?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
An antibacterial peptide gene (designated as accA) was found in Bacillus amyloliquefaciens CMW1. The accA gene consists of 336 nucleotides and encodes 111 amino acids. The accA gene product, amylocyclicin CMW1, consists of a leader peptide and a mature peptide. The genome analysis indicated that the accA gene constitutes a gene cluster together with five open reading frames (accBCDEF genes) that are predicted to play roles in amylocyclicin CMW1 biosynthesis. Amylocyclicin CMW1 is similar to precursor proteins of amylocyclicin FZB42 and uberolysin A, which are known as circular hydrophobic bacteriocins. To investigate the antimicrobial activity of the accA gene product, we constructed recombinant Escherichia coli cells expressing amylocyclicin CMW1, His-tagged amylocyclicin CMW1 or His-tagged mature amylocyclicin CMW1. The culture supernatant, the cell-free extract and debris fraction were prepared with recombinant E. coli cells expressing amylocyclicin CMW1. Antibacterial activity against Bacillus subtilis JCM10629 was detected in the debris fraction, but not in the culture supernatant or cell-free extract. His-tagged mature amylocyclicin CMW1 inhibited Gram-positive bacteria but not Gram-negative bacteria. Unlike amylocyclicin FZB42, His-tagged mature amylocyclicin CMW1 exhibited antibacterial activity against Staphylococcus aureus. Electrospray ionization mass spectrometry of His-tagged mature amylocyclicin CMW1 indicated that the peptide is a linear rather than a circular peptide.
Introduction
Circular bacteriocins constitute a group of ribosomally synthesized antimicrobial peptides and are of great interest due to their potential industrial applications as novel antibiotics [Citation1–3]. Circular bacteriocins are synthesized as linear precursor proteins, containing a leader peptide of variable size that is cleaved off during maturation. In the classification of Gram-positive bacteriocins, circular bacteriocins have been considered to be class II non-modified peptides and are often assigned to subclass IIc and IId [Citation4–6]. So far, it has been reported that circular bacteriocins are produced by lactic acid bacteria, for example, uberolysin A (GenBank accession no. DQ650653) from Streptococcus uberis strain 42 [Citation7], garvicin ML (GenBank accession no. EKF52513) from Lactococcus garvieae DCC43 [Citation8] and leucocyclicin Q (GenBank accession no. BAL14584) from Leuconostoc mesenteroides TK41401 [Citation9]. However, a few bacteriocins from Bacillus sp. bacteria have been reported. To our knowledge, only amylocyclicin FZB42 was reported to belong to the unique cyclic bacteriocins produced by Bacillus amyloliquefaciens FZB42 [Citation10]. Amylocyclicin FZB42 inhibits most bacilli, but not Staphylococcus aureus, Escherichia coli K-12, Klebsiella terrigena, Pseudomonas sp. or Erwinia carotovora. Amino acid sequence analysis indicated that amylocyclicin FZB42 is distinct from other bacteriocins produced by lactic acid bacteria, and genome analysis indicated that this type of bacteriocin gene is widespread among the Bacillus taxa [Citation4,Citation11,Citation12]. Although the bacteriocins from Bacillus sp. are considered to play crucial roles in antimicrobaial activities among the bacteria, there are many unexplained points concerning functions of the circular bacteriocins. Genome mining of Bacillus sp. bacteria let us obtain a novel bacteriocin that is different from the bacteriocins produced by lactic acid bacteria.
