 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
This study was performed to screen Lactobacillus and evaluate its inhibiting effects on Helicobacter pylori (Hp). Lactobacillus was isolated from fermented food in northeastern China. After identification and physiological characterization, the identified Lactobacillus was used to interact with Hp. The aggregation ability of Lactobacillus and Hp was determined. Moreover, the inhibiting effects of Lactobacillus on Hp urease enzyme activity and antioxidant ability were assessed. The results showed that four Lactobacillus strains including Lactobacillus sake, Lactobacillus plantarum, Lactobacillus rhamnosus and Lactobacillus brevis were isolated from fermented food in northeastern China. All four strains of Lactobacillus could inhibit Hp growth in different extent. The interaction with Hp was statistically analyzed with significant differences. The Lactobacillus isolated from fermented food in Northeast China, had an inhibitory effect on Hp growth. This study provided a foundation for development of probiotic preparations and exploring the inhibitory effect of Lactobacillus on Hp.
Introduction
Helicobacter pylori (Hp) is a micro-aerobic, helical gram-negative bacillus that can colonize gastric mucosa. It is known as the only microorganism surviving in human stomach with an infection rate of up to 50% worldwide. In 1994, the International Agency for Research on Cancer (IARC) of the World Health Organization (WHO) listed Hp as a first class carcinogen, which has been identified to be associated with the development of some diseases such as chronic gastritis, peptic ulcer, gastric cancer and gastric mucosa-associated lymphoid tissue lymphoma [Citation1]. Therefore, eradication of Hp has become an important means for treatment and prevention of these diseases. With the development of microecology, the antagonistic effect of probiotics on Hp in vivo and in vitro has been investigated [Citation2–5]. And a previous study has shown that addition of probiotics containing Lactobacillus acidophilus to standard triple therapy increased the eradication success rate [Citation6]. In this study, we aimed to isolate new lactobacilli from fermented food in Northeast China and investigate their possible role in the inhibition of Hp growth.
Materials and Methods
Bacterial isolates and materials
The brine of Northeast pickle and spicy cabbage, yoghurt, MRS liquid and solid medium, calcium carbonate, and HBI lactobacillus biochemistry measuring band (GB) were purchased from Qingdao Haibo Company. Genomic DNA Extraction Kit was supplied by Tian'en Co., Ltd. DL2,000 DNA Marker and Premix Taq, pepsin, xylene, Hp standard strain (ATCC 43504), Colombian medium, and brain infusion were bought from Qingdao Haibo. Sterile sheep blood, fetal bovine serum (FBS) and diphenylpicrylhydrazyl (DPPH) were provided from Shanghai Jinsui Biotechnology Co., Ltd.
Instruments and equipment
Incubator GHP-9095 was purchased from Shanghai Heng Technology Co., Ltd. The biological safety cabinet BSC-1600IIB2 was bought from Shanghai Su net Industrial Co., Ltd. Automatic autoclave pot LDZX-50KBS was provided from Shanghai Shen'an medical equipment factory. Magnetic microscope BA310-T was obtained from Mike Audi Chemical Industry Group Co., Ltd. ETC-811PCR instrument (Dongsheng), DYY-III regulator steady flow electrophoresis instrument was offered by Beijing Liuyi Biotechnology Co., Ltd. Thermo microplate reader, 96-well plate, Oxford Cup, anaerobic incubator was purchased from Qingdao Haibo Co., Ltd.
Isolation of Lactobacillus strains
Appropriate amount of samples of Northeast pickle and spicy cabbage brine together with fermented yogurt were added to 0.9% sterile saline, respectively. Then 10-fold serial dilution was undertaken. Samples of different gradient dilutions were taken with an L bar to smear in the calcium carbonate-MRS solid medium plate [Citation7], and then they were cultured in the incubator at 37 °C for 48 h.
