Abstract
Traditional diagnostic methods cannot meet the requirements for rapid diagnosis of Mycoplasma pneumoniae. Therefore, developing a serological diagnostic method with high specificity and sensitivity is urgent. In this study, we expressed and purified the recombinant P1’ protein, which comprises amino-acid residues 1160–1498 of the P1 protein. Thereafter, after immunising New Zealand rabbits, we purified the anti-P1’ polyclonal antibody using cyanogen bromide-activated sepharose 4B. By using this purified antibody as target molecule, we then performed a 4-round biopanning for a phage display random 12-mer peptide library. After sequencing analysis, we found that the polypeptide HLQMRLTKLRMP was a 12-mer peptide specifically binding to the anti-P1' polyclonal antibody and had 75% homology with the 1266-1272 amino acid (QVRTKLR) of the M. pneumoniae P1 protein. Besides, we further confirmed that the representative phage 1 which displayed this peptide could specifically bind to the anti-P1’ polyclonal antibody by dot immunobinding assay. Indirect ELISA showed that the synthesized 12-mer peptide could specifically bind to the serum of M. pneumoniae positive patients. The sensitivity and the specificity were 81.87% and 95%, respectively. These results indicated that HLQMRLTKLRMP might be a dominant epitope of the P1 and has the potential for serological diagnosis of M. pneumoniae.
Introduction
Mycoplasma pneumoniae is the main pathogen of community-acquired pneumonia and can cause multiple systemic diseases [Citation1]. However, the isolation and culture of M. pneumoniae is cumbersome and time consuming. Moreover, the isolation rate of M. pneumoniae is too low [Citation2], which makes it difficult to make early diagnosis. At present, the most commonly used clinically diagnostic methods are polymerase chain reaction (PCR) (for DNA detection) and enzyme-linked immunosorbent assay (ELISA) (for M. pneumoniae IgM antibodies detection) [Citation3], but they both have low specificity and sensitivity [Citation4]. Therefore, it is particularly important to find a more efficient method and to screen the dominant epitope of M. pneumoniae for serological diagnosis of M. pneumoniae.
Epitopes, or antigenic determinants, locate on the surface of antigens and are easy to induce antibody response. The dominant epitope, which is the key site for antigen–antibody binding, is usually only 5–7 amino acids in length [Citation5, Citation6]. However, using a full protein as a diagnosis antigen usually has some disadvantages such as low affinity, high background, and high cross-reactivity. The mimotope is a conformational epitope formed by a spatial conformation of the discontinuous amino acids. But it may or may not have homology with the protein itself [Citation7]. Thus, mimotopes have many advantages, such as low background, low cross-reactivity and ease of preparation, over traditional protein antigens in serological diagnosis [Citation8].
The P1 protein, one of the most dominant antigens of M. pneumoniae, binds to the neuraminic receptors on the membrane of respiratory mucosal epithelial cells, helping M. pneumoniae to adhere to the host cell surface [Citation9]. Studies have shown that the C-terminus of P1 has a functional domain that adheres to host cells [Citation10, Citation11]. Moreover, P1 protein also has strong immunogenicity and its corresponding antibody can be detected in the serum of infected patients [Citation12]. Rastawicki et al. [Citation11] used a recombinant protein containing amino acids 1160–1521 of the C-terminus of the P1 protein for serological diagnosis and found that it could react with 70% of M. pneumoniae-positive sera, but its specificity and sensitivity were still unsatisfactory.
In our previous study, two heptapeptides (TVNFKLY and LPQRLRT) targeting M. pneumoniae-positive serum were screened from a phage display random 7-mer peptide library. The two synthesized heptapeptides had potential serological diagnosis value for M. pneumoniae infection [Citation13]. In this study, we expressed and purified the recombinant P1’ protein, which contains a fragment of rich hydrophilic amino acids from the P1 protein and has good antigenicity. The dominant epitopes of this recombinant protein were then screened from a phage display random 12-mer peptide library using the corresponding anti-P1’ polyclonal antibody as target molecule. Finally, the value of the biopanned epitopes in the serological diagnosis of M. pneumoniae infection was evaluated.