B. amyloliquefaciens CMW1 (16S rRNA gene GenBank accession no. AB983212) is a unique strain that was isolated from a Japanese fermented soybean paste [Citation13]. The strain CMW1 tolerates growth in the presence of 10% (vol/vol, 631 mM) 1-butyl-3-methylimidazolium chloride ([BMIM]Cl) [Citation14]. CMW1 belongs to the ionic liquid-tolerant bacteria. The whole genome (3,908,571 bp) of B. amyloliquefaciens CMW1 was sequenced and a total of 9,175 protein-coding genes were predicted. Bioinformatic analysis using the amino acid sequence of uberolysin A (GenBank accession no. DQ650653) indicated a novel bacteriocin gene (accA gene) that belongs to the class IId bacteriocins in the whole genome sequence of strain CMW1. RNA-seq analysis of strain CMW1 indicated that the accA gene was transcribed (Kurata, unpublished data). The predicted amino acid sequence of the accA gene from strain CMW1 is similar to that of amylocyclicin FZB42, so we named the accA gene product ‘amylocyclicin CMW1’. At present, there is a demand for the development of bacteriocins as novel antibiotics. Therefore, the objective of this study was to elucidate the properties of amylocyclicin CMW1.
Materials and methods
Bacterial strains and culture conditions
Strain CMW1 was cultivated aerobically at 37 °C for 24 h in the medium containing 1% yeast extract (Becton Dickinson, MD, USA), 0.5% tryptone (Becton Dickinson, MD, USA), 1% NaCl and 1% skim milk (Morinaga Milk, Japan) at pH 7.0 [Citation14]. Cell growth was measured based on the optical density at 600 nm. The following strains were used in this study: Gram-positive bacteria: Bacillus subtilis JCM10629, Bacillus licheniformis JCM2505T, Bacillus pumilus JCM2508T, Staphylococcus aureus NBRC12732 and Staphylococcus carnosus JCM6067T, and Gram-negative bacteria: Escherichia coli NBRC3301, Enterobacter asburiae JCM6051T and Pseudomonas putida JCM13063T. The bacterial strains were cultured on Luria–Bertani (LB) medium (Becton Dickinson, USA) at 28–37 °C.
Identification of amylocyclicin CMW1 gene cluster
The whole genome of strain CMW1 (GenBank accession no. DF836084-DF836091) was sequenced with paired-end and mate-paired libraries using a Miseq sequencer [Citation13]. A local BLAST analysis of the whole genome sequence of strain CMW1 with the uberolysin A gene cluster (GenBank accession no. DQ650653.1) determined the DNA fragment encoding amylocyclicin CMW1 (accA gene) using Geneious software (Biomatters Ltd., New Zealand). The acc gene cluster comprising accB, accA, accC, accD, accE and accF is indicated in using Genetyx software (Genetyx, Japan) and included in scaffold 2 (1,073,793 bp) in the whole genome sequence of strain CMW1[Citation13]. The accA gene sequence of strain CMW1 was submitted to the NCBI GenBank database (accession no. LC373979).
Figure 1. Comparison of the bacteriocin gene clusters. (A) A putative amylocyclicin CMW1 gene cluster from B. amyloliquefaciens CMW1, (B) Uberolysin A gene cluster from S. uberis strain 42 (ublABCDE genes, GenBank accession no. DQ650653 [Citation7]).
