Abstract
Radix Bupleuri (root of Bupleurum spp.) is an important medicinal herb. Its lateral root number is one of the decisive factors that influence the content of a major bioactive component, saikosaponin. To identify genes associated with content and total yield of saikosaponin, it is of key importance to select stable references in gene expression analyses using quantitative real-time polymerase chain reaction (qRT-PCR). In this study, 18 candidate reference genes were selected and evaluated through their expression stability during the lateral root development in tissue samples from B. chinense DC., B. falcatum L. and B. scorzonerifolium Willd. The GeNorm, NormFinder and Bestkeeper methods were used for selecting stably expressed internal controls in the three Bupleurum species. These results revealed that, among these 18 candidate reference genes, ADF7 showed the best performance in all the experimental systems. ADF5 and ADF1b could also be proposed as suitable reference genes for gene expression studies. This study supplied more candidate reference genes to monitor the content and yield of saikosaponin during lateral roots growth in the Bupleurum genus.
Introduction
In biological research, real time quantitative reverse transcription-PCR (qRT-PCR) is increasingly being used in gene expression analysis due to its technical ease, low reagent cost, less hands-on time, reproducibility and high throughput [Citation1]. However, multiple factors such as sample amount, RNA recovery, RNA integrity, cDNA quality and tissue or cell activities can affect the quantitative measurement of gene expression [Citation2]. To achieve accurate and stable results, normalization is required to correct for these variations. For normalization, one or several reference genes should serve as internal controls to normalize and monitor the expression variation between samples and reactions.
Theoretically, an ideal reference gene is stably expressed in various samples across different experimental conditions or treatments. However, no gene is universally stable among different plant species and differing experimental conditions. Hundreds of reference genes have been validated in plants including Oryza sativa, Arabidopsis thaliana, Zea mays, Brassica juncea, Brassica napus, Solanum tuberosum, Solanum lycopersicum L., Setaria italic L., Brachypodium beauv, Hordeum vulgare, Sorghum bicolor, Triticum aestivum, Glycine max, Vitis vinifera, Cucumis sativus, Nicotiana tabacum, Phyllostachys edulis and Bupleurum chinense DC. [Citation3–5]. These genes have been validated across different tissues and different treatments by comparative delta Ct method, Bestkeeper, NormFinder and GeNorm [Citation2].
Radix Bupleuri (root of Bupleurum spp.) is one of the most important medicinal herbs in Eurasia and North Africa as treatment for fever, chronic hepatitis, nephrotic syndrome, inflammatory diseases, menstrual disorders and digestive ulcers [Citation6–9]. Among the 190 species of the Bupleurum genus, only B. chinense DC. and B. scorzonerifolium Willd. are officially recorded as the source species of Radix Bupleuri in the Chinese Pharmacopoeia [Citation10–12]. In the Japanese Pharmacopoeia, the official botanical origin of Radix Bupleuri is the root of B. falcatum L. [Citation13]. It is believed that saikosaponins are responsible for the pharmaceutical properties of Bupleuri Radix, especially the oleanane-saponins, saikosaponin a and saikosaponin d [Citation14]. According to previous studies, the content and total yield of saikosaponin depend on the type of tissue, the growth period, the root structure and the environmental conditions such as drought, fertilizer treatment and light deficiency [Citation15–17]. A previous study characterized 11 candidate genes for their suitability as reference genes in B. chinense DC.; however, it did not evaluate these genes in relation to saikosaponin content and yield [Citation5].
In this study, 18 genes, including Ubiquitin-protein ligase gene (UBC), Actin depolymerizing factor (ADF), Actin (ACT), Eukaryotic translation initiation factor (eIF) and Eukaryotic translational elongation factor (EF), were selected as candidate reference genes for evaluation based on the analyses by three software programs (Bestkeeper, NormFinder and GeNorm) in B. chinense DC. B. scorzonerifolium Willd. and B. falcatum L. This research analyzed eight samples and aimed to select the well-founded gene which could potentially be used as a candidate reference gene in Bupleurum genus experiments in different tissues with various treatments.