Identification of biochemical reactions
HBI lactic acid bacteria biochemical identification was used to measure biochemiacl reactions [Citation8]. The selected single colonies were inoculated into the following medium: aesculin, cellobiose, maltose, mannitol, salicyl, sorbitol, sucrose, raffinose, and inulin, lactose. After the inoculation was finished, they were cultured at 37 °C for 48 h.
16SrDNA molecular biology identification
Fresh bacteria were obtained by inoculating MRS liquid culture medium at 37 °C and centrifuged at 13000g for 14–16 h. Genomic DNA of each lactobacillus strain was extracted by the bacterial genomic DNA extraction kit strictly according to the manufacturer’s protocol. The concentration and purity of genomic DNA were detected by ND-1000 micro-spectrophotometer. Then 25 μL reaction system containing 1 μL DNA template, 12.5 μL Premix Taq, 0.5 μL primer 27f, 0.5 μL primer 1542r, and 10.5 μL sterile double distilled water was used for polymerase chain reaction (PCR) amplification. The program of the PCR reaction was as follows: 94 °C pre-degeneration for 4 min, followed by 30 cycles of 94 °C denaturation for 30 s, 55 °C annealing for 45 s, and 72 °C extension for 60 s, and final extention at 72 °C for 10 min. The primers were synthesised by Beijing BGI Biotechnology Co., Ltd. The sequences were: 5′-AGAGTTTGATCCTGGCTCAG-3 and 5′-AAGGAGGTGATCCAGCCGCA-3′. The purity of the 16S rDNA amplification products was tested and then the products were sequenced by Beijing BGI Gene Research Center. The results of the sequencing were blasted with GenBank.
Physiological characteristics of Lactobacillus
To detect the tolerance to artificial gastric juice, 1.5 × 108 CFU/mL Lactobacillus (150 r/min, 6 h) was activated with MRS liquid medium and inoculated in artificial gastric juice (pH 3.0). After growth at 37 °C for 48 h, the viable lactobacilli counts was determined by counting [Citation9] with a sterile saline treated group as the control group [Citation10].
To the measure the hydrophobicity, Lactobacillus (150 r/min, 6 h) was activated with MRS liquid medium and centrifuged at 2000 r/min. After washing 3 times with PBS, they were adjusted to OD600 = 0.40 ± 0.05. Then, 2 mL Lactobacillus and 2 mL xylene were mixed. The mixture was shaken for 5 min. Immediately, the water phase was measured by the absorbance as A0. And then the samples were placed in a temperature incubator at 37 °C for 1 h and the absorbance of the water phase as At was measured [Citation11]. The following formula was used: Hydrophobic force (%), where A0 was 0 h absorbance value and At was the absorbance at the t-th hour. As for self-aggregation ability calculation, the formula used was: Self-aggregation ability (%)
, where A0 was 0 h absorbance value and At was the absorbance at the t-th hour.
Aggregation of Lactobacillus and Hp
Lactobacillus and Hp were cultured and centrifuged at 2000 r/min. They were washed three times with PBS and adjusted to OD600 = 0.40 ± 0.05 with the concentration of 107–108 colony forming units (CFU)/mL. The absorbance value of Lactobacillus and Hp supernatant was measured, respectively, as Ax and Ay. Then equal counts of Lactobacillus and Hp were mixed. The mixture was shaken for 5 min, and then cultured in a 37 °C incubator. The OD600 absorbance value of the upper liquid at different time points was determined and recorded as Amix. Interactive Agglomeration Force (%), where: Ax is 0 h Lactobacillus absorbance value, Ay is 0 h Hp absorbance value, Amix was the Lactobacillus and Hp mixed absorbance value [Citation12].