![Figure 1. Comparison of the bacteriocin gene clusters. (A) A putative amylocyclicin CMW1 gene cluster from B. amyloliquefaciens CMW1, (B) Uberolysin A gene cluster from S. uberis strain 42 (ublABCDE genes, GenBank accession no. DQ650653 [Citation7]).](/cms/asset/22fb66b9-b07f-43bf-a8fe-35a0b778922d/tbeq_a_1627246_f0001_b.jpg)
Expression of amylocyclicin CMW1 and derivatives with recombinant E. coli cells
The DNA fraction of strain CMW1 was extracted from a bacterial cell pellet using the Qiagen DNA Genomic-Tip Kit (QIAGEN, Germany) according the manufacturer’s recommendations. DNA fragments coding for amylocyclicin CMW1 and derivatives were amplified by PCR, the genomic DNA of B. amyloliquefaciens CMW1 as a template with primers 5′-CACCATGAACTTAGTAAAATCTAATA-3′/5′-TTACCAAGCAGCTGCGTAT-3′ for amylocyclicin CMW1, 5′-CACCATGAACTTAGTAAAATCTAATA-3′/5′-CCAAGCAG CTGCGTATTTTTT-3′ for amylocyclicin CMW1 His-tag and 5′-CACCATGTTAGCTTCGACTCTTGGCA-3′/5′-CCAAGCAGC TGCGTATTTTTT-3′ for mature amylocyclicin CMW1 His-tag. The programme was as follows: 30 cycles of 98 °C for 10 s, 55 °C for 15 s and 72 °C for 30 s. The amplified products were cloned into the expression vector pET101/D-TOPO (Invitrogen, CA) downstream of the T7lac promoter. Each plasmid was introduced into E. coli BL21(DE3). Each of the recombinant E. coli BL21(DE3) cells was grown in LB medium containing 100 μg/mL ampicillin at 37 °C. When the absorbance at 600 nm reached 0.6, isopropyl-1-thio-d-galactopyranoside (IPTG) was added to the culture medium to a final concentration of 2 mM. After cultivation for 20 h at 20 °C in the presence of IPTG, the cells were harvested and the culture supernatant was obtained. Each recombinant cell was suspended in 500 μL of 10 mM potassium phosphate buffer (pH 7.0) and disrupted by sonication. The cellular debris was removed by centrifugation, and each supernatant was obtained as the cell-free extract. The cellular debris was resuspended in 500 μL of 10 mM potassium phosphate buffer (pH 7.0) to prepare the debris fraction.
Agar spot assay for antibacterial activity
Using the culture supernatant, cell-free extract and debris prepared from recombinant E. coli cells, antimicrobial activities against B. subtilis JCM10629, B. licheniformis JCM2505T, B. pumilus JCM2508T, S. aureus NBRC12732, S. carnosus JCM6067T, E. coli NBRC3301, E. asburiae JCM6051T and P. putida JCM13063T were evaluated by agar-spot assays with a penicillin cup (small stainless-steel column, 8 mm inner diameter). LB agar plate (20 mL) was mixed with 1.0 × 107 cfu/mL of one of the indicated strains described above (OD at 600 nm, 1.0). Then, 50 μL of the culture supernatants, 50 μL of the cell-free extracts at 16 mg/mL or 50 μL of debris fractions at 41 mg/mL, which were prepared from recombinant E. coli cells harbouring each expression vector, were applied to a penicillin cup and incubated for 44 h at 22 °C. Inhibitory activity appeared as a clear zone under the penicillin cup. All experiments for evaluation of the antimicrobial activity were carried out in triplicate.
Analysis of mature amylocyclicin CMW1 His-tag by mass spectrometry
Electrospray ionization mass spectrometry (ESI–MS) analysis was performed to determine the molecular weight of mature amylocyclicin CMW1. The debris fraction at 50 μg/mL in 50% MeOH with 10 mM potassium phosphate buffer (pH 7.0) was prepared from recombinant E. coli cells harbouring the expression vector for mature amylocyclicin CMW1 His-tag and was introduced into a mass spectrometer (Waters Q-TOF Premier, Micromass MS Technologies, UK) at a flow rate of 10 μL/min with the capillary voltage of 2.5 V, collision energy of 5.0 V and source temperature of 100 °C. The molecular ion of mature amylocyclicin CMW1 was detected in the positive ion mode.
Results and discussion
Finding of acc gene cluster involved in amylocyclicin CMW1 biosynthesis
In the whole genome (3,908,571 bp) of B. amyloliquefaciens CMW1, a total of 9,175 protein-coding genes were predicted [Citation13]. As shown in , we found a putative amylocyclicin CMW1 gene cluster (, named accABCDEF genes) in strain CMW1 genomic DNA using nucleotide sequences of uberolysin A gene cluster (, ublABCDE genes, GenBank accession no. DQ650653 [Citation7]) from S. uberis strain 42. In , the bacteriocin precursor gene, membrane protein gene, ATPase and immunity protein are indicated by a closed arrow, open arrow, shaded arrow and dashed arrow, respectively.