Materials and methods
Plant materials
The three experimental materials, Zhongchai No. 2, Zhonghongchai No. 1 and B1, were from three species, B. chinense DC, B. scorzonerifolium Willd. and B. falcatum L., respectively. All of them were bred by systemic selection and purification selection from farmholding populations. Zhongchai No. 2 and Zhonghongchai No. 1 were provided by Professor Jianhe Wei from the Institute of Medicinal Plant Development (IMPLAD), Chinese Academy of Medical Sciences & Peking Union Medical College. B1 was provided by Professor Da-Bin Hou from the School of Life Science and Engineering, Southwest University of Science and Technology.
For each genotype, 10 plants with similar growth vigour were selected to harvest as whole plants before lateral root germination. Tissue samples of the leaves and roots were taken after the first lateral root germination at the seedling stage. During the fruiting period, five plants of similar height and structure were selected for harvesting of their roots, stems, leaves, blossoms and fruit. Another replication of each tissue sample was collected from the same experimental plot at both the seedling and fruiting stage. All tissue samples were wrapped in tinfoil, immediately flash-frozen in liquid nitrogen and then kept at −80 °C until RNA isolation.
RNA isolation and cDNA synthesis
RNAprep Pure Plant Kit (DP441) (TIANGEN BIOTECH (BEIJING) CO., Beijing, China) was used in the RNA isolation and genomic DNA elimination of 16 tissue samples following the manufacturer’s instructions. The integrity of the RNA samples was checked by agarose gel electrophoresis, and the concentration and quality were examined by NanoDrop 2000 (Thermo, USA) at 230, 260 and 280 nm. Synthesis of cDNA was performed using the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Canada) following the manufacturer's protocol [Citation2].
qRT-PCR
A total of 18 candidate reference genes were selected for qRT-PCR (). Real-time PCR was carried out using the Trans-AQ111-02 Green qPCR SuperMix UDG (TransGen Biotech, Beijing, China) and the ABI CFX96 Touch™ Real Time PCR System (Applied Biosystems, Foster City, CA, USA). A reaction mixture of a total volume of 10 μL in each well in an optical 96-well plate was employed for qRT-PCR. This reaction mixture contained 5 μL of Trans-AQ111-02 Green qPCR SuperMix, 5 pmol/L of each primer, 5 ng of final cDNA and 3.4 μL of RNase-free water. The PCR procedures were described previously [Citation5]. The ABI CFX Manager Software V3.1 was used for visualizing and analyzing the data, including the quantification cycle values, PCR efficiency and correlation coefficients.
Table 1. Oligonucleotide primers for amplification of 18 candidate reference genes.
Data analysis
Data are presented as mean values with standard deviation (±SD). After collecting and converting the quantification cycle (Cq) data, Cq average values were calculated statistically by SPSS 16.0 software (http://www.spss.com/). To obtain reliable results, the software programs Bestkeeper, NormFinder and GeNorm were used to analyse the expression stability of reference genes (RefFinder, http://150.216.56.64/referencegene.php). Pearson correlation coefficients were generated for ranking results from four different algorithms using Minitab 15 software (http://www.minitab.com/).
Results and discussion
Expression profile of candidate reference genes
QRT-PCR has become a standard method for detection and quantification of RNA targets, because of its sensitivity, specificity and accuracy [Citation18]. However, due to the potential systematic variation introduced by total RNA, first-strand cDNA synthesis and qRT-PCR assay, there is a need to normalize the raw expression data by expressing internal controls for accurate and reliable results [Citation19,Citation20]. Previous studies have shown that the expression of such controls could significantly change the stability in the tested plant tissues under differing experimental conditions. Therefore, no single control is appropriate for all experimental treatments [Citation21,Citation22]. In addition, with a variable reference gene, there could occur nearly 100-fold variations in the quantified expression of the target gene. This could eventually result in misinterpretation of the expression pattern and faulty understanding of the mechanisms under study [Citation23]. Therefore, it is generally suggested to select suitable internal controls prior to use for normalization of specific experimental conditions.
