ABSTRACT
Food handlers are important component in assessment and maintenance of food quality as they are carriers of food pathogens causing spoilage. Food spoilage is attributed to quorum sensing (QS) controlled development of biofilms. Therefore, there is an urgent need to develop novel QS and biofilm inhibitors to prevent spoilage of food products. In the present study, toxin producing biofilm forming methicillin-resistant Staphylococcus aureus (MRSA) were isolated from food handlers. Further, eugenol was screened for its QS and anti-biofilm properties. Analysis of nasal and hand swabs revealed the presence of seven toxigenic and biofilm forming MRSA strains. Eugenol demonstrated significant anti-QS activity in CVO26 and also reduced the QS-regulated production of elastase, protease, chitinase, pyocyanin and exopolysaccharide (EPS) in PAO1 considerably. Eugenol demonstrated 17%–86%, 24%–69%, 30%–91%, 9%–94% and 4%–89% reduction in biofilm biomass of S. aureus ATCC 25923 and MRSA strains FSA3, FSA11, FSA13 and FSA32, respectively. Sub-inhibitory concentrations of eugenol also decreased the metabolic activity in biofilm cells. Molecular docking analysis showed high binding affinity of eugenol that represents its biofilm inhibitory activity. This is the first report on the carriage of toxigenic drug-resistant biofilm forming S. aureus by food handlers and inhibition of their biofilms in the Kingdom of Saudi Arabia. The findings give a clear insight into the food safety hazards associated with the carriage of S. aureus and present eugenol as a broad-spectrum anti-QS and anti-biofilm agent.
Introduction
Foodborne illness is one of the major health concerns worldwide and according to the estimates of World Health Organization (WHO) around 30% population in the developed countries suffers from food-related health hazards annually, whereas in the third world countries mortality of two million is estimated per year [Citation1]. Food handlers are a common and persistent cause of the spread of foodborne diseases [Citation2] and Staphylococcus aureus is one of the important pathogens often transmitted via food contaminated by an infected food handler.
Staphylococci are ubiquitously found in nature and are frequently isolated from food and environmental sources. Staphylococcus aureus is a foodborne pathogen that can cause localized and invasive infections in humans due to consumption of contaminated food [Citation3]. The infection causing ability of S. aureus is attributed primarily to the production of staphylococcal enterotoxins (SEs) and toxic shock syndrome toxin (TSST) [Citation4]. Although toxin production is the major cause of the foodborne illness caused by the S. aureus, other factors like biofilm formation and antibiotic resistance also contribute to its pathogenicity. Drug-resistant staphylococci are a major public health concern as the bacteria can circulate easily in the environment through the food chain [Citation5]. In the past two decades, infection caused by methicillin-resistant Staphylococcus aureus (MRSA) has increased considerably across the globe. Multi-drug resistant strains of S. aureus have been frequently recovered from food stuff and food handlers [Citation6]. Staphylococci can also form biofilms on various surfaces and impart resistance to the pathogen against antibiotics and sanitizers [Citation7]. Therefore, biofilm formation is a marker of pathogenicity for drug-resistant S. aureus [Citation8]. Biofilm formation has been extensively studied in clinical settings, but limited information is available regarding staphylococcal biofilm in the food industry.
Food industry is currently facing severe problem of food spoilage and biofilm formation by foodborne pathogens and even with the application of modern preservation techniques, losses due to microbial spoilage of food are still huge [Citation9]. Foodborne pathogens like Pseudomonas spp., E. coli, Staphylococcus spp., Salmonella spp., Campylobacter jejuni and Yersinia enterocolitica have been identified to form biofilms [Citation10]. Attachment of these foodborne pathogens to the food product or the processing surfaces leads to mass food spoilage, severe public health risk and economic losses [Citation10]. Recently, bacterial quorum sensing (cell-to-cell communication) has received huge attention due to its role in the attachment, growth and survival of pathogenic bacteria in biofilm mode in foods [Citation11]. Since quorum sensing (QS) in bacteria is mediated by the production of small signal molecules called autoinducers, therefore, any interference in the QS signalling system can effectively alter the microbial gene expression related to food spoilage and biofilm formation.
Since the discovery of marine alga (Delisea pulchra) as QS inhibitor, many synthetic and natural compounds have been investigated for their QS and biofilm inhibitory activity. However, the stability and toxicity associated with these compounds limit their use in food industries [Citation12]. This has led to search for a novel stable and non-toxic QS inhibitor for their potential use in the prevention of food spoilage. Eugenol is a major constituent of clove oil and is known to possess several biological properties, including antimicrobial, antioxidant, anti-inflammatory, anticarminative and antispasmodic activities. It is also important industrially and has been used as a flavouring agent in food and cosmetic products [Citation13]. Recently, biofilm inhibition and eradication properties of eugenol against MRSA and methicillin-sensitive S. aureus (MSSA) clinical strains have been reported [Citation14]. However, the effect of eugenol on QS and biofilm of MRSA strains isolated from food handler's is not studied yet.
