ABSTRACT
In the Erwinia amylovora genome, the hrp gene cluster containing the dspA/E/EB/F operon plays a crucial role in mediating the pathogenicity and the hypersensitive response (HR) in the host plant. The role of the dspA/E gene derived from E. amylovora was investigated by monitoring the expression of the β-glucuronidase (GUS) reporter system in transgenic Arabidopsis thaliana cv. Pri-Gus seedlings. A mutant ΔdspA/E strain of E. amylovora was generated to contain a deletion of the dspA/E gene for the purpose of this study. Two-week-old seedlings of GUS transgenic Arabidopsis were vacuum-infiltrated with the wild-type and the mutant (ΔdspA/E) E. amylovora strains. The Arabidopsis seedlings were fixed and stained for GUS activity after 3–5 days following infiltration. The appearance of dense spots with blue staining on the Arabidopsis leaves indicated the typical characteristic of GUS activity. This observation indicated that the wild-type E. amylovora strain had induced a successful and efficient infection on the A. thaliana Pri-Gus leaves. In contrast, there was no visible GUS expression on leaf tissues which were inoculated with the ΔdspA/E mutant E. amylovora strain. These results indicate that the dspA/E gene is required by the bacterial cells to induce HR in non-host plants.
Introduction
Fire blight is a destructive and economically significant disease affecting apples, pears and many other Rosaceae family plants. It is usually caused by the bacterial plant pathogen Erwinia amylovora [Citation1]. Genetic analyses have identified a multiple gene system of E. amylovora that functions in inducing hypersensitive reaction (HR) in non-host plants and pathogenicity in host plants [Citation2,Citation3]. Most Gram-negative phytopathogenic bacteria have hrp genes essential for pathogenesis and HR. Some mutations in the hrp genes of E. amylovora inhibit the pathogenicity in susceptible host plants and impair the ability to elicit HR in resistant hosts or non-host plants [Citation4]. In the E. amylovora hrp system, two regulatory components, namely hrpX and hrpY, have been identified [Citation5]. Thus, the understanding on the regulation of hrp genes is considered important in elucidating the molecular mechanism of HR elicitation and pathogenesis [Citation6]. The hrp gene cluster region of E. amylovora contains the 6.6-kb dspA/EB/F operon, which is homologous to avrE of Pesudomonas syringae pv. tomato. The disease-specific effector DspA/E is a large protein (198 kDa), whereas the dspB/F-encoded protein is much smaller (16 kDa) [Citation4,Citation7–9]. Indirect evidence for DspA/E translocation into the plant cell was obtained by using a DspA/E-cya fusion protein [Citation10] and it was further demonstrated that the transient expression of DspA/E can induce cell death in Malus domestica, Nicotiana tabacum, Nicotiana benthamiana, and yeast [Citation11,Citation12]. The E. amylovora DspE/A protein interacts with an apple-like receptor protein kinase, indicating its involvement in the suppression of host defences [Citation13].
The GUS gene fusion system (Escherichia coli β-D-glucuronidase gene) described by Jefferson is often used in assessing the gene activities and in developing the molecular genetic analysis in transgenic plants [Citation14,Citation15]. The GUS system offers several advantages over other potential markers, as it is a sensitive assay system in quantifying or localizing the enzyme expression in tissues and has minimal activity interfering with the assay in most plant species [Citation16]. Common substrates for the enzyme include methylumbelliferylglucuronide, which is broken down to give an easily quantified fluorogenic compound, methlyumbelliferone, and the compound 5-bromo-4-chloro-3-indoyl-glucuronide (X-gluc), which is broken down to give an indigo dye that can represent a gene expression site. The versatility in the use of this system has been shown in animals, plants, bacterial, and fungal cells [Citation15,Citation17,Citation18]. Arabidopsis thaliana, a well-known model plant with established lab culture conditions, is very suitable for gene expression studies [Citation19]. In this study, the role of the dspA/E gene of E. amylovora was investigated by monitoring the expression of the β-glucuronidase (GUS) reporter gene in transgenic A. thaliana cv. Pri-Gus leaves.
Materials and methods
Plant material
Arabidopsis thaliana cv. Pri-Gus seedlings (Arabidopsis Biological Resource Center, Columbus, OH, USA) were used in this study. Seeds were sterilized overnight (18–20 h) by means of a vapour-phase sterilization method [Citation20]. Seedlings of A. thaliana were grown on Murashige and Skoog medium (15 g⋅L−1 sucrose and 8 g⋅L−1 Phytagar) in Petri dishes for two weeks at 18–20 ºC, 70% relative humidity and a 16/8 h photoperiod [Citation21].