Oxford Cup antibacterial test
Colombian medium blood plate and Hp bacteria solution were prepared using the previous method [Citation13]. Then diluted Hp bacteria solution (1 × 108 CFU/mL) was evenly coated on the fresh antibiotic-free Colombian culture plate three times. Four sterilized Oxford Cups were put into the blood plate and each Oxford Cup was added 50 μL lactic acid bacteria liquid culture, 50 μL Lactobacillus supernatant, levofloxacin drug sensitive paper and amoxicillin drug sensitive paper. The plate was placed in an anaerobic culture bag at 37 °C for 72 h, and then the size of the inhibition zone was measured.
Urease activity of Hp by Lactobacillus
After incubation of Hp with brain–heart leaching broth culture medium, Hp bacteria were washed with PBS buffer twice to adjust the concentration to 1 × 108 CFU/mL. Then three groups were added in a 96-well plate: 40 μL Hp bacteria and 10 μL MRS liquid medium; 40 μL Hp bacteria and 10 μL lactic acid bacteria cell-free supernatant; and 40 μL Hp bacteria and 10 μL Lactobacillus bacteria. The 96-well plates were added to the anaerobic culture bag and incubated at 37 °C for 48 h. After that, 150 μL urease indicator was added and the absorbance at 540 nm was measured.
Antioxidant capability of Lactobacillus
Measurement of antioxidant capability was assigned to three groups: sample group: 1 mL bacterial solution and 1 mL DPPH ethanol solution (0.2 mmol/L); control group: 1 mL MRS liquid medium and 1 mL DPPH ethanol solution; blank group: 1 mL MRS liquid medium and 1 mL absolute ethanol. Then the mixtures were placed in darkness at room temperature for 30 min. The supernatant was centrifuged at 6000 r/min for 10 min and the absorbance at 517 nm was measured. The following formula was used: Scavenging rate (%), where A1 is the absorbance value of the sample group, A2 is the absorbance value of the control group, and A3 is the absorbance value of the blank control.
Statistical analysis
The data analysis was performed using SAS9.4 statistical analysis software (SAS Institute Inc., Cary, NC, USA). The tolerance of Lactobacillus to artificial gastric juice was analyzed using t test. Rank sum test was used for analyses of lactobacillus hydrophobicity and self-aggregation ability. The two pair comparison was performed by Bonferroni method. Determination of the aggregation ability of Lactobacillus and Hp was carried out by repeated measures analysis of variance. Urease activity and Lactobacillus scavenging DPPH used rank sum test.
Results and discussion
Isolation of Lactobacillus
Lactobacillus colonies were round uplift, smooth, moist, easy to pick, and neat edge colonies. They were milky white, opaque and shiny, with a diameter of 1–3 mm (). The colloidal bacteria were picked and Gram stained. Gram-positive bacilli were arranged in a single, double or short chain shape. The screening four Lactobacillus stained by Gram is shown in . The results from the biochemical assays of the four screened strains for fermentation of aescinate, cellobiose, maltose, mannitol, salicylline, sorbitol, sucrose, raffinose, inulin and lactose are shown in .
Figure 1. Four Lactobacillus isolates. Colony morphology: Lactobacillus sakei, No. 1 (A); Lactobacillus plantarum, No. 2 (B); Lactobaeillus rhamnosus, No. 3 (C); Lactobacillus brevis, No. 4 (D). Gram staining: Lactobacillus sakei, No. 1 (E); Lactobacillus plantarum, No. 2 (F); Lactobaeillus rhamnosus, No. 3 (G); Lactobacillus brevis, No. 4 (H).
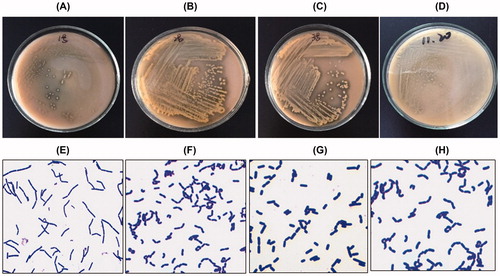
Table 1. Biochemical assays of screened Lactobacillus isolates.