In Bupleurum genus, saikosaponins are the most important bioactive components due to their pharmacological properties [Citation11]. Previous studies have reported the biosynthetic pathways of saikosaponins in B. falcatum L. B. kaoi, B. chinense DC. and B. scorzonerifolium Willd. [Citation24–28]. Genes involved in the biosynthesis of saikosaponins such as squalene epoxidase, β-amylase, cytochrome P450 and uridine diphosphate glycosyltransferases were cloned and identified by their expression profiles in B. kaoi, B. falcatum L. and B. chinense DC [Citation25,Citation29–31]. However, only a few of these genes were associated with the content and total yield of saikosaponins, and none of them have been utilized in the metabolic pathway in saikosaponin production. Previous histochemical studies on B. chinense DC., B. falcatum L. and B. scorzonerifolium Willd. have demonstrated that saikosaponins are mainly found in the epidermal areas of the roots [Citation15,Citation32–34]. This was confirmed as plants with more lateral roots showed higher saikosaponin content than plants with less lateral roots. So the lateral root number is one of the decisive factors that influences the saikosaponin content.
To examine the genetic mechanism of lateral root development in B. chinense DC., B. falcatum L. and B. scorzonerifolium Willd., a total of 18 candidate reference genes were selected for determining the most stable one at various developmental stages and tissues. Amplification of each reference gene in 24 samples (2 replicates per sample) produced 48 Cq values, and samples with missing Cq values or inconsistencies between replicates (Cq differences >0.5 cycle) were removed from the analysis. Based on the standard curves using a serial dilution of cDNA samples, the efficiency of gene amplification ranged from 91.08% to 108.33%. The observed correlation coefficient R2 values for most of the genes varied in the range of 0.989–1.000.
Over all, the Cq values of the 18 candidate reference genes varied over a wide range, and the mean Cq values of these genes varied in the samples from 16.82 to 30.17 (). Among these candidate reference genes, UBC13 was the most abundantly expressed gene (mean Cq ± SD =16.82 ± 1.27) followed by ADF1b (mean Cq ± SD =16.84 ± 0.91), whereas eIF6 was the least abundantly expressed gene (mean Cq ± SD =30.17 ± 6.60). All candidate reference genes showed small standard deviations (SD) from 0.63 to 1.80, with the exception of eIF6, which presented the largest variation among the Cq values (6.60). An individual value plot was used to evaluate and to compare all the samples. The results showed that all the genes had a similar distribution or trend except for eIF6 (). eIF6 is an essential component of ribosome biogenesis, so its gene is ubiquitously expressed. This gene is involved both in ribosome biogenesis and protein synthesis, which might induce great expressional changes in different organs and species. Similar results have been reported in Arabidopsis and Oryza sativa [Citation35].
Figure 1. Expression levels of candidate reference genes tested using the qPCR cycle threshold values (Cq).
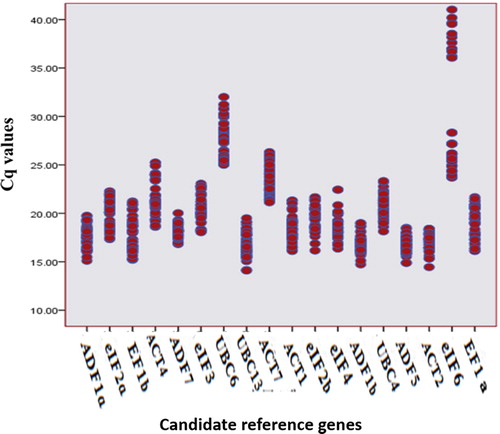
Table 2. Expression levels of 18 reference genes in B. chinense DC., B. falcatum L. and B. scorzonerifolium Willd.
Expression stability of the eighteen candidate reference genes
In order to analyse the expression of the candidate reference genes in greater detail, the 24 samples were divided into four experiment sets. Set 1 to 3 consisted of 8 samples from B. chinense DC., B. falcatum L. and B. scorzonerifolium Willd. individually. In set 4, all 3 sample sets were included. Three software packages, Bestkeeper, NormFinder and GeNorm, which use different algorithms were used to analyse and evaluate the stability of the candidate reference genes in the four experiment sets.
GeNorm analysis
Based on the expression stability analysis of the 18 candidate reference genes by GeNorm (), the five top ranked genes in B. chinense DC. were ADF1b = ADF5 > ADF7 > eIF2b > ACT2. The top ranked genes in B. falcatum L. were ACT2 > UBC4 > ADF5 > ADF1b > ADF7, and those in B. scorzonerifolium Willd. were ADF5 > ACT2 > ADF7 > eIF2b > eIF2a. The top five genes that were consistently expressed in set 4 were ADF7 = ADF1b > ADF5 > ACT2 > UBC4. In all four sets, ADF5, ADF7 and ACT2 were always determined as three of the top five most stable reference genes.