Bacterial strain
The wild-type strain of E. amylovora from our collection was used in this experiment. This strain was grown on nutrient glucose agar (NGA: 3.0 g⋅L−1 beef extract; 5 g⋅L−1 peptone; 2.5 g⋅L−1 glucose; 15.0 g⋅L–1 agar) at 26–28 ºC.
Mutation of dspA/E
The DNA fragment of the wild-type E. amylovora containing dspA/E was purified by polymerase chain reaction (PCR). The purified fragment was cloned in a pCR®4 vector unit using a TOPO TA Cloning® Kit (Invitrogen, Paisley, UK). This was transformed into electroSHOX competent Escherichia coli cells (Bioline, Taunton, MA, USA) by following the manufacturer's instructions. The plasmid containing the dspA/E gene insert was extracted using a plasmid DNA midiprep kit (Qiagen, Hilden, Germany). The presence of the inserts was confirmed by agarose gel electrophoresis run in 1.5% w/v Ultrapure™ agarose (Invitrogen, Paisley, UK). Sequencing of the purified plasmid was done in an automated DNASTAR Analyser (Madison, WI, USA). The purified pCR4-TOPO vector/dspA/E plasmid was prepared from competent E. amylovora, according to Sambrook et al. [Citation22]. The purified pCR4-TOPO vector/dspA/E plasmid obtained from competent E. coli was digested with an excess of EcoRV (Invitrogen, Paisley, UK) in order to perform deletion of the dspA/E gene. The effectiveness of the digestion procedure was monitored by gel electrophoresis. Then, the digested pCR4-TOPO vector/ΔdspA/E plasmid was transformed into competent E. amylovora by electroporation with a Bio-Rad™ Micro Pulser unit (Hercules, CA, USA). The transformation of the digested pCR4-TOPO vector/ΔdspA/E was assayed by using competent E. amylovora cells prepared as follows: 2 μL of digested pCR4-TOPO vector/ΔdspA/E was mixed with 25 μL of competent E. amylovora. Transformation was completed by using standard procedures [Citation23]. The cells were streaked onto NGA agar plates supplemented with kanamycin in order to score the formation of transformed colonies (kanamycin resistance, Kmr).
PCR amplification
The quality and quantity of the DNA were measured in a NanoDrop® ND-1000 spectrophotometer (Thermo Fisher Scientific,Wilmington, MA, USA). PCR amplifications were carried out in a 100 μL reaction volume, containing 100 ng of genomic DNA; 0.2 μmol⋅L−1 of each dspA/E primer (dspA/E/iF: 5'-ATGGAATTAAAATCACTGGGAACTGAA-3' (forward); dspA/E/iR; 5'-TTAGCTCTTCATTTCCA GCCCTTCCTT-3' (reverse)), 0.25 mmol⋅L−1 of each deoxynucleoside triphosphate (dNTP; Invitrogen, Paisley, UK), 2 mmol⋅L−1 MgCl2, 1×Taq buffer and 2.5 U of Taq DNA polymerase for identification of E. amylovora. PCR was performed in a MyCycler™ thermal cycler (BioRad, Hercules, CA, USA) with initial denaturation at 95 ºC for 4 min, then 10 cycles using a touch-down strategy (an initial cycle of 94 ºC for 30 s, 65 ºC for 30 s, 72 ºC for 2 min, then lowering the annealing temperature for each cycle by 1 ºC during the following 9 cycles), followed by 25 cycles at 95 ºC for 30 s, 56 ºC for 30 s, and 72 ºC for 2 min. Amplified PCR products were electrophoresed on 1.5% w/v Ultrapure™ agarose gels (100 V).
Infiltration of bacterial population
Arabidopsis thaliana cv. Pri-Gus leaves were infiltrated with the wild-type and the ΔdspA/E mutant strain of E. amylovora for GUS activity staining. The wild type of E. amylovora grown on NGA and the ΔdspA/E mutant strain grown on NGA with Km were incubated at 26–28 °C. These isolates were then suspended in sterile distilled water at 107 cfu⋅mL−1, corresponding to optical density at 600 nm (OD600) of 0.1. Sterile distilled water was used as a negative control on plant leaves. Bacterial suspensions were injected with a 1 mL syringe, without a needle, into the leaves through the stomatal pores on the abaxial surface. In this assay, the wild-type and the ΔdspA/E mutant strain of E. amylovora were inoculated in the leaves of the A. thaliana Pri-Gus cultivar to elicit HR.
