank accession number: GU373667). ATG indicates start codon; TAA indicates stop codon; * indicates the stop codon.
Further BLAST analysis of this protein revealed that this protein has high homology with the motile sperm domain-containing protein 2 (MOSPD2) of five species: horse (accession number: XP_001917228; 89%), human (accession number: NP_689794; 90%), chimpanzee (accession number: XP_520943; 89%), rhesus monkey (accession number: XP_001100061; 89%) and mouse (accession number: NP_084006; 85%) ().
Figure 3. Alignment of the MOSPD2 proteins from pig, horse, human, chimpanzee, rhesus monkey and mouse.

From the sequence analysis described earlier, this gene can be defined as the porcine MOSPD2 gene. This novel porcine gene was assigned GeneID: 100153601. Based on the result of sequence comparison, it can be seen that the porcine MOSPD2 protein is highly homologous with the MOSPD2 proteins of horse, humans, chimpanzee and other mammals. This suggests that the MOSPD2 genes are highly conserved in these mammals and these mammal MOSPD2 genes might have similar functions. We also found that the pig MOSPD2 protein does not show complete identity to horse (89%), human (90%), chimpanzee (89%) or other mammals. This implies that the functions of the porcine MOSPD2 gene might have some differences to those of other mammals.
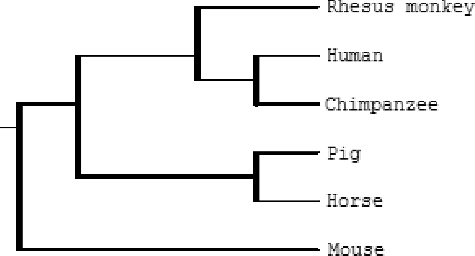
Based on the results of the alignment of the MOSPD2 gene from 30 different species, a phylogenetic tree was constructed using the ClustalW software, as shown in . The phylogenetic tree analysis revealed that the pig MOSPD2 gene has a closer genetic relationship with the horse MOSPD2 gene than with those of human, chimpanzee, rhesus monkey and mouse. This suggests that the horse may be selected as a model animal in studies on the functions of the pig MOSPD2 gene.
To obtain the genomic DNA of the MOSPD2 gene, the publicly available pig genome database was screened using the ORF cDNA sequence of the MOSPD2 gene as a seed. A bacterial artificial chromosome clone (Sus scrofa chromosome X clone CH242-290D21, GenBank accession no. CU487221), which encompasses the entire MOSPD2 gene, was identified by BLASTGen analysis. The pig MOSPD2 gene (nucleotides 14,681–76,953 in the Sus scrofa chromosome X clone CH242-290D21) is 62,273 bp in length and consists of 15 exons. All exon–intron splice junction sequences conform to the GT–AG rule ().
With the establishment of the pig EST database and the pig genome database along with convenient bioinformatics analysis tools, useful ESTs or pig genomic sequences that are highly homologous to the coding sequence of some human genes can be found [Citation10,Citation11]. Based on these porcine sequences, we can obtain the complete coding sequences of some unknown porcine genes through some modern experimental methods such as RACE and RT-PCR (Reverse Transcription-Polymerase Chain Reaction). From the cloning of the porcine MOSPD2 gene, it could be seen that this is an effective method to isolate some novel porcine genes.
Tissue expression profile
The qRT-PCR analysis of the tissue expression profile was carried out using the tissue cDNAs of one adult pig as the templates. The tissue expression analysis indicated that the porcine MOSPD2 gene is generally and differentially expressed in the spleen, muscle, skin, kidney, lung, liver, fat and heart ().
Figure 6. Tissue expression profile analysis of the porcine MOSPD2 gene: 1, skin; 2, kidney; 3, heart; 4, spleen; 5, fat; 6, muscle; 7, liver; 8, lung.
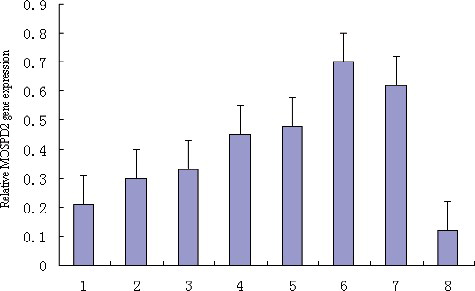
The MOSPD1 gene has been reported to be generally strongly expressed in mesenchymal tissues [Citation3]. Interestingly, the tissue distribution analysis in our experiment revealed that the expression of the porcine MOSPD2 gene was generally detected in the spleen, muscle, skin, kidney, lung, liver, fat and heart, with particularly strong expression in muscle and liver. Therefore, the relationship between the function and expression of the MOSPD2 gene deserves further study in order to explain the observed differences between the tissue expression profiles of the MOSPD1 gene and the porcine MOSPD2 gene.
Conclusions
To the best of our knowledge, this is the first report of isolation of the porcine MOSPD2 gene. The sequence analysis and tissue transcription profile analysis established the primary foundation for further insight into this novel porcine gene.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- King KL, Stewart M, Roberts TM, et al. Structure and macromolecular assembly of two isoforms of the major sperm protein (MSP) from the amoeboid sperm of the nematode, Ascaris suum. J Cell Sci. 1992;101(Pt 4):847–857.
- Bullock TL, Roberts TM, Stewart M. 2.5 Å resolution crystal structure of the motile major sperm protein (MSP) of Ascaris suum. J Mol Biol. 1996;263(2):284–296.
- Thaler R, Rumpler M, Spitzer S, et al. MOSPD1, a new player in mesenchymal versus epidermal cell differentiation. J Cell Physiol. 2011;226(10):2505–2515.
- Velez Edwards DR, Naj AC, Monda K, et al. Gene-environment interactions and obesity traits among postmenopausal African-American and Hispanic women in the Women's Health Initiative SHARe Study. Hum Genet. 2013;132(3):323–336.
- Gupta GD, Coyaud É, Gonçalves J, et al. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 2015;163(6):1484–1499.
- Liu YG, Gao SZ. A novel sheep gene, MMP7, differentially expressed in muscles from black-boned sheep and local common sheep. J Appl Genet. 2009;50:253–256.
- Liu YG, Gao SZ. Molecular cloning, sequence identification and tissue expression profile of three novel sheep (Ovis aries) genes – BCKDHA, NAGA and HEXA. Biol Res. 2009;42:69–77.
- Liu YG. A novel HADHA gene differentially expressed in muscle and other tissues from black-boned vs. ordinary sheep. Anim Sci Pap Rep. 2009;27:127–137.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Lim D, Cho YM, Lee KT, et al. The Pig Genome Database (PiGenome): an integrated database for pig genome research. Mamm Genome. 2009;20:60–66.
- Uenishi H, Eguchi-Ogawa T, Shinkai H, et al. PEDE (Pig EST Data Explorer) has been expanded into Pig Expression Data Explorer, including 10 147 porcine full-length cDNA sequences. Nucl Acids Res. 2007;35(Database issue):D650–653.