Subjects and methods
Reagents
The Ph.D.-12 phage display peptide library, the Escherichia coli ER2738 (the host bacteria of M13 phage), the sequencing primer and horseradish peroxidase (HRP)-labelled anti-M13 pIII monoclonal antibody were from New England Biolabs (NEB, Beverly, MA). CNBr activated Sepharose 4B was purchased from Pharmacia (Peapack, NJ). (NH4)2SO4 and Complete Freund's adjuvant were purchased from Sigma-Aldrich (St. Louis, MO).
Animals
New Zealand rabbits (n = 4) were purchased from the Animal Experimental Center of the University of South China (Hengyang, China). They were kept in standard conditions.
Patients
A total of 160 patients infected with M. pneumoniae were randomly enrolled from Chenzhou City First People's Hospital. The cohort included 72 males and 88 females, with an average age of 7.4 (range 2–9). The patients were diagnosed as M. pneumoniae infection by the SERODIA-MYCOII Diagnostic Kit (FUJIREBIO, Tokyo, Japan) and the antibody titre was higher than 1:160. For negative control, 20 cases of age and gender matched healthy individuals were recruited, including 9 males and 11 females, with an average age of 7.2 (range 4–8).
Ethics statement
All animal experiments were conducted according to the ethical guidelines of the Ethics Committee of the University of South China.
Prior written and informed consent were obtained from every patient and the study was approved by the ethics review board of Chenzhou City First People's Hospital.
P1' expression and purification
The 3601–4617 bases of the p1 gene, coding the amino acids 1160–1498 of the P1 protein, which is named P1', was amplified and cloned into a pET-30a (+) vector, which contains a His tag in the C-terminal. The correctly constructed plasmid was verified by restriction enzyme digestion using EcoRI and XhoI at 37 °C for 90 min and then analysed by 1% agarose electrophoresis. The recombinant plasmid was then transformed into E. coli BL21 (DE3). The recombinant E. coli BL21 (DE3) was induced by 1 mmol/L IPTG (isopropyl ß-d-1-thiogalactopyranoside) at 30 °C for 4 h to express the P1' protein and was then broken by ultrasonication (300w, 4 s, interval 6 s, altogether 50 times). Then the expressed protein was identified by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue. The recombinant P1' protein was purified using a protein purification system (GE, AKTA Pure, New York City, NY) and identified by Western blot.
Western blot analysis
The purified recombinant protein was electrophoresed by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. After blocking overnight at 4 °C with 5% skim milk, the primary antibodies of mouse anti-His tag monoclonal antibody (1:5 000, AH367, Beyotime, Beijing, China) and P1' protein-immunised New Zealand rabbit serum were added and incubated for 2 h at 37 °C. After washing, HRP-labelled goat anti-mouse IgG (1:5 000, Cat#A-11004, Invitrogen, Carlsbad, CA) and HRP-labelled goat anti-rabbit IgG (1:5 000, Cat#A-27036, Invitrogen, Carlsbad, CA) were added and incubated for 1 h at 37 °C. Finally, colour development imaging was performed. The bovine serum albumin (BSA) was used as control.