Bacterial population density
Bacterial populations of both the wild-type and the ΔdspA/E mutant E. amylovora strain on the leaves of the A. thaliana cv. Pri-Gus were assessed at 0, 24, 48, and 72 h after infiltration. At each sampling point, cells were recovered from necrotic tissues cuts by suspension in NGA broth and plating on NGA agar medium at appropriate dilution. The bacterial growth following two days of incubation at 26—28 ºC was scored as colony-forming units per millilitre (CFU/mL). This was carried out in a completely randomized plot design, and the infiltration assay was performed twice.
GUS activity assay
This assay was done according to Allen et al. [Citation24] to monitor the expression of the GUS reporter gene in A. thaliana cv. Pri-Gus leaves. Leaves infiltrated with the wild-type and ΔdspA/E mutant strain were collected and rinsed in 50 mmol⋅L−1 Na-phosphate buffer (pH 7.0). Then, these leaves were stained with 2 mmol⋅L−1 of 5-bromo-4-chloro-3-indolyl glucuronide (X-gal from Biosynth, Staad, Switzerland) in 50 mmol⋅L−1 of Na-phosphate buffer (pH 7.0), followed by brief vacuum infiltration and were then placed at 37 ºC in darkness overnight. After staining, the leaves were rinsed extensively in ethanol to remove chlorophyll before examination. GUS activity was evaluated on the basis of X-gluc staining visualized under a stereomicroscope. The percentage of GUS-positive seedlings was calculated by scoring the number of seedlings with X-gluc staining compared to the unstained seedlings.
Data analysis
Logarithmic transformation of the colony-forming units (CFU) was used to normalize the data distribution. All data were analysed by the two-way analysis of variance procedure of the SPSS 15.0 statistical software package (SPSS Inc., Cary, NC, USA). Means were separated by using Turkey's multiple comparison test and differences were considered significant at P < 0.05 for all tests.
Results and discussion
Mutation of dspA/E
Erwinia amylovora harbouring the DspA/E virulent factor in their genome cause major economic losses in the agriculture produce from Rosaceae family plants. As a first step in this study, a 670-bp PCR product corresponding to the dspA/E gene of E. amylovora was successfully amplified by using the dspAEiF/dspAEiR primer pair. This fragment was successfully cloned into a pCR4 vector. Then, the dspA/E region of the purified plasmid was digested by using EcoRV in order to compare the digested plasmid contents of the mutant ΔdspA/E strain to that of the wild type. The restriction of the large plasmids present in the wild type showed a band profile different from that of the ΔdspA/E mutant strain (result not shown). The analysis of the plasmid contents of the ΔdspA/E mutant strain transformed with TOPO vector showed that the mutant strains were stable and the deletion was maintained after repeated sub-culturing on NGA supplemented with Km.
GUS activity assay
In this study, the role of the E. amylovora dspA/E gene for induction of HR in the transgenic A. thaliana plants was demonstrated by the expression of the GUS reporter gene. To determine whether the expression of the GUS reporter gene could be affected by the dspA/E gene of E. amylovora, A. thaliana cv. Pri-Gus leaves were infiltrated with the wild-type E. amylovora strain carrying the DspA/E effector and also with the ΔdspA/E mutant strain of E. amylovora deficient in DspA/E. Dense spots on the leaves revealed the typical blue staining characteristic of GUS activity, indicating that infiltration of dspA/E was very efficient in A. thaliana Pri-Gus leaves. In contrast, no GUS-expressing tissues were observed on the leaves inoculated with sterile distilled water used as a control (). This indicates that the infiltration of the dspA/E gene was successful and efficient in A. thaliana Pri-Gus leaves. Thus, we demonstrated that the GUS activity assay system proved to be a quick and easy method to test for reporter gene constructs in Arabidopsis seedlings prior to stable transformation. It is also considered useful in understanding the effect of gene regulation in plant resistance.
Figure 1. Arabidopsis leaves stained for GUS activity. Leaves infiltrated with: sterile water (negative control) (a), mutant ∆dspA/E strain (b), and wild-type E. amylovora strain (c).
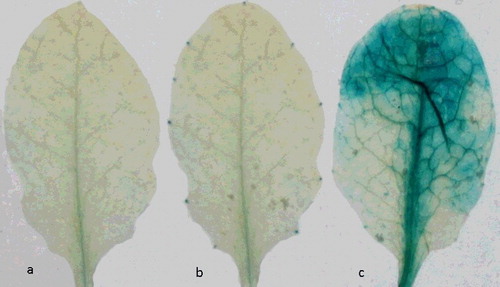
In our experiments, the observation of strong and spotty staining on the leaves showed that the efficiency of infection in Pri-Gus leaves was higher, whereas there was no observable GUS expression on leaves inoculated with the mutant ΔdspA/E strain. The blue spots appeared on the infected leaves over the same time scale as the necrotic spots appeared, indicating that dspA/E was a major cell-death inducer in the non-host plant A. thaliana.