Table 3. Expression stability of 18 candidate reference genes as calculated by GeNorm.
NormFinder analysis
NormFinder analysis revealed that ADF7 and ADF1b were always two of the top five most stable reference genes in all four sets (). Gene ACT7 had the best stability of the 18 candidate reference genes in set 3, and occupied the top 6th and 3rd place in the 1st and 4th sets, respectively. However, this gene ranked 11th in set 2. ADF5 was one of the top five most stable reference genes in set 1, 2 and 4 but ranked 6th in set 3. ACT2 was rated as the 5th and 4th most stable reference gene in set 1 and 3, respectively.
Table 4. Expression stability of 18 candidate reference genes as calculated by NormFinder.
BestKeeper analysis
Average Cq values were used to calculate the coefficient of variance (CV) and SD for each of the reference genes in BestKeeper analysis. Genes with higher variation were classified as less stable, whereas genes with lower variation were more stable. Based on this analysis, ADF7, UBC13, ADF1b and ADF5 always ranked as four of the top five most stably expressed genes across all the datasets ().
Table 5. Expression stability of 18 candidate reference genes as calculated by BestKeeper.
Comprehensive analysis of expression stability
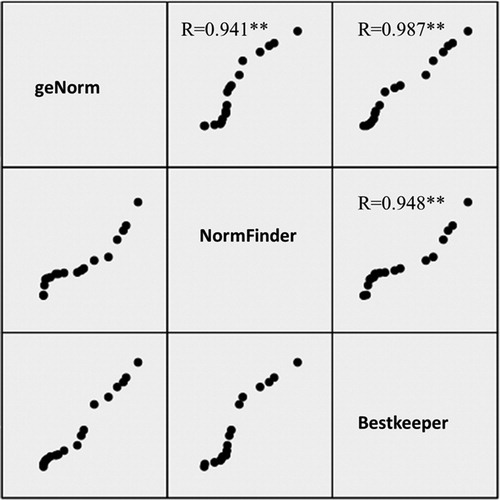
For a comprehensive judgment of the suitable reference genes in the four sets, Pearson correlations were calculated using the ranks from the most stable to the least stable among the three methods (comparative GeNorm, NormFinder and Bestkeeper) used in this study. The Pearson correlations for the three stability tests showed a significant or extremely significant positive correlation in all of the four sets (). This indicated that the ranking results from all of the three methods were nearly identical.
Figure 2. Comparison of the ranking results from BestKeeper, NormFinder and GeNorm. The correlation was evaluated for the ranking results of 18 candidate reference genes in all samples, by Bestkeeper, NormFinder and GeNorm. Correlation coefficient (r) values are shown (*p < 0.05, **p < 0.01).
In previous studies, GAPDH, ACT, EF, eIF, UBC and 18S rRNA have already been used as reference genes for expression studies in many plant species [Citation2,Citation36,Citation37]. Different reference genes have been used in Bupleurum species. Actin has been used as the internal control in B. kaoi [Citation26]; the genes for β-tubulin and actin, as reference genes in B. chinense DC; and β-actin, as the reference gene in B. falcatum L. [Citation5,Citation24,Citation31]. In this study, we observed similar rankings of stability among three Bupleurum species. In GeNorm analysis, ADF5, ADF7 and ACT2 were three of the top five most stable reference genes in all three species. In NormFinder analysis, ADF7, ADF1b and eIF2b were three of the top five most stable reference genes in all three species. In BestKeeper analysis, ADF7, UBC13, ADF1b and ADF5 were ranked as four of the top five most stably expressed genes in all three species. The differences in these stability rankings could be attributed to the fact that the computational programs use different approaches and algorithms, rather than to species differences. Taking into account the results from all the three software analysis methods (), the gene ADF7 was considered as one of the most suitable reference genes for normalization of all the samples of the three species of Bupleurum L. It is also worthy to note that all the three programs showed similar stability rankings on the least stable genes such as eIF6 in B. falcatum L. and ACT4 in B. scorzonerifolium Willd.
Conclusions
In this study, the gene ADF7 performed optimally in all the experiments. Genes ADF5 and ADF1b could be also proposed as good starting points for gene expression studies. However, it is recommended to choose more than one reference gene for normalization, such that each of the chosen genes is involved in different biological functions and pathways.