Since human carriers are the most important cause of transmission of foodborne illness, there is a need to investigate the association between food handlers and transmission of food-related illness. Therefore, the present study was aimed at assessing the prevalence of toxigenic, biofilm forming MRSA isolated from the nose and hands of food handlers employed in the restaurants of King Saud University, Riyadh. Additionally, the goal was also to investigate the anti-QS and biofilm inhibitory activity of eugenol against isolated MRSA strains. Bacterial characterisctics like antibiotic resistance and biofilm formation were also studied as limited data is available on these aspects, especially in the Arab world.
Materials and methods
Sample collection
Samples were collected from the hands (interdigital region, index fingers, thumbs and palms of both right and left hands) and anterior nares of 100 male food handlers employed in the King Saud University, Riyadh, restaurants and cafeterias during meal preparation. One swab was used for each region.
Swabs were streaked on Baird–Parker plates (Oxoid, UK) immediately (within 1 h) after collection and the plates were incubated at 35 °C for 48 h in our laboratory.
Staphylococcal isolation and identification
After incubation, five black colonies from each plate (presence and absence of halo) were identified. Gram staining, production of catalase and coagulase were used as screening tests. The coagulase-positive strains were submitted to the kit ‘Staphytect Test Dry Spot’ (Oxoid, UK). The two positive clumping species were subjected to the Voges–Proskauer test (S. aureus positive).
Detection of toxins
Only one identified isolate, per positive individual, was investigated using an immunological technique to verify its ability to synthesize SE(s). The strains were grown in 10 mL of Tryptone Soya Broth (Oxoid, UK) by shaking aerobically for 16–18 h at 37 °C. After centrifugation at 9000 × g for 20 min at 4 °C, production of SEs A, B, C, D and TSST-1 was determined by reversed passive latex agglutination using two commercial kits, SET-RPLA and TST-RPLA (Oxoid, UK), according to the manufacturer's protocols. Negative controls were used with all the tested samples.
Screening for methicillin resistance
Resistance to oxacillin was determined in all S. aureus isolates using an oxacillin broth screening test recommended by the British Society for Antimicrobial Chemotherapy (BSAC).
Microtitre plate assay
The ability of the strains to form a biofilm was investigated in flat-bottom 96-well polystyrene microtitre plates according to the protocol of Stepanovic´ et al. [Citation15]. For each strain, four wells of a microtitre plate were filled with 200 µL of bacterial suspension in Tryptone Soya Broth (Oxoid, UK) supplemented with 0.5% glucose, and the plates were incubated aerobically at 37 °C for 24 h under static conditions. Then, the content of each well was aspirated, and each well was washed three times with 250 µL of sterile physiological saline. The attached bacteria were fixed with methanol for 15 min, and then the plates were emptied and left to dry. The plates were stained for 5 min with 200 µL of crystal violet per well. Excess stain was rinsed-off with tap water, and after the plates were airdried, the dye bound to biofilm was resolubilized with 160 µL of ethanol. The OD of each well was measured at 570 nm by using a Multiskan Microplate Reader (Thermo Scientific).
Congo red agar method
Congo red agar (CRA) method was adopted for screening of biofilm formation by MRSA isolates as described by Freeman et al. [Citation16]. Briefly, plates were inoculated and incubated aerobically for 24–48 h at 37 °C. Positive result was indicated by black colonies with a dry crystalline consistency.
Determination of minimum inhibitory concentration (MIC)
MIC of eugenol was determined against Chromobacterium violaceum CVO26, Pseudomonas aeruginosa PAO1, S. aureus ATCC 25923 and four MRSA strains by broth macrodilution method (CLSI, 2007). Sub-MICs were selected for the assessment of anti-virulence and anti-biofilm activity in the above test strains. Eugenol used in the present study was purchased from Hi-Media Laboratories, India.
Quantitative estimation of violacein
Extent of violacein production by C. violaceum (CVO26) in the presence of sub-MICs of eugenol was studied by extracting violacein and quantifying photometrically using the method of Husain et al. [Citation17].
Effect on virulence factor production
Effect of sub-MICs of eugenol on virulence factors of P. aeruginosa PAO1 such as lasB elastase, protease, pyocyanin, chitinase, EPS extraction and quantification was assessed as described previously [Citation18].
Analysis of lasB transcriptional activity in E. coli
Measurements of β-galactosidase luminescence in E. coli MG4/pKDT17 were done by adopting the method described by Pearson et al. [Citation19].
Assay for biofilm inhibition
Wells of a microtitre plate were filled with 200 µL of bacterial suspension in Tryptone Soya Broth (Oxoid, UK) supplemented with 0.5% glucose, sub-MICs of eugenol and the plates were incubated aerobically at 37 °C for 24 h under static conditions. Further, the protocol of Stepanovic´ et al. [Citation15] was followed as described above.