Bacterial density
Although there were no differences between the leaves infiltrated with the wild-type and those infiltrated with the ΔdspA/E mutant at 0 h, the growth of the mutant ΔdspA/E strain was significantly higher compared to the wild type of E. amylovora at 48 h after the bacterial inoculation (P < 0.05). In addition, there was a marked increase in the growth of the mutant strain on the leaves at 72 h post inoculation (). The growth area affected by the wild-type strain was lesser than that affected by the mutant strain and this was most likely due to the stimulation of the immune response by the effector protein expressed in the wild-type strain (). Conversely, the growth area affected by the mutant strain was greater within the tissues, which could possibly be attributed to lack of stimulation of the immune response by this mutant strain.
Figure 2. Bacterial growth in leaves of A. thaliana after inoculation of E. amylovora strains (OD600 = 0.1) or sterile distilled water (control).
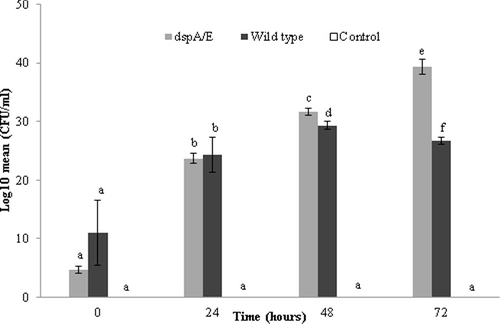
Figure 3. Bacterial growth in Arabidopsis leaves infiltrated with sterile water (negative control) (a), mutant ∆dspA/E strain (b), and wild-type E. amylovora strain (c).

The observed bacterial growth reduction may be attributable to the major role of the dspA/E gene, which is known to induce HR efficiently in tobacco and Arabidopsis spp. [Citation25,Citation26]. This is in agreement with previous reports that dspA/E mutants trigger HR in tobacco [Citation6,Citation27,Citation28]. Moreover, in a study by Laura et al. [Citation28], E. amylovora was shown to be unable to satisfy its arginine demand from host tissues, either at the beginning of infection or later when the disease is at an advanced stage, indicating that E. amylovora needs an intact arginine biosynthesis pathway to cause fire blight disease in apple trees and immature apple and pear fruits.
Our results that a highly noticeable leaf area was affected by the growth of the ΔdspA/E mutant strain could be considered evidence for the lack of immune stimulation upon infection with this strain. Conversely, the smaller leaf area affected by the growth of the wild-type strain was possibly due to the immune system triggered by the effector DspA/E, which is present in the wild-type strain. Based on these results, it could be speculated that the mutated strain had failed to positively regulate or induce protein-mediated resistance in the non-host plant against the pathogenic strain, reflecting the role of the host plant immune response in limiting bacterial growth.
Conclusions
The results from this study suggested that the virulence factor DpsA/E was highly essential and involved in the stimulation of the plant immune system in transgenic A. thaliana. Based on three investigated parameters, i.e. HR induction, bacterial growth in plant tissues, and GUS expression in the infiltrated leaves, it was observed that the GUS reporter gene was induced after infitration of E. amylovora cells harbouring the dspA/E gene into A. thaliana Pri-Gus leaves. However, the presence of the DspA/E virulence factor of E. amylovora seemed to suppress the invading bacterial growth due to activation of HR. In contrast, the ΔdspA/E mutant strain deficient in the virulent factor DspA/E was unable to show GUS expression on A. thaliana Pri-Gus leaves. Due to the lack of immune stimulation, the growth area affected by the ΔdspA/E mutant strain was found to be larger within the infected tissues. This study demonstrated that the virulence factor DspA/E is essential in E. amylovora pathogenicity in non-host plants. The pathogenicity or outcome of the invasion could depend on the intricate balance between virulence factor expression, the immune response of the host, and the growth of the invading pathogen.
Acknowledgments
We would like to thank Assoc. Dr Soner Cankaya of Ondokuz Mayis University, Samsun, Turkey for statistical analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Lagonenko AL, Sadovskaya O, Valentovich LN, et al. Characterization of a new ViI-like Erwinia amylovora bacteriophage phiEa2809. FEMS Microbiol Lett [Internet]. 2015 [cited 2016 Feb 17];362(7):fnv031. Available from: http://femsle.oxfordjournals.org/content/362/7/fnv031.