As shown in , a single open reading frame encoding amylocyclicin CMW1, which started from an ATG codon at nucleotide +1 and ended with a TAA codon (asterisk) at nucleotide +336, was found in the sequence and named accA. The G + C content of the accA gene was 40%. The deduced amino acids are indicated by the single-letter codes under the nucleotide sequence. A putative ribosome-binding site (RBS) of 5′-GGAGAT-3′ was found 3 bp upstream from the initiation codon ATG. A putative promoter sequence 5′-GAGAGAGTCAA-3′, as the potential –35 region, and 5′-TATTAT-3′, as the potential –10 region, with 13 bp spacing was located 28 bp upstream from the initiation codon. An inverse-repeat sequence (convergent arrows) was found 21 bp downstream of the TAA stop codon.
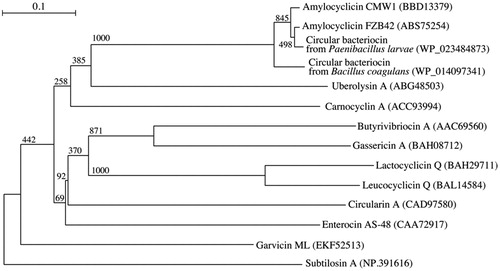
Amylocyclicin CMW1 consists of 111 amino acids. The results of the phylogenetic analysis based on amino acid sequence data are shown in ; amylocyclicin CMW1 clearly falls within the group of circular bacteriocins from Bacillus and related bacteria and is distinct from circular bacteriocins from lactic acid bacteria. Bootstrap values were calculated from multiple resamplings of the sequence data set, which are the basis for multiple tree topologies. GenBank accession numbers are given in parentheses.
Figure 3. Phylogenetic tree with amylocyclicin CMW1 and related circular bacteriocins based on mature amino acid sequences. Note: Bar 0.1 nucleotide substitutions per site.
Amylocyclicin CMW1 is similar to precursor proteins of uberolysin A (UblA, DQ650653 [Citation7]) and amylocyclicin FZB42 (AcnA, ABS75252 [Citation10]) with 28% and 98% amino acid identities, respectively. Comparison with amylocyclicin FZB42 indicated that amylocyclicin CMW1 is composed of two domains, a leader peptide (47 amino acids, Met1 to Glu47) and a bacteriocin class IId cyclical uberolysin-like protein (64 amino acids, Leu48 to Trp111) as shown in . The leader peptide of amylocyclicin CMW1 probably works as a molecular chaperone for a correct circularization; subsequently, it might be cleaved off and circularized to give an active mature bacteriocin [Citation10].
The accB gene is not found in the uberolysin A gene cluster and the protein AccB is similar to AcnB (94% amino acid identities), which might be an ABC transporter of amylocyclicin FZB42 [Citation10]. AccB might be involved in export of amylocyclicin CMW1; AccC is similar to UbelB (20% amino acid identities), which probably functions in the maturation and circularization of uberolysin A [Citation7]; AccD is similar to UbelD (27% amino acid identities), which might be the ATP-binding protein of the ABC transporter involved in uberolysin A biosynthesis [Citation7]. In the case of amylocyclicin CMW1, AccC and AccD might be involved in each of similar functions. AccE is similar to UbelC (31% amino acid identities) and has a weak homology to LcyD of L. mesenteroides TK41401 (21% amino acid identities). LcyD is an integral membrane protein belonging to the DUF95 superfamily and acts as an immunity-associated transporter as well as a secretion-supporting protein in the biosynthesis of gene cluster of leucocyclicin Q [Citation15]. This might be the case for AccE. AccF is similar to UbelE (22% amino acid identities). UblE might be involved in the self-immunity of uberolysin A [Citation7]. Similarly, AccF might be involved in the similar self-immunity of amylocyclicin CMW1. Bioinformatics analysis revealed that the structure of amylocyclicin CMW1 gene cluster is similar to those of uberolysin A and amylocyclicin FZB42 gene clusters. For this reason, it was assumed that the accABCDEF gene cluster might be involved in the synthesis and processing of amylocyclicin CMW1.