Identification of 16SrDNA
In order to identify the species to which the isolates belong, genomic DNA of the Lactobacillus isolates was extracted. The molecular weight of the amplified bands of 16S rDNA was 1515 bp. Based on the sequencing data, the four strains of Lactobacillus were identified as: Lactobacillus sakei (No. 1; ), Lactobacillus plantarum (No. 2; ), Lactobaeillus rhamnosus (No. 3; ) and Lactobacillus brevis (No. 4; ).
Tolerance to artificial gastric juice
Four strains of Lactobacillus were treated in artificial gastric juice at pH 3.0 for 4 h to determine their tolerance to artificial gastric juice. PBS treated culture was used as the control group. Statistical analysis showed that the four strains of lactobacilli were resistant to pH 3.0 artificial gastric juice, and the differences were not statistically significant between the saline group and the gastric juice group for the four strains of Lactobacillus (P > 0.05; ).
Table 2. Tolerance of the four isolates to artificial gastric juice.
Hydrophobicity of Lactobacillus
To explore the physiological characteristics of lactobacillus, the hydrophobicity of the Lactobacillus isolates was detected. The results showed that the hydrophobicity in the four strains of lactobacilli was statistically significant (P < 0.05). Among the four strains of lactobacilli, L. rhamnosus (0.234 ± 0.020) had the highest hydrophobicity, followed by L. sakei (0.131 ± 0.009), L. brevis (0.056 ± 0.025) and L. plantarum (−0.144 ± 0.097) (all P < 0.01; ).
Table 3. Measurements of the hydrophobicity (M ± Q) of the four Lactobacillus isolates.
Self-aggregation ability of Lactobacillus
In order to unraveal the potential probiotic properties of the identified bacteria, the self-aggregation ability of the Lactobacillus isolates at 37 °C was determined. The results showed that the aggregation abilities of the four strains of lactobacilli increased in a time-dependent manner and the aggregation ability was highest at 24 h (). At one time point, such as 24 h, L. brevis (0.494 ± 0.014) had the highest self-aggregation ability, followed by L. rhamnosus (0.477 ± 0.011), L. plantarum (0.452 ± 0.012) and L. sake (0.420 ± 0.00). The statistical analysis showed that the differences were statistically significant (all P < 0.05).
Table 4. Self-aggregation ability (M ± Q) of the four Lactobacillus isolates at different points of times.
Aggregation of Lactobacillus and Hp
To study the interaction of Lactobacillus and Hp, the aggregation ability was evaluated at 37 °C. The four strains of lactobacilli and Hp showed strong aggregation ability, and the aggregation ability was also elevated with the advancement of the incubation time. The aggregation of different lactobacilli was different. Compared with the Bonferroni method, L. gasseri was used to compare the aggregation ability of four lactobacilli and Hp at different time points. The aggregation rate of L. brevis was higher than that of the other three lactobacilli (P < 0.05). The duration of the action could affect the aggregation of Lactobacillus, additionally, the aggregation of different lactobacilli varied with time.
Oxford Cup antibacterial test
In order to investigate the susceptibility of the identified bacteria against antibacterial agents, the Oxford Cup bacteriostatic test was performed. Levofloxacin was used as a negative control and amoxicillin was used as a positive control. The results in showed that the four strains of Lactobacillus and their supernatant had inhibitory effects on Hp growth. The results showed that L. plantarum and L. sake had the highest and the lowest inhibition, zone respectively, which indicated that L. plantarum had the strongest effect on Hp, and L. sake had the weakest effect.
Table 5. Anti-Hb activity. Inhibition zone (diameter, mm).
Urease activity of Hp by Lactobacillus
Hp urease activity was determined to further explore the effect of Lactobacillus on Hp. Ammonia can raise the pH value and then the colour of mixed bacterial liquid culture hence changes from yellow to red. The bacterial biomass and supernatant of L. sakei and L. plantarum could inhibit the activity of Hp urease, and the difference was statistically significant (P < 0.05; ). Additionally, L. brevis and L. rhamnosus were more effective than the supernatant in the inhibition of Hp urease activity (P < 0.05).