Light microscopy analysis of biofilm inhibition
Glass cover slips (18 mm diameter) were placed in six well tissue culture plates. Wells of the plates were dispensed with 1 mL of tryptic soy broth (TSB) media containing sub-MICs of eugenol. Further, 1 mL of standardized cell suspension was added to each well and incubated at 37 °C for 48 h. Well without test agents served as positive control. After the formation of biofilm, medium was discarded, and discs were washed three times with sterile phosphate-buffered saline (PBS) and stained with 0.1% crystal violet and incubated at 37 °C for 10 min. The discs were visualized under a bright field light microscope (Olympus, Japan) at 40 X.
Biofilm metabolic activity measurement
The metabolic (respiratory) activity of biofilm was determined by using (XTT) reduction assay. Biofilm formation was done as described above. Following incubation (24 h), the contents of each well were removed, and wells were washed three times with PBS to remove loosely attached cells. The sodium salt of XTT (2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)- 2H-tetrazolium-5-carboxanilide) was dissolved in PBS to 1 mg/mL concentration, filter sterilized and stored at −80 °C. Menadione was dissolved in acetone to 1 mmol/L and sterilized immediately before each measurement. Working solution of XTT/menadione reagent was freshly prepared before each assay in the ratio of 12.5:1. Following washing, 100 mL PBS was added to each well of a 96-well microtitre plate. 13.5 mL of XTT/menadione mixture was then added to each well; the plate was gently shaken, then covered (in darkness) and incubated at 37 °C for 3 h. Following incubation, the absorbance was measured at 490 nm. Blank well, negative and positive controls were performed as described above. Each assay was performed in triplicate [Citation20].
In silico analysis
Preparation of enzyme and molecular docking
The three-dimensional (3D) crystal structure of sarA protein structure (PDB ID code 2FNP) was extracted from Research Collaboratory for Structural Bioinformatics (RCSB) protein databank. All the hetero atoms as well as solvent were removed. The 3D structure of eugenol and c-di-GMP were retrieved from pubchemcompound database (CID3314 and CID6323195). AutoDock tools 4.0 [Citation21] was used to carry out docking of eugenol and c-di-GMP within the binding site of sarA. Lamarckian genetic algorithm, a combination between the genetic algorithm and the local search Pseudo-Solis and Wets algorithm, was used as a parameter for the molecular docking. The grid box dimension was set to 60 × 60 × 60 Å around active site of sarA making sure that eugenol and c-di-GMP can freely rotate inside the grid. The number of docking runs was set to 15. The final poses were selected on the basis of their binding energies. The final complexes were subsequently visualized using PyMol [Citation22].
Results and discussion
Prevalence of toxigenic MRSA
Due to inadequate personal hygiene and cross contamination, food handlers are vector of foodborne disease. Therefore, in this study we have collected samples from hands and nasal cavity of 100 male food handlers employed in different restaurants and cafeterias of King Saud University, Riyadh. Of the total collected samples, 43% of nose samples and 15% of hand samples were found to be carrying S. aureus (). Our results on nasal and skin carriage of S. aureus by food handler's (43% and 15%, respectively) were in agreement to the previous report by Loeto et al. [Citation23] and El-Shenawy et al. [Citation24] demonstrating the prevalence of S. aureus among food handlers of Botswana and Egypt, respectively. Carriage rates of S. aureus reported by several investigators differ and these variations may be due to the ecological differences of the studied population.
Table 1. Prevalence of nasal and hand carriage of coagulase positive Staphyloccocus aureus among food handlers working in KSU restaurants.
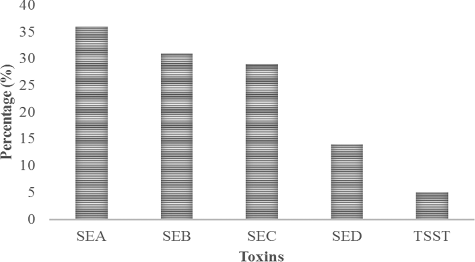
Staphylococcal food poisoning is an intoxication caused by the ingestion of food containing pre-formed SE. Among different types of SEs, Enterotoxin A is most commonly associated with staphylococcal food poisoning and to a lesser extent to Enterotoxins D and B which are also known to cause poisoning [Citation25]. Food handlers carrying toxigenic S. aureus in their noses or hands are causes for food contamination via direct contact or through respiratory secretions [Citation26]. Results of the present investigation showed that among the 58 S. aureus isolates tested for SEs and TSST by reversed passive latex agglutination, 36 (62%), 2 (3.4%) and 1 (1.7%) were positive for SE, TSST or both, respectively (). Among the enterotoxins detected, prevalence of Enterotoxin A (SEA) was highest (36%) followed by SEB (31%), SEC (29%) and SED (14%) as depicted in . TSST was detected in 5% of the isolated S. aureus strains. Our findings are in agreement with those of the Fueyo et al. [Citation27] who reported 1.2%, 3.7% and 3.7% prevalence of SE, TSST-1 or both among S. aureus isolates from nasal swabs of human carriers.