Antibacterial activity against B. subtilis
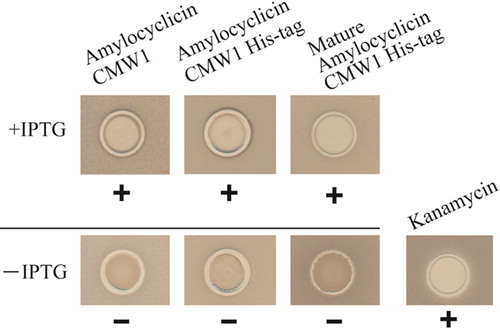
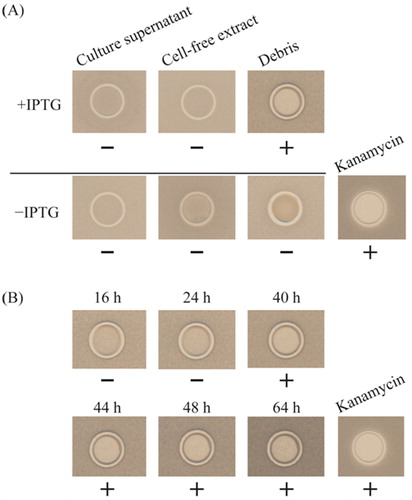
Amylocyclicin FZB42, a circular bacteriocin, was reported to be an antibacterial substance acting against B. subtilis 168 [Citation10]. Therefore, we investigated the antibacterial effect of recombinant amylocyclicin CMW1 against B. subtilis JCM10629. The accA gene was cloned and amylocyclicin CMW1 containing a leader peptide was expressed in recombinant E. coli cells (). As shown in , after spotting the samples with penicillin cups at 44 h, the antibacterial activities were compared. Kanamycin (50 µL) at 9.8 µg/mL was used as the positive control. Growth inhibition of B. subtilis by the debris fraction (+IPTG) was detected (), but not by culture supernatant (+IPTG) and cell-free extract (+IPTG). In contrast, no inhibitory activity was detected without the addition of IPTG (–IPTG). The antibacterial activities at 16, 24, 40, 44, 48 and 64 h after spotting 50 µL of debris at 41 mg/mL is shown in . The inhibitory activity was detected after a 40-h incubation.
Figure 4. Domain structures of amylocyclicin CMW1 and its derivatives. Note: Amino acid numbers shown above the diagram are the same as in .
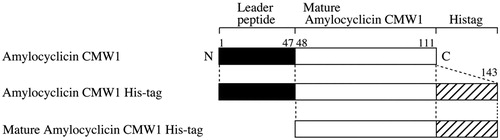
Figure 5. Antibacterial activity against B. subtilis JCM10629 by the agar spot test. (A) Detection of antibacterial activities with the culture supernatant, cell-free extract, and debris. (B) Time course of the antibacterial activity of amylocyclicin CMW1. Note: +, inhibited; –, not inhibited.
Additionally, we investigated whether the leader peptide is required for the antibacterial activity or not, and whether addition of a His-tag can affect the antibacterial activity. Domain structures of amylocyclicin CMW1 and its derivatives are shown in . The amylocyclicin CMW1 was fused at the C-terminus to a His-tag sequence. The modification bears several advantages [Citation16,Citation17]: the fusion of the comparatively small amylocyclicin CMW1 protein to a larger protein prevents any proteolytic denaturation within the recombinant E. coli cells, which is often observed when small recombinant proteins are expressed. Furthermore, the His-tag domain allows a simple purification of the recombinant amylocyclicin CMW1 [Citation17]. As shown in , regardless of the addition of leader peptide and His-tag, the antibacterial effect against B. subtilis was detected with the recombinant peptides (+IPTG), but not in the cases of –IPTG. Kanamycin (50 µL) at 9.8 µg/mL was used as the positive control.