Disclosure statement
The authors declare that they have no conflict of interest.
Funding
This work was supported by the National Natural Science Foundation of China under grant number 81603223, China Agriculture Research System under grant number CARS-21, CAMS Innovation Fund for Medical Sciences (CIFMS) under grant number 2016-I2M-2-003 and the Crop and livestock Breeding Project in Sichuan under grant number 2016NYZ0020.
References
- Kubista M, Andrade JM, Bengtsson M, et al. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27:95–125.
- Long X, He B, Gao X, et al. Validation of reference genes for quantitative real-time PCR during latex regeneration in rubber tree. Gene. 2015;563:190–195.
- Zhang K, Niu SF, Di DP, et al. Selection of reference genes for gene expression studies in virus-infected monocots using quantitative real-time PCR. J Biotechnol. 2013;68:7–14.
- Nakayama TJ, Rodrigues FA, Neumaier N, et al. Reference genes for quantitative real-time polymerase chain reaction studies in soybean plants under hypoxic conditions. Genet Mol Res.. 2014;13:860–871.
- Dong L, Sui C, Liu Y, et al. Validation and application of reference genes for quantitative gene expression analyses in various tissues of Bupleurum chinense. Mol Biol Rep.. 2011;38:5017–5023.
- Pistelli L, Bertoli A, Bilia AR, et al. Minor constituents from Bupleurum fruticosum roots. Phytochemistry. 1996;41:1579–1582.
- Guo YJ, Matsumoto T, Kikuchi Y, et al. Effects of a pectic polysaccharide from a medicinal herb, the roots of Bupleurum falcatum L. on interleukin 6 production of murine B cells and B cell lines. Immunopharmacology. 2000;49:307–316.
- Ikegami F, Sumino M, Fujii Y, et al. Pharmacology and toxicology of Bupleurum root-containing Kampo medicines in clinical use. Hum Exp Toxicol. 2006;25:481–494.
- Mabberley DJ. Mabberley’s plant-book: a portable dictionary of plants, their classification and uses. New York: Cambridge University Press; 2008.
- State Pharmacopoeia Committee. Pharmacopoeia of the People's Republic of China. Beijing (China): China Medical Science and Technology Press; 2015. p. 280.
- Ashour ML, Wink M. Genus Bupleurum: a review of its phytochemistry, pharmacology and modes of action. J Pharm Pharmacol. 2011;63:305–321.
- Yang F, Dong X, Yin X, et al. Radix Bupleuri: a review of traditional uses, botany, phytochemistry, pharmacology, and toxicology. Biomed Res Int. 2017;2017:7597596. [cited 2018 Nov 28]. DOI:10.1155/2017/7597596
- The Society of Japanese Pharmacopoeia. The Japanese Pharmacopoeia 16th ed (English Version). Tokyo (Japan): The Ministry of Health, Labor and Welfare; 2011; p. 1613–1614.
- Zhu Z, Liang Z, Han R, et al. Growth and saikosaponin production of the medicinal herb Bupleurum chinense DC. under different levels of nitrogen and phosphorus. Industrial Crops and Products. 2009;29:96–101.
- Tan LL, Cai X, Hu ZH, et al. Localization and dynamic change of saikosaponin in root of Bupleurum chinense. J Integr Plant Biol. 2008;50:951–957.
- Zhu Z, Liang Z, Han R. Saikosaponin accumulation and antioxidative protection in drought-stressed Bupleurum chinense, DC. plants. Environ Exp Bot. 2009;66:326–333.
- Gong J, Liu M, Xu S, et al. Effects of light deficiency on the accumulation of saikosaponins and the ecophysiological characteristics of wild Bupleurum chinense DC. in China. Industr Crop Product. 2017;99:179–188.
- Galeano E, Vasconcelos TS, Ramiro DA, et al. Identification and validation of quantitative real-time reverse transcription PCR reference genes for gene expression analysis in teak (Tectona grandis Lf). BMC Res Notes. 2014;7:464. [cited 2018 Nov 28]. DOI:10.1186/1756-0500-7-464
- Chandna R, Augustine R, Bisht NC. Evaluation of candidate reference genes for gene expression normalization in Brassica juncea using real-time quantitative RT-PCR. PLoS One. 2012;7:e36918. [cited 2018 Sep 28]. DOI:10.1371/journal.pone.0036918.