Table 6. Ability of the four Lactobacillus isolates to inhibit Helicobacter pylori urease activity (M ± Q).
Antioxidant capacity of Lactobacillus
The stronger the antioxidant ability of Lactobacillus, the longer it will survive in the environment of reactive oxygen free radicals, and consequently will be able to exert better probiotic functions. To understand the probiotic function of the Lactobacillus isolates, we assayed their antioxidant capacity. The results showed that the scavenging rate of DPPH by L. sake (79.22 ± 11.69)% was highest compared with that of L. rhamnosus (73.28 ± 11.47)%, L. plantarum (69.87 ± 12.90)% and L. brevis (62.34 ± 11.69)% (P < 0.05; ). According to the results, we concluded that the antioxidant ability of L. sake was the strongest and that of L. brevis was the weakest.
Table 7. DPPH scavenging rate (M ± Q) of the four Lactobacillus isolates.
Potential application
With the increase in drug resistance, long-term indiscriminate application of antibiotics can cause gastrointestinal disorders, imbalance of the gastrointestinal flora and other adverse reactions [Citation14]. Micro-ecological therapy is a new point of view to solve a variety of problems in the traditional therapy [Citation14]. Probiotics not only have anti-infective function, but also can regulate the immune function, balance the normal gastrointestinal tract flora, and reduce the side effects of antibiotics [Citation15]. Therefore, the application of probiotics has important significance in the prevention of Hp related diseases. In this study, four Lactobacillus isolates from fermented food in Northeast China that were identified as L. sakei, L. plantarum, L. rhamnosus and L. brevis based on biochemical assays and 16SrDNA sequencing [Citation16–18] were shown as potentially beneficial for the human body. The acidic environment in the stomach (pH 1.5–3.0) can kill bacteria unless they are sufficiently acid resistant to retain their viable state [Citation19]. Currently, the ability to tolerate real gastric juice is commonly assessed by measuring the growth of Lactobacillus in simulated artificial gastric juice environment, which provides evidence of whether a studied Lactobacillus strain can play a health-promoting and probiotic role in the gastrointestinal tract. The four strains of Lactobacillus studied by us showed resistance to pH 3.0 artificial gastric juice, indicating that they have the potential to maintain their viable state in acidic environment.
Among the four strains, L. rhamnosus had the highest hydrophobicity, whereas L. plantarum had the weakest hydrophobicity. The results were in contrast to the study performed by Agaliya and Jeevaratnam [Citation20], who reported that the hydrophobicity of L. plantarum was higher than that of L. rhamnosus isolated from fermented idli batter. The difference between the two studies may be due to the fact that the strains were isolated from different sources. Strong hydrophobic lactobacilli strains might be due to the presence of protein-like substances on their surface, and hence they had better self-protection. Hydrophobicity is a non-specific interaction between bacteria and gastric epithelial cells. Therefore, the stronger the hydrophobicity of the bacteria, the stronger the effect on the mucosal cells [Citation20].
It is documented that adhesion in the gastrointestinal tract is more solid and the beneficial effect they play is more potent when Lactobacillus strains show some aggregation. The self-aggregation ability of a strain is positively correlated with its adhesion in the intestinal tract. The self-aggregation of probiotic strains might be one of the reasons for maintaining the vitality of the strain and inhibiting the invasion of the upper gastrointestinal resistant pathogens. In addition, self-aggregation can cause intestinal stomata to form. In this study, we found that the self-aggregation ability of Lactobacillus from strong to weak was L. brevis, L. rhamnosus, L. plantarum and L. sake. Interactive aggregation plays an important role in removing pathogens from the intestine. The self-aggregation ability of Lactobacillus can block the adhesion of pathogens and colonization of the intestine. The interaction of Lactobacillus with pathogens can make them more easily excreted from the gut [Citation21]. Therefore, the aggregation of probiotics and intestinal pathogens is also a very important evaluation index. We found that the duration of action can affect the aggregation of Lactobacillus, and the aggregation of different lactobacilli varies with time. Additionally, the aggregation rate of L. brevis was higher than that of the other three lactobacilli, which indicated that L. brevis may have an important role in removing pathogens.