Inhibitory activity of mature amylocyclicin CMW1 His-tag
We prepared a debris fraction from the bacterial cell pellet and examined the antimicrobial activity. As shown in , 50 µL of the debris at 41 mg/mL exhibited antimicrobial activities against the Gram-positive bacteria B. subtilis JCM10629, B. licheniformis JCM2505T, S. aureus NBRC12732 and S. carnosus JCM6067T, but not against the Gram-negative bacteria E. coli NBRC3301, E. asburiae JCM6051T and P. putida JCM13063T. Unlike amylocyclicin FZB42, mature amylocyclicin CMW1 His-tag inhibited the growth of S. aureus [Citation7]. Antimicrobial activities of the solvent against test strains were not detected by the agar spot assay.
Table 1. Inhibitory activities of mature amylocyclicin CMW1 His-tag.
In general, class II bacteriocins contain cationic and hydrophobic amino acids [Citation1,Citation2]. Thus, the bacteriocins have amphiphilic structures, which would insert themselves into the membrane of the target bacterial cells [Citation1,Citation2,Citation4]. As a result, the circular bacteriocins probably act to disrupt the integrity of the membrane, leading to death and lysis of the cells. As a result of the comparison between the amino acid sequences of amylocyclicin CMW1 and amylocyclicin FZB42, six kinds of amino acids might disrupt the integrity of the cell membrane: cationic amino acids: four of Lys, and hydrophobic amino acids: one of Leu, five of Ala, seven of Ile, one of Val and one of Trp.
Detection of amylocyclicin CMW1 His-tag with LC-MS analysis
Antibiotic peptides, such as amylocyclicin FZB42 from B. amyloliquefaciens FZB42 [Citation10], plantaricyclin A from Lactobacillus plantarum NI326 [Citation18] and antifungal peptide from Aneurinibacillus sp. YR247 [Citation19] have been detected using mass spectral analysis. We investigated whether the recombinant amylocyclicin CMW1 is a circular peptide or a linear peptide using mature amylocyclicin CMW1 His-tag. If mature amylocyclicin CMW1 His-tag is a linear peptide, the predicted molecular weight is 10,040.6. Using 10 mM potassium phosphate buffer (pH 7.0) and MeOH, a 50 μg/mL debris fraction was prepared. In order to detect mature amylocyclicin CMW1 His-tag, LC-MS analysis with the debris fraction was carried out [Citation19,Citation20]. The molecular weight of mature amylocyclicin CMW1 His-tag was estimated to be 10,041.2 because the molecular ion peaks at m/z 327.1, 348.3 and 559.9 probably correspond to the charged ions [M + 31H]31+, [M + 29H]29+ and [M + 18H]18+, respectively. The calculation of molecular weight is indicated as follows:
The molecular weight was estimated as 10,080.3 using the average of the values (10,109.1, 10,071.7 and 10,060.2). The estimated molecular weight (10,080.3) probably consists of mature amylocyclicin CMW1 His-tag (molecular weight 10,041.2) and a potassium ion (molecular weight 39.1) [Citation19]. Therefore, recombinant mature amylocyclicin CMW1 His-tag is considered to be a linear peptide, not a circular peptide. We demonstrated that the linear amylocyclicin CMW1 exhibits an antimicrobial activityagainst Bacillus sp. as well as the circular amylocyclicin FZB42.
Conclusions
Our study demonstrated that a circular bacteriocin gene was expressed by recombinant E. coli cells and that the linear recombinant peptide has antimicrobial activity. At present, there is a demand for development of an efficient method to detect the biological activities of novel circular bacteriocins. Heterologous production offers the most cost-effective means for development of new bacteriocins. We plan to develop better methods for the production and purification of mature amylocyclicin CMW1 His-tag and to investigate the stability of the purified recombinant bacteriocin.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3:777–788.
- Sánchez-Hidalgo M, Montalbán-López M, Martínez-Bueno M, et al. Natural ribosomally synthesized circular proteins. In: Mendez-Vilas A, editor. Communicating current research and educational topics and trends in applied microbiology. Badajoz: FORMATEX; 2007. p. 283–294.