Preparation and purification of anti-P1' polyclonal antibody
The recombinant protein P1' (200 μg) was fully emulsified with Freund's complete adjuvant and was subcutaneously injected into New Zealand rabbits on the dorsal skin. Booster immunisation was performed every 2 weeks, and the rabbits were immunised a total of three times. Blood was collected from the heart on day 14 after the last immunisation. The serum was subjected to IgG precipitation using a saturated (NH4)2SO4 method [Citation14]. After precipitation, the IgG were submitted to dialysis to remove the ammonium sulphate. The pre-purified rabbit serum was then subjected to affinity chromatography with cyanogen bromide-activated sepharose 4B to obtain purified polyclonal antibody [Citation14]. Briefly, cyanogen bromide-activated sepharose 4B was mixed with recombinant protein P1' and conjugated at 4 °C overnight. After blocking with 0.1 mol/L Tris-HCl (pH 8.0), the coupling medium was washed 4 times with a coupling buffer (0.5 mol/L NaCl, pH 8.3) and an acetate buffer (0.1 mol/L NaAc, 0.5 mol/L NaCl, pH 4.0). Thereafter, the column was washed, and pre-purified serum was added, and after washing several times with PBS, the target antibody was eluted with 0.1 mol/L of glycine-HCl buffer (pH = 2.2). Finally, the elute was analyzed by SDS-PAGE.
Biopanning of phage display random 12-mer peptide library
Biopanning of the phage display random 12-mer peptide library was performed as previously described [Citation15]. Briefly, in the first round of panning, we coated the ELISA plate with 100 μg/mL purified anti-P1' antibody diluted in 0.1 mol/L NaHCO3 (pH 8.6) at 4 °C. After blocking for 3 h with 5 mg/mL BSA, the plate was washed 10 times with 0.1% TBST (TBS containing 0.1% Tween 20; TBS, Tris-buffered saline: 50 mmol/L Tris–HCl, 150 mmol/L NaCl, pH 7.5). Then approximately 4 × 1010 pfu of the original phage library was added. The plate was incubated for 1 h with shaking, and then washed 8 times with 0.1% TBST. Then samples were eluted by using 100 μL of elution buffer (0.2 mol/L glycine-HCl, pH = 2.2) for 10 min. After that, reverse-adsorption was carried out using empty ELISA plates and negative sera as target, respectively, to exclude non-specific binding phages. The subsequent procedures of the three rounds of biopanning were the same as the first round, except that the coating concentration of the anti-P1' polyclonal antibody was appropriately reduced to 60 μg/mL, while the TBST concentration was increased to 0.5%. The final obtained phages were subjected to titre measurement and then were amplified for further usage. The yield of each round of biopanning was calculated based on the phage titre of the output and input.
DNA sequencing and sequence analysis
The products after the fourth round of screening were plated, and 32 clones were randomly picked from the plates with less than 100 plaques. After amplification, the single-stranded DNAs of phages were extracted by the sodium iodide method and sequenced by Shanghai Sangon Biotech Co., Ltd. (Shanghai, China). The exogenous DNA sequences displayed by the phages were deduced and translated into the corresponding amino acid sequence for BLAST alignment.
Dot immunoblotting
The specific binding of the representative phage 1 to the anti-P1' polyclonal antibody was verified by dot immunoblotting as previously described [Citation15]. Briefly, about 2 × 1010 pfu phages were spotted on PVDF membrane, and the anti-P1' polyclonal antibody was used as a positive control; negative serum and wild-type vcsM13 phage were set as negative controls. After the liquid was absorbed, the PVDF membrane was incubated overnight at 4 °C with the purified anti-P1' polyclonal antibody. After washing, HRP-labelled goat anti-rabbit IgG (1:5 000, Cat#A-27036, Invitrogen, Carlsbad, CA) was added and incubated for 1 h at 37 °C. The membrane was washed 8 times with TBST before colour development.
Peptide synthesis
Since the representative phage P1 had the highest frequency, the peptide 1 as listed in was synthesised using the solid phase method, purified by reversed phase high performance liquid chromatography (HPLC) and then identified by mass spectrometry by Shanghai Jier Biochemical Company (Shanghai, China).
Table 1. Input, output, and yield of phages after each round of screening.