Hp is able to survive in the low acidity of the stomach as it can produce urease. Urease can decompose urea in the body to NH3 and CO2, and then the pH in the environment around Hp can rise, which may form a micro-environment where Hp can survive and proliferate [Citation22]. Therefore, if probiotics can inhibit Hp urease activity, this will case Hp to lose its ability to create a favourable environment for its survival of; thereby the colonization and growth of Hp in the stomach will be inhibited. The four strains of Lactobacillus assayed in this study could inhibit Hp urease activity. And L. brevis and L. rhamnosus were more effective than the supernatant in the inhibition of Hp urease activity.
Oxidation is a necessary process for most living organisms, but excessive oxidation can cause damage to biological macromolecules. In the environment of reactive oxygen species, the survival time of Lactobacillus with antioxidant activity is longer than without antioxidant activity [Citation23]. Most lactobacilli remove hydroxyl free radicals and hydrogen peroxide by producing antioxidant enzymes and glutathione [Citation23, Citation24]. The results from the DPPH-scavenging assay showed that L. sake can scavenge the DPPH most effectively of all four strains.
We also found that all four strains of Lactobacillus could inhibit Hp growth to a different extent. The acid resistance, the interaction with Hp and its antioxidant ability were the strongest for L. sake. L. plantarum had the strongest inhibitory effect on Hp and Hp urease activity by Oxford Cup antibacterial test. The hydrophobicity of the L. rhamnosus was relatively the strongest and the self-aggregation of L. brevis was the strongest. Taken together, the obtained data suggest that the four strains isolated from fermented food in Northeast China have potential as probiotics in the food industry.
Conclusions
In this study, four Lactobacillus strains identified as Lactobacillus sake, Lactobacillus plantarum, Lactobacillus rhamnosus and Lactobacillus brevis were isolated from fermented food in northeastern China. We found that the self-aggregation ability of Lactobacillus from strong to weak was L. brevis, L. rhamnosus, L. plantarum and L. sake. All these four strains of Lactobacillus could inhibit Hp growth to a different extent. Thus, the Lactobacillus isolated from fermented food in Northeast China has an inhibitory effect on Hp growth and may have the potential to be used as probiotic.
Disclosure statement
All authors declare no financial competing interests and no non-financial competing interests.
Additional information
Funding
References
- Loprieno N. International Agency for Research on Cancer (IARC) monographs on the evaluation of carcinogenic risk of chemicals to man: Relevance of data on mutagenicity. Mutat Res. 1975;31(3):201.
- Fujimura S, Watanabe A, Kimura K, et al. Probiotic mechanism of Lactobacillus gasseri OLL2716 strain against Helicobacter pylori. J Clin Microbiol. 2012;50(3):1134–1136.
- Jung JH, Cho IK, Lee CH, et al. Clinical outcomes of standard triple therapy plus probiotics or concomitant therapy for Helicobacter pylori infection. Gut Liver. 2018;12(2):165–172.
- Alvi S, Javeed A, Akhtar B, et al. Probiotics for cure of Helicobacter pylori infection: A review. Int J Food Prop. 2017;20(10):2215–2222.
- Wang F, Feng J, Chen P, et al. Probiotics in Helicobacter pylori eradication therapy: systematic review and network meta-analysis. Clin Res Hepatol Gastroenterol. 2017;41(4):466–475.