Further, out of the 58 isolates screened, 7 (12%) were found to be MRSA. Results obtained from investigation of MRSA isolates were in accordance with the findings of Dagnew et al. [Citation6] who reported 9.8% methicillin resistance among the isolated S. aureus from food handlers in Ethiopia. Detection of MRSA in the present study is vital because food handlers carrying MRSA have been identified as major cause of community-acquired foodborne outbreaks [Citation28,Citation29]
Screening for biofilm formation
Biofilm formation capacity of the MRSA strains was evaluated by CRA method and microtiter plate method ((A,B)). In CRA method, six S. aureus isolates were positive for biofilm formation, showing black coloration of the colony. However, usingmicrotiter plate (MTP) method, four MRSA strains formed strong biofilm, while one of the strains formed moderate biofilm and two test strains demonstrated weak biofilm forming capability as shown in . Staphylococcus aureus ATCC 25923 was used as positive control for biofilm formation. Since little information is available regarding staphylococcal biofilms in the food environment, isolated MRSA were screened for biofilm formation. Our findings find support from the observation made by Marino et al. [Citation29] who showed moderate to high biofilm formation among staphylococci isolated from food samples and food handlers. The ability of food pathogens like S. aureus to adhere to an abiotic surface and form a biofilm in food environments may ultimately lead to food poisoning [Citation30].
Figure 2. Screening of MRSA strains for biofilm formation: (A) Congo red agar (CRA) assay and (B) microtitre plate assay.
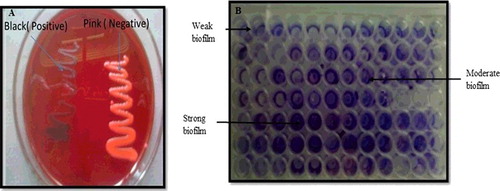
Table 2. Screening of MRSA strains for biofilm formation by Congo red agar and microtitre plate method.
Determination of minimum inhibitory concentration (MIC)
MIC of eugenol was determined against C. violaceum CVO26, P. aeruginosa PAO1 and biofilm forming MRSA strains. Eugenol demonstrated a minimum inhibitory concentration of 160 µg/mL against CVO26 and PAO1. MIC of eugenol against S. aureus ATCC 25923 and four biofilm forming MRSA strains ranged from 42 to 665 µg/mL. Assessment of QS and biofilm inhibitory activity of eugenol was done at sub-inhibitory concentrations, i.e. concentrations below the MICs.
Effect of eugenol on quorum sensing regulated traits
Significant problems in the food industry are due to the microbial spoilage of foods and biofilm formation by foodborne bacteria. Micro-organisms adhere to surfaces to survive the cleaning process [Citation9]. Moreover, biofilms are more resistant to antimicrobials compared to planktonic cells, and this makes their elimination from food processing facilities a big challenge. Several studies have shown that bacterial spoilage of foods is regulated by cell-to-cell communication or QS mechanism that plays a major role in various stages of biofilm formation in bacteria [Citation31]. Therefore, the present study was focused on the anti-QS and biofilm inhibitory effect of eugenol.
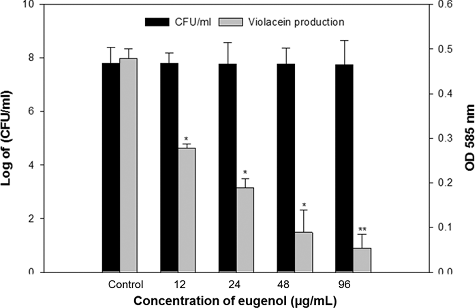
The QS inhibitory potential of eugenol was screened using the C. violaceum biosensor strain CVO26. The CviIR-dependent QS system of C. violaceum coordinates the production of violacein pigment. CviR (QS receptor) in C. violaceum binds to the autoinducer (AHL) at high cell density. This CviR-AHL complex binds to DNA and activates expression of the vio genes required for the production of violacein. The compounds inhibiting the violacein pigment without significant reduction of the bacteria are termed as QS inhibitors. Eugenol exhibited a concentration-dependent decrease in QS-regulated violacein production, and statistically significant inhibition was recorded at all tested concentrations (). Maximum reduction of 80% was recorded at 96 µg/mL concentration while lowest of 41% decrease over control was observed at 12 µg/mL eugenol concentration without any significant decrease in growth of the bacteria. Similar QS inhibitory properties of menthol and methyl eugenol based on violacein inhibition in CVO26 have been reported previously [Citation17,Citation32]. QS inhibitory activity of phytochemicals is attributed to the similarity in their chemical structure to those of AHL signal molecules and also because of their ability to degrade signal receptors (LuxR/LasR) [Citation33,Citation34]. Therefore, the reduction of QS-dependent violacein production in C. violaceum by the eugenol is plausibly due to inhibition of the CviIR-dependent QS signalling either by prevention of AHL signal reception or decrease in the expression of the AHL family of synthases.