- Vanneste JL, Paulin JP, Expert D. Bacteriophage Mu as a genetic tool to study Erwinia armylovora pathogenicity and hypersensitive reaction on tobacco. J Bacteriol. 1990;172:932–941.
- Bauer DW, Beer SV. Further characterization of an hrp gene cluster of Erwinia amylovora. Mol Plant Mic Interac. 1991;4:493–499.
- Mor H, Manulis S, Zuck M, et al. Genetic organization of the hrp gene cluster and dspAE/BF operon in Erwinia herbicola pv. gypsophilae. Mol Plant Mic Interac. 2001;14:431–436.
- Núria P, Miñana-Galbis D, Merino S, et al. Virulence factors of Erwinia amylovora: a review. Int J Mol Sci. 2015;16:12836–12854.
- Wei ZM, Laby RJ, Zumoff CH, et al. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science. 1992;257:85–88.
- Lorang JM, Keen NT. Characterization of avrE from Pseudomonas syringae pv. tomato: a hrp-linked avirulence locus consisting of at least two transcriptional units. Mol Plant Mic Interac. 1995;8:49–57.
- Gaudriault S, Brisset MN, Barny MA. HrpW of Erwinia amylovora, a new Hrp-secreted protein. FEBS Lett. 1998;428:224–228.
- Bogdanove AJ, Bauer DW, Beer SV. Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the Hrp (type III secretion) pathway. J Bacteriol. 1998;180:2244–2247.
- Bocsanczy AM, Nissinen RM, Oh CS, et al. HrpN of Erwinia amylovora functions in the translocation of DspA/E into plant cells. Mol Plant Pathol. 2008;9:425–434.
- Boureau T, El Maarouf-Bouteau H, Garnier A, et al. DspA/E, a type III effector essential for Erwinia amylovora pathogenicity and growth in planta, induces cell death in host apple and non-host tobacco plants. Mol Plant Mic Interac. 2006;19:16–24.
- Oh CS, Martin GB, Beer SV. DspA/E, a type III effector of Erwinia amylovora is required for early rapid growth in Nicotiana benthamiana and causes NbSGT1-dependent cell death. Mol Plant Pathol. 2007;8:255–265.
- Meng X, Bonasera JM, Kim JF, et al. DspE of Erwinia amylovora interacts with receptor kinases of apple. Acta Horticult. 2002;590:463–466.
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405.
- Jefferson RA. The GUS reporter gene system. Nature. 1989;342:837–838.
- Kaya Y, Maraklı S, Gozukirmizi N, et al. Herbicide tolerance genes derived from bacteria. J Anim Plant Sci. 2013;23(1):85–91.
- Roberts IN, Oliver RP, Punt PJ, et al. Expression of Eschenchia coli, B-glucuronidase gene in industrial and phytopathogenic filamentous fungi. Cur Gen. 1989;15:177–180.
- Bunkers GJ. Expression of the Escherichia coli 3-glucuronidase gene in Pseudocercosporella herpotrichoides. Appl Environ Microbiol. 1991;57:2896–2900.
- Yu-ling H, Wang YS, Yen JH. Enantioselective effects of herbicide imazapyr on Arabidopsis thaliana. J Environ Sci Health B. 2014;49(9):646–653.
- Clough SJ, Bent A. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743.
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1989.
- Bauer DW. Molecular genetics of pathogenicity of Erwinia amylovora: techniques, tools and their applications [dissertation]. Ithaca (NY): Cornell University; 1990.
- Allen GC, Hall GE, Childs LC, et al. Scaffold attachment regions increase reporter gene expression in stably transformed plant cells. Plant Cell. 1993;5(6):603–613.
- Gaudriault S, Paulin JP, Barny MA. The DspB/F protein of Erwinia amylovora is a type III secretion chaperone ensuring efficient intrabacterial production of the Hrp-secreted DspA/E pathogenicity factor. Mol Plant Pathol. 2002;3:313–321.
- Kim JF, Beer SV. HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. J Bacteriol. 1998;180:5203–5210.
- Barny MA. Erwinia amylovora hrpN mutants blocked in harpin synthesis express a reduced virulence on host plant and elicit variable hypersensitive reactions on tobacco. Eur J Plant Pathol. 1995;101:333–340.
- Ramos LS, Lehman BL, Peter KA, et al. Mutation of the Erwinia amylovora argD gene causes arginine auxotrophy, nonpathogenicity in apples, and reduced virulence in pears. Appl Environ Microbiol. 2014;80(21):6739–6749.