ELISA
Indirect ELISA was performed to detect the reactivity of the synthesised peptide with different clinical sera, including 160 cases of M. pneumoniae positive serum and 20 cases of M. pneumoniae negative serum. BSA served as the negative control. Briefly, the ELISA plate was coated with 200 μL synthesised peptide (20 μg/mL) at 4 °C overnight and then blocked with 5% skim milk for 3 h at room temperature. Then, 50 μL of diluted different serum (1:10) was added and incubated at 37 °C for 45 min. After washing five times with 0.5% PBST, the HRP-conjugated goat anti-human IgG antibody (1:5000) (Cat#62-8420, Invitrogen, Carlsbad, CA) was added and the reaction was incubated for 30 min at 37 °C. After washing, the reaction was terminated and colour development was performed. Then the optical density (OD) value was measured at a wavelength of 450 nm by a microplate reader (Tecan Infinite F50).
Statistical analysis
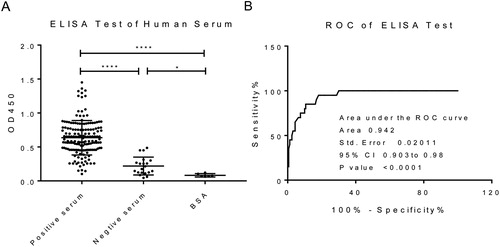
The sensitivity was calculated as true positive number/(true positive number + false negative number) ×100%, and the specificity was calculated as true negative number/(true negative number + false positive number) ×100%. The receiver operating characteristic (ROC) curve was calculated using GraphPad Prism software (GraphPad Software, La Jolla, CA) to analyse the sensitivity and specificity. The area under the ROC curve (AUC) was calculated and compared. When AUC > 0.9, the accuracy is high, and the ELISA results have reference significance.
Results and discussion
Expression and purification of P1’ protein
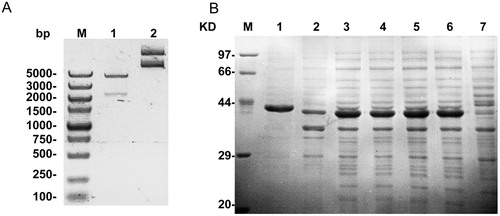
The bioinformatics method is commonly used to predict the epitope based on the hydrophilicity, accessibility and flexible regions of the amino acid sequence to ensure the antigenicity of the recombinant protein [Citation16–18]. In general, the N-terminus and C-terminus of proteins are good regions for antigenic determinants [Citation19]. Studies have shown that M. pneumoniae P1 protein is an important serum diagnostic marker [Citation20], and its main antigenicity is located at the C-terminus [Citation10]. In this study, the recombinant protein that contains amino acids 1160-1498 of the P1 protein with high antigenicity and hydrophilicity was expressed. Firstly, the prokaryotic expression plasmid encoding 1160–1498 amino acids of the P1 protein was constructed. Gel electrophoresis indicated that the plasmid was constructed correctly (). The recombinant protein (named P1') was then expressed and analysed by SDS-PAGE. As shown in , an obvious protein band with approximately 38 kDa was observed in lanes of 2–6. After purification, only a relatively single band appeared in the first lane (). The above results indicated that the recombinant P1' protein was successfully expressed and purified.
Figure 1. Plasmid construction and protein expression. (A) Lane M, DNA Marker (Takara, Tokyo, Japan); Lane 1, plasmid digested by EcoRI and XhoI; lane 2, plasmid DNA. (B) Protein expression was analysed by SDS-PAGE. Lane M, protein marker (Takara, Tokyo, Japan); lane 1, purified P1’ protein; lane 2, supernatant of bacteria by ultrasonication; Lane 3, mixed sediment of three positive clones by ultrasonication; lanes 4–6, Three positive clones containing pET-30a (+)/P1’protein; lane 7: empty bacteria without plasmid.