Figure 3. Quantitative assessment of violacein inhibition in CVO26 by sub-MICs of eugenol. All of the data are presented as mean ± SD.
To ascertain the anti-QS property of eugenol, it was also tested against QS-regulated virulence factors of P. aeruginosa PAO1 at its respective sub-MICs. Pseudomonas aeruginosa has three QS systems, namely, las, rhl and pqs. The las and the rhl systems are LuxI/LuxR homologues of LasI/LasR and of RhlI/RhlR, respectively [Citation35]. Virulence factors such as elastase, protease, pyocyanin production, EPS production and biofilm formation in P. aeruginosa are regulated by three above-mentioned QS systems. Effect of eugenol on QS-regulated functions of P. aeruginosa PAO1 was determined as shown in . Significant reduction (p ≤ 0.001) in elastase (47%–82%), total protease activity (44%–87%) and pyocyanin production (45%–85%) of PAO1 was recorded at tested sub-MICs (21–84 µg/mL) of eugenol. Chitinase activity was lowered significantly (39.8%–63.2%) at sub-MICs ranging from 0.2 to 0.8% v/v eugenol. Similarly, decrease in EPS production was also dose-dependent but statistically significant reduction (49.1%) was observed at 84 µg/mL concentration. Considering las, rhl and pqs systems regulate the expression of numerous virulence-related genes, using eugenol to inhibit these QS systems would significantly decrease P. aeruginosa virulence.
Table 3. Effect of sub-MICs of eugenol on inhibition of quorum sensing regulated virulence factors in P. aeruginosa PAO1.
Effect of eugenol on β-galactosidase activity
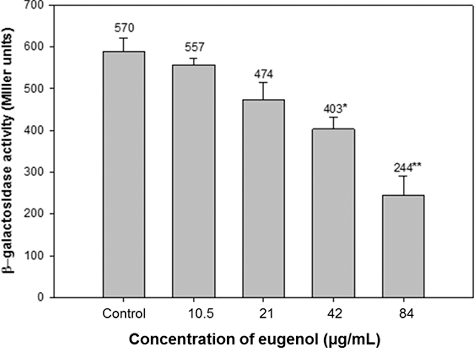
Effect of eugenol on AHL production was also assessed using β-galactosidase assay in E. coli MG4/pKDT17. Untreated PAO1 produced 590 miller units of AHL whereas at 10.5, 21, 42 and 84 µg/mL concentration of eugenol, quantity of AHL produced was 551, 474, 403 and 244 miller units, respectively. The results obtained demonstrated a significant decrease of 31.6% and 58.6% at 42 and 84 µg/mL eugenol concentrations (). Reduced AHL levels of treated PAO1 demonstrates that eugenol decreases both elastase activity and the transcriptional activation of lasB in E. coli, which indicates that eugenol inhibits the las system as reported by Zhou et al. [Citation36]
Effect of eugenol on biofilm formation of MRSA
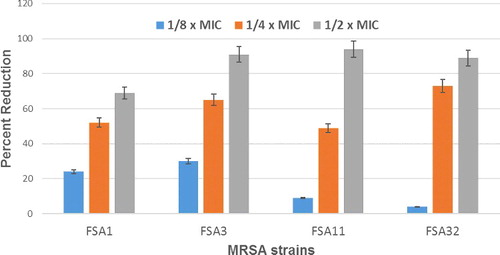
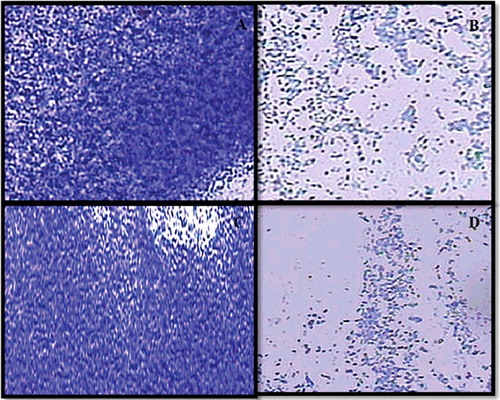
Four MRSA strains that formed strong biofilms along with S. aureus ATCC 25923 were selected to study the biofilm inhibitory property of eugenol at sub-inhibitory concentrations (1/2 × MIC-1/8 × MIC). In the quantitative microtiter plate assay, eugenol exhibited a dose-dependent reduction in the biofilm biomass of MRSA strains. Eugenol demonstrated 17%–86%, 24%–69%, 30%–91%, 9%–94% and 4%–89% reduction in biofilm biomass of S. aureus ATCC 25923 and MRSA strains FSA3, FSA11, FSA13 and FSA32, respectively (). The architecture of S. aureus (ATCC 25923) and MRSA biofilms grown with and without eugenol (0.5 × MIC) was analysed by light microscopy. The light microscopic images show significantly impaired biofilm growth in eugenol-supplemented sample in comparison to the untreated samples (). This is the first report on the biofilm inhibition of MRSA strains isolated from hands and nose of food handlers. In MRSA, biofilm formation is ica-independent and is mediated by proteins binding to proteins of the extracellular matrix [Citation37]. As the biofilm experiments were carried-out in TSB medium supplied with 0.5% glucose, it has been reported previously that glucose can induce ica-independent biofilm formation in MRSA [Citation37]. Therefore, it is envisaged that eugenol is effective against ica-independent biofilm formation. Similar findings were reported with eugenol in inhibition of biofilms of MRSA strains of clinical origin [Citation14,Citation38].