- Field D, Ross RP, Hill C. Developing bacteriocins of lactic acid bacteria into next generation biopreservatives. Curr Opin Food Sci. 2018;20:1–6.
- Gabrielsen C, Brede DA, Nes IF, et al. Circular bacteriocins: biosynthesis and mode of action. Appl Environ Microbiol.. 2014;80:6854–6862.
- Perez RH, Zendo T, Sonomoto K. Circular and leaderless bacteriocins: biosynthesis, mode of action, applications and prospects. Front Microbiol. 2018;9:2085.
- Bédard F, Biron E. Recent progress in the chemical synthesis of class II and S-glycosylated bacteriocins. Front Microbiol. 2018;9:1–4.
- Wirawan RE, Swanson KM, Kleffmann T, et al. Uberolysin: a novel cyclic bacteriocin produced by Streptococcus uberis. Microbiology (Reading, Engl). 2007;153:1619–1630.
- Gabrielsen C, Brede DA, Hernández PE, et al. Genome sequence of the bacteriocin-producing strain Lactococcus garvieae DCC43. J Bacteriol. 2012;194:6976–6977.
- Masuda Y, Ono H, Kitagawa H, et al. Identification and characterization of leucocyclicin Q, a novel cyclic bacteriocin produced by Leuconostoc mesenteroides TK41401. Appl Environ Microbiol. 2011;77:8164–8170.
- Scholz R, Vater J, Budiharjo A, et al. Amylocyclicin, a novel circular bacteriocin produced by Bacillus amyloliquefaciens FZB42. J Bacteriol. 2014;196:1842–1852.
- van Heel AJ, Montalban-Lopez M, Oliveau Q, et al. Genome-guided identification of novel head-to-tail cyclized antimicrobial peptides, exemplified by the discovery of pumilarin. Microb Genom. 2017;3:1–9.
- Elshaghabee FM, Rokana N, Gulhane RD, et al. Bacillus as potential probiotics: status, concerns, and future perspectives. Front Microbiol. 2017;8:1490.
- Kurata A, Hirose Y, Misawa N, et al. Draft genome sequence of the ionic liquid-tolerant bacterium Bacillus amyloliquefaciens CMW1. Genome Announc. 2014;2012:e01051–01014. DOI: 10.1128/genomeA.01051-14
- Kurata A, Senoo H, Ikeda Y, et al. Properties of an ionic liquid-tolerant Bacillus amyloliquefaciens CMW1 and its extracellular protease. Extremophiles. 2016;20:415–424.
- Mu F, Masuda Y, Zendo T, et al. Biological function of a DUF95 superfamily protein involved in the biosynthesis of a circular bacteriocin, leucocyclicin Q. J Biosci Bioeng. 2014;117:158–164.
- Klocke M, Mundt K, Idler F, et al. Heterologous expression of enterocin A, a bacteriocin from Enterococcus faecium, fused to a cellulose-binding domain in Escherichia coli results in a functional protein with inhibitory activity against Listeria. Appl Microbiol Biotechnol.. 2005;67:532–538.
- Li Y. Recombinant production of antimicrobial peptides in Escherichia coli: a review. Protein Expres Purif. 2011;80:260–267.
- Borrero J, Kelly E, O'Connor PM, et al. Plantaricyclin A, a novel circular bacteriocin produced by Lactobacillus plantarum NI326: purification, characterization, and heterologous production. Appl Environ Microbiol. 2018;84:e01801–01817.
- Kurata A, Yamaura Y, Tanaka T, et al. Antifungal peptidic compound from the deep-sea bacterium Aneurinibacillus sp. YR247. World J Microb Biot. 2017;33:1–8.
- Zhao X, Zhou Z-J, Han Y, et al. Isolation and identification of antifungal peptides from Bacillus BH072, a novel bacterium isolated from honey. Microbiol Res. 2013;168:598–606.