- Chan PL, Rose RJ, Murad AMA, et al. Evaluation of reference genes for quantitative real-time PCR in oil palm elite planting materials propagated by tissue culture. PLoS One. 2014;9:e99774. [cited 2018 Sep 28]. DOI:10.1371/journal.pone.0110079.
- Li Q, Fan CM, Zhang XM, et al. Validation of reference genes for real-time quantitative PCR normalization in soybean developmental and germinating seeds. Plant Cell Rep. 2012;31:1789–1798.
- Xiao D, Zhang NW, Zhao JJ, et al. Validation of reference genes for real-time quantitative PCR normalisation in non-heading Chinese cabbage. Functional Plant Biol.. 2012;39:342–350.
- Gutierrez L, Mauriat M, Pelloux J, et al. Towards a systematic validation of references in real-time RT-PCR. Plant Cell. 2008;20:1734–1735.
- Kim YS, Cho JH, Ahn J, et al. Upregulation of isoprenoid pathway genes during enhanced saikosaponin biosynthesis in the hairy roots of Bupleurum falcatum. Mol Cells. 2006;22:269–274.
- Kim YS, Cho JH, Park S, et al. Gene regulation patterns in triterpene biosynthetic pathway driven by overexpression of squalene synthase and methyl jasmonate elicitation in Bupleurum falcatum. Planta. 2011;233:343–355.
- Chen LR, Chen YJ, Lee CY, et al. MeJA-induced transcriptional changes in adventitious roots of Bupleurum kaoi. Plant Sci. 2007;173:12–24.
- Sui C, Zhang J, Wei JH, et al. Transcriptome analysis of Bupleurum chinense focusing on genes involved in the biosynthesis of saikosaponins. BMC Genomics. 2011;12:539. [cited 2018 Sep 28]. DOI:10.1186/1471-2164-12-539.
- Sui C, Chen M, Xu J, et al. Comparison of root transcriptomes and expressions of genes involved in main medicinal secondary metabolites from Bupleurum chinense and Bupleurum scorzonerifolium, the two Chinese official Radix Bupleuri source species. Physiol Plantarum. 2015;153:230–242.
- Liu WY, Peng PH, Lin TY. Cloning and characterization of beta-amyrin synthase from Bupleurum kaoi. 8th International Congress of Plant Molecular Biology. Book of Abstracts, ISPMB, 2016 Aug 20–25, POS-TUE-121; Adelaide, Australia; 2006.
- Sui C, Wei JH, Chen SL, et al. Construction of a full-length enriched cDNA library and analysis of 3111 ESTs from root of Bupleurum chinense DC. Bot Stud. 2010;51:7–16.
- Gao K, Xu JS, Sun J, et al. Molecular cloning and expression of squalene epoxidase from a medicinal plant, Bupleurum chinense. Chinese Herbal Med. 2016;8:67–74.
- Du XW, Liu MY. Studies of histochemistry of saikosaponins. China J Chin Mater Med. 1992;17:261–263.
- Zhao X, Zheng L, Si J, et al. Immunocytochemical localization of saikosaponin-d in vegetative organs of Bupleurum scorzonerifolium Willd. Bot Stud. 2013;54:32. [cited 2018 Sep 28]. DOI:10.1186/1999-3110-54-32
- Liang Z, Oh K, Wang Y, et al. Cell type-specific qualitative and quantitative analysis of saikosaponins in three Bupleurum species using laser microdissection and liquid chromatography–quadrupole/time of flight-mass spectrometry. J Pharm Biomed Analysis. 2014;97:157–165.
- Kato Y, Konishi M, Shigyo M, et al. Characterization of plant eukaryotic translation initiation factor 6 (eIF6) genes: The essential role in embryogenesis and their differential expression in Arabidopsis and rice. Biochem Biophys Res Commun. 2010;397:673–678.
- Goulao LF, Fortunato AS, Ramalho JC. Selection of reference genes for normalizing quantitative real-time PCR gene expression data with multiple variables in Coffea spp. Plant Mol Biol Rep. 2012;30:741–759.
- Reddy PS, Reddy DS, Sharma KK, et al. Cloning and validation of reference genes for normalization of gene expression studies in pearl millet [Pennisetum glaucum (L.) R. Br.] by quantitative real-time PCR. Plant Gene. 2015;1:35–42.