Biofilm metabolic activity (XTT assay)
XTT reduction assay was used to evaluate the metabolic activity of cells in biofilm mode after 24 h. Eugenol at sub-MICs demonstrated a decrease of 28%–72% in the respiratory activity of S. aureus 25923 cells growing in biofilm mode. Similar dose-dependent reduction in the metabolic activity of MRSA strains was recorded at respective sub-MICs as depicted in . XTT reduction assay showed inhibition percentage range of 13%–64%, 24%–90%, 54%–88% and 45%–93% in biofilm cells of FSA3, FSA11, FSA13 and FSA32, respectively.
Figure 7. Percent reduction in metabolic activity of MRSA cells in biofilm mode at sub-MICs of eugenol.
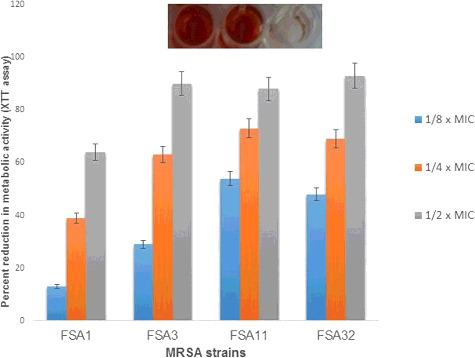
The results of inhibition of biofilm biomass and metabolic activity by sub-MICs of eugenol were almost similar. Moreover, the XTT reduction assay confirmed the inhibition of biofilm by sub-MICs of eugenol. However, results of XTT reduction assay could not be correlated with crystal violet assay as in strains FSA13 and FSA32 reduction in metabolic activity was significant as compared to the decrease in biofilm biomass of the two strains at tested sub-MICs. Reduced metabolic activity of biofilm cells is mainly attributed to the decreased nutrient and oxygen supply and such reduction as a physiological change can account for the resistance of biofilms to antimicrobial agents [Citation39,Citation40]. Also, many reports have indicated an inverse correlation or no correlation between biofilm biomass and metabolic activity for plant extract or essential oil [Citation40–42]. However, our results showed that the eugenol had similar results in inhibition of biofilm formation and metabolic activity at higher sub-MICs tested.
In silico study
Molecular docking
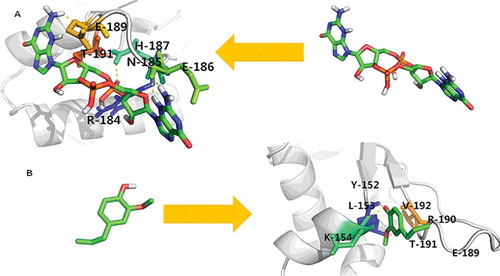
The molecular docking was complementary applied to gain a better insight of the interaction of eugenol within the binding site of sarA. sarA has been reported to be an important regulator responsible for biofilm development in S. aureus. Previous findings have reported the upregulation of sarA transcript in biofilm [Citation43]. It has also been reported earlier that targeting sarA inhibition would be an effective way of inhibiting the biofilm in S. aureus [Citation44,Citation45]. Therefore, sarA was taken as a molecular target for eugenol and structure–activity relationship of eugenol was assessed against sarA binding site. As a reference model, the known inhibitor, c-di-GMP was docked in ligand binding domain of sarA. Earlier studies have demonstrated that c-di-GMP treatment can be used as a novel intervention strategy to control S. aureus biofilm formation and virulence [Citation46]. Literature shows that c-di-GMP inhibits the biofilm formation in S. aureus clinical isolates, including MRSA strains [Citation46]. The molecular docking scores of eugenol and c-di-GMP against sarA are depicted in and . It was revealed through molecular docking that eugenol was equally effective against sarA as compared to c-di-GMP. Eugenol was found to interact with the binding affinity of −5.09Kcal/mol against sarA. In this study, we also demonstrated that some key amino acid residues are involved in the positioning of eugenol within their respective binding site of sarA. It was found that the binding of eugenol within the active site of sarA was equally contributed by hydrogen bonding as well as hydrophobic interaction of the active site residues. Here, L153, K154, V192, Y152, E189, R190, T191 and L193 were the key residues involved in accommodating eugenol within the binding site of sarA. L153 and K154 were the common amino acids found to be involved in making hydrogen bonding as well as hydrophobic interaction with eugenol ( and ). The importance of these active site residues of sarA has already been discussed in previous studies [Citation47]. The molecular docking scores of eugenol and c-di-GMP against sarA are shown in . Docking analysis further confirms that eugenol can interfere with QS and its regulated virulence and biofilm. Hence, it is envisioned that eugenol can be exploited as a broad-spectrum QS and biofilm inhibitor.