- Jang YN. AODWE-001 The impact of the probiotics of lactobacillus acidophilus on the outcomes of Helicobacter pylori eradication treatment. Gut. 2017;66(2):A120–A120.
- Zhao Y. Screening and preliminary identification of lactic acid bacteria in sauerkraut in Chifeng area. China Brewing. 2013;32(5):35–37. Chinese.
- Ding M, Liu Y, Ge Pingzhen, et al. Fermented acid in the cholesterol-lowering lactic acid bacteria screening, identification and cholesterol-lowering effect of the preliminary study. Food Sci. 2014;1(3):1–7. Chinese.
- Yang XY, Zhang QJ, Yu CL, et al. Advances in animal and medicine resistance of lactic acid bacteria from different sources. Adv Anim Med. 2014;35(2):73–77. Chinese.
- Zhou XX, Pan YJ, WYB, et al. In vitro assessment of gastrointestinal viability of two photosynthetic bacteria, Rhodopseudomonas palustris and Rhodobacter sphaeroides. J Zhejiang Univ Sci B. 2016;8(9):686–692.
- Collado MC, Meriluoto J, Salminen S. Adhesion and aggregation properties of probiotic and pathogen packages. Eur Food Res Technol. 2008;226(5): 1065–1073.
- Gong H, Wang HX, Ma ZT, et al. Adhesion of lactic acid bacteria and biofilm, hydrophobicity and self-agglutination characteristics. Chin J Micro. 2016;28(9):1026–1028,1033. Chinese.
- Zhou QR, Liu Y, Liu X, et al. Studies of the factors linked to the bacteriostatic ability of Lactobacillus in human vagina. Wei Sheng Yan Jiu. 2006,35(3): 310–313. Chinese.
- Yang JX, Yang JC. Probiotics on intestinal epithelial cell protection mechanism of the research progress. World Chin J Dig. 2015;23(4):577–583. Chinese.
- Fu LJ, Hao JF, Song YL, et al. Study on the effect of probiotics on intestinal mucosal immunity. Chi J Animal Sci and Vet Med. 2013;40(2):113–116. Chinese.
- Jonasson J, Olofsson M, Monstein H J. Classification, identification and subtyping of bacteria based on pyrosequencing and signature matching of 16S rDNA fragments. APMIS. 2002;110(3):263–272.
- Woo P C Y, Lau S K P, Teng J L L, et al. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect. 2008;14(10):908–934.
- Lee I M, Hammond R W, Davis R E, et al. Universal amplification and analysis of pathogen 16S rDNA for classification and identification of mycoplasmalike organisms. Phytopathology. 1993;83(8):834–842.
- Hu AH, Ao Xiaolin, Chen Cen, et al . Lactic acid bacteria resistance to acid and bile salt mechanism research progress. Food Sci Technol. 2015;36(8): 380–383. Chinese.
- Agaliya P J, Jeevaratnam K. Screening of Lactobacillus plantarum isolated from fermented idli batter for probiotic properties. Afr J Biotechnol. 2012;11(65): 12856–12864.
- Jin C. Screening and identification of high adhesion lactic acid bacteria and its surface hydrophobic characteristics [Dissertation]. Yangzhou: Yangzhou University; 2013. Chinese.
- Bhandari P, Prabha V. Evaluation of profertility effect of probiotic Lactobacillus plantarum 2621 in a murine model. Indian J Med Res. 2015;142(1):79–84.
- Palizban A, Saghaie L. Synthesis and evaluation of the complex-forming ability of hydroxypyranones and hydroxypyridinones with Ni (II) as possible inhibitors for urease enzyme in Helicobacter pylori. Res Pharm Sci. 2016;11(4):332–342.
- Lasrado LD, Gudipati M. Antioxidant property of synbiotic combination of Lactobacillus sp. and wheat bran xylo-oligosaccharides. J Food Sci Technol. 2015;52(7): 4551–4557.