Table 4. Binding efficacy of eugenol against sarA and the amino acid residues involved in their complex formation.
Conclusion
The study provides an insight on the carriage of S. aureus among food handlers. Findings of the current investigation give information regarding the prevalence of different types of toxins, prevalence of methicillin resistance and biofilm formation in the pathogen. The results of the present study demonstrate that eugenol has significant anti-QS and biofilm inhibitory activity. To the best of our knowledge, this is the first time that the biofilm inhibitory activity of eugenol has been reported against biofilm formed by MRSA isolated from food handlers. These results assume great importance as the above-studied traits influence bacterial survival and contamination of food and food processing environment. Specific QS and biofilm inhibitors can find application in food industry to enhance safety and quality of foods. Our observations should help in the better management of food products and food handlers to enhance food safety and safety of the consumers not only in KSU restaurants but also across the whole Kingdom of Saudi Arabia.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through the Research project number NFG-15-03-16.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- World Health Organization: Food safety – food bornediseases and value chain management for food safety, “Forging links between Agriculture and Health”. CGIAR on agriculture and health meeting in WHO/HQ; 2007.
- Zain M, Naing N. Sociodemographic characteristics of food handlers and their knowledge, attitude and practice towards food sanitation: a preliminary report. Southeast Asian J Trop Med Pub Health. 2002;33:410–417.
- Rodriguez M, Nunez F, Cordoba JJ, et al. Gram-positive, catalase-positive cocci from dry cured Iberian ham and their enterotoxigenic potential. Appl Environ Microbiol. 1996;62:1897–1902.
- Greig JD., Todd ECD, Bartleson CA, et al. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 1. Description of the problem, methods, and agents involved. J Food Prot. 2007;70:1752–1761.
- Angulo FJ, Nargund VN, Chiller TC. Evidence of an association between use of antimicrobial agents in food animals and antimicrobial resistance among bacteria isolated from humans and the human health consequences of such resistance. J Vet Med B. 2004;51:374–379.
- Dagnew M, Tiruneh M, Moges F, et al. Survey of nasal carriage of Staphylococcus aureus and intestinal parasites among food handlers working at Gondar University, Northwest Ethiopia. BMC Pub Health. 2012;12:837.
- Cloete TE. Resistance mechanisms of bacteria to antimicrobial compounds. Int Biodeterioration Biodegrad. 2003;51:277–282.
- Jain A., Agarwal A. Biofilm production, a marker of pathogenic potential of colonizing and commensal staphylococci. J Microbiol Meth. 2009;76:88–92.
- Bai A.J, Rai VR. Bacterial quorum sensing and food industry. Comp Rev Food Sci Food Saf. 2011;10:183–193.
- Bridier A, Sanchez-Vizuete P, Guilbaud M, et al. Biofilm-associated persistence of food-borne pathogens. Food Microbiol. 2015;45:167–178.
- Chmielewski RAN, Frank JF. Biofilm formation and control in food processing facilities. Comp Rev Food Sci Food Saf. 2003;2:22–32.
- Gopu V, Meena CK, Shetty PH. Quercetin influences quorum sensing in food borne bacteria: in-vitro and in-silico evidence. PLoS One. 2015;10(8):e0134684.
- Pramod K, Ansari SH, Ali J. Eugenol: a natural compound with versatile pharmacological actions. Nat Prod Comm. 2010;5:1999–2006.
- Yadav MK, Chae SW, Im GJ, et al. Eugenol: a phyto-compound effective against methicillin-resistant and methicillin-sensitive Staphylococcus aureus clinical strain biofilms. PLoS One. 2015;10(3):e0119564
- Stepanovic´ S, Vukovic´ D, Dakic´ I, et al. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Meth. 2000;40:175–179.
- Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol. 1989;42:872–874.
- Husain FM, Ahmad I, Khan MS, et al. Sub-MICs of Mentha piperita essential oil and menthol inhibits AHL mediated quorum sensing and biofilm of Gram negative bacteria. Front Microbiol. 2015;6:420.
- Husain FM, Ahmad I, Asif M, et al. Influence of clove oil on certain quorum sensing regulated functions and biofilm of Pseudomonas aeruginosa and Aeromonas hydrophila. J Biosci. 2013;38:835–844.
- Pearson JP, Gray KM, Passador L, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. PNAS USA. 1994;91:197–201.
- Chaieb K, Kouidhi B, Jrah H, et al. Antibacterial activity of thymoquinone, an active principle of Nigella sativa and its potency to prevent bacterial biofilm formation. BMC Compl Alter Med. 2011;11:29.
- Morris GM, Goodsell DS, Halliday RS, et al. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem. 1998;19:1639–1662.
- DeLano WL. The PyMOL molecular graphics system. 2002. Available from: htttp://www.pymol.org
- Loeto D, Matsheka MI, Gashe BA. Enterotoxigenic and antibiotic resistance determination of Staphylococcus aureus strains isolated from food handlers in Gaborone, Botswana. J Food Prot. 2007;70:2764–2768.
- El-Shenawy M, Tawfeek M, El-Hosseiny L, et al. Cross sectional study of skin carriage and enterotoxigenicity of Staphylococcus aureus among food handlers. Open J Med Microbiol. 2014;4:16–22.
- Pinchuk IV, Beswick EJ, Reyes VE. Staphylococcal enterotoxins. Toxins. 2010;2:2177–2197.
- Argudín MA, Mendoza MC, Rodicio MR. Food poisoning and Staphylococcus aureus enterotoxins. Toxins. 2010;2:1751–1773.
- Fueyo JM, Mendoza MC, Martín MC. Enterotoxins and toxic shock syndrome toxin in Staphylococcus aureus recovered from human nasal carriers and manually handled foods: epidemiological and genetic findings. Microbes Infect. 2005;7:187–194.
- Jones TF, Kellum ME, Porter SS, et al. An outbreak of community-acquired foodborne illness caused by methicillin-resistant Staphylococcus aureus. Emerg Infect Dis. 2002;8:82–84.
- Marino M, Frigo F, Bartolomeoli I, et al. Safety-related properties of staphylococci isolated from food and food environments. J Appl Microbiol 2011;110:550–561.
- Brooks JD, Flint SH. Biofilms in the food industry: problems and potential solutions. Int J Food Sci Technol. 2008;43(12):2163–2176.
- Simões M, Simoes, LC, Vieira MJ. A review of current and emergent biofilm control strategies. LWT – Food Sci Technol 2010:43:573–583.
- Packiavathy IASV, Agilandeswari P, Musthafa KS, et al. Antibiofilm and quorum sensing inhibitory potential of Cuminum cyminum and its secondary metabolite methyl eugenol against gram negative bacterial pathogens. Food Res Int. 2012;45:85–92.
- McLean RJC, Pierson III, LS, Fuqua C. A simple screening protocol or the identification of quorum sensing signal antagonists. J Microbiol Meth. 2004;58:351–360.
- Teplitski M, Mathesius U, Rambaugh KP. Perception and degradation of N-acyl homoserine lactone quorum sensing signals by mammalian and plant cells. Chem Rev. 2011;111:100–116.
- Dekimpe V, Deziel E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 2009;155:712–723.
- Zhou L, Zheng H, Tang Y, et al. Eugenol inhibits quorum sensing at sub-inhibitory concentrations. Biotechnol Lett. 2013;35:631–637.
- Vergara-Irigaray M, Valle J, Merino N, et al. Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect Immun. 2009;77:3978–3991.
- Jafri H, Husain FM, Ahmad I. Antibacterial and antibiofilm activity of some essential oils and compounds against clinical strains of Staphylococcus aureus. J Biomed Therapeutic Sci. 2014;1:65–71.
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322.
- Sandasi M, Leonard CM, Viljoen AM. The in vitro antibiofilm activity of selected culinary herbs and medicinal plants against Listeria monocytogenes. Lett Appl Microbiol. 2009;50:30–35.
- Budzyńska A, Wieckowska-Szakiel M, Sadowska B, et al. Antibiofilm activity of selected plant essential oils and their major components. Pol J Microbiol. 2011;60:35–41. Available from: https://www.ncbi.nlm.nih.gov/pubmed/21630572
- Bazargani MM, Rohloff J. Antibiofilm activity of essential oils and plant extracts against Staphylococcus aureus and Escherichia coli biofilms. Food Cont. 2016;61:156–164.
- Beenken KE, Mrak LN, Griffin LM, et al. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS One. 2010;5(5):e10790.
- Arya R, Ravikumar R, Santhosh R, et al. SarA based novel therapeutic candidate against Staphylococcus aureus associated with vascular graft infections. Front Microbiol. 2015;6:416. doi:10.3389/fmicb.2015.00416.
- Balamurugan P, Hema M, Kaur G, et al. Development of a biofilm inhibitor molecule against multidrug resistant Staphylococcus aureus associated with gestational urinary tract infections. Front Microbiol. 2015;6:832. doi:10.3389/fmicb.2015.00832.
- Karaolis DK, Rashid MH, Chythanya R, et al. c-di-GMP (3′-5′-cyclic diguanylic acid) inhibits Staphylococcus aureus cell-cell interactions and biofilm formation. Antimicrob Agents Chemother. 2005;49:1029–1038.
- Balamurugan P, Hema M, Kaur G. Development of a biofilm inhibitor molecule against multidrug resistant Staphylococcus aureus associated with gestational urinary tract infections. Front Microbiol 2015;6:832. doi:10.3389/fmicb.2015.00832.