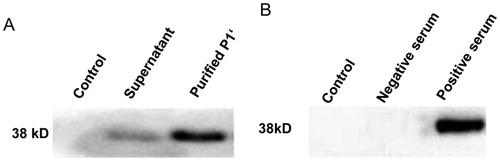
Western blot analysis of recombinant protein P1'
Western blotting was performed to identify the purified recombinant protein. As shown in , a single clear specific band around 38 kDa was observed. No band was observed for the controls. The experimental results showed that the target protein was expressed successfully and can induce the production of corresponding polyclonal antibodies. And the corresponding antibody from the serum of the recombinant protein immunized New Zealand rabbits could specifically bind to the recombinant protein. Thus, it exhibits good immunoreactivity and may provide suitable target molecules for the biopanning of phage display 12-mer peptide library.
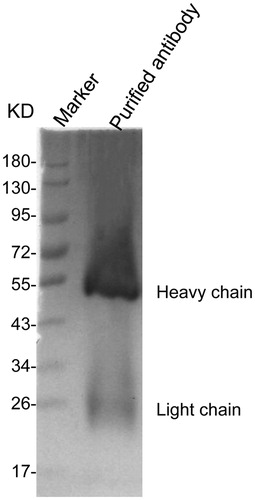
The purification of polyclonal antibody to P1’
To purify the polyclonal antibody of the target protein, the P1’-immunised rabbit serum was preliminarily purified by a saturated (NH4)2SO4 method, and further purified by a cyanogen bromide-activated sepharose 4B method, and the product was subjected to SDS-PAGE electrophoresis. As shown in , there was a band at a position near a molecular weight of about 55 kDa, which is the heavy antibody chain, and a weak band at 26 kDa, which is the light antibody chain. These results showed that the P1' protein-specific polyclonal antibody was successfully purified.
Phages enrichment
To enrich the P1' antibody-specific phages, we performed 4-round biopanning of the phage display random 12-mer peptide library using the purified P1' antibody as a target molecule. The phage input, output and yield of each round of panning are shown in . As the times of biopanning increased, the output and yield of the phage gradually increased, indicating that the phages specific for anti-P1' antibody were successfully enriched.
Phage display is a technique that inserts the DNA fragment encoding the exogenous polypeptide into the capsid protein gene of the phage, thus displaying the exogenous polypeptide on the phage surface [Citation20]. Phage display technology is based on the ‘adsorption-elution-amplification’ biopanning mode, which allows the phages specifically bound to the target molecule to be gradually enriched, and the mimotope can be obtained even if we do not know the structure of the antigen [Citation21]. A total of 4 rounds of biopanning were performed in this study, and negative panning was performed using both negative serum and empty ELISA plates to remove non-specifically bound phages as much as possible. At the same time, during the biopanning, we gradually reduced the concentration of coated serum and increased the concentration of Tween-20 in the washing solution to enrich the highly specific phage clones [Citation22].
Sequencing and derivation of dominant epitopes
To investigate the exogenous polypeptide sequences inserted into the enriched phages, we randomly picked 32 monoclonal plaques from the 4th round of panning and extracted their single-stranded DNA. Then the DNA sequences were translated into amino acids, and the results indicated that a total of four different sequences were displayed by all 32 phages, namely peptides 1–4 displayed by representative phages 1–4 (). Representative phage 1 that displayed HLQMRLTKLRMP sequence was repeated 23 times with a frequency of 71.87%. Thereafter, we performed a BLAST alignment for the exogenous peptide 1 displayed by the representative phage 1. It was found that Q-R-TKLR had 75% homology with the 1266–1272 amino acids (QVRTKLR) of M. pneumoniae P1 protein. The above results indicated that the 12-mer peptide displayed by representative phage 1 might be the dominant epitopes of the P1 protein.
Table 2. Peptide sequences displayed by representative phages.
The exogenous polypeptides displayed on the surface of phages can maintain relatively independent spatial structure and biological activity. Therefore, the phage display peptide library technique makes it possible to rapidly obtain the immunodominant epitopes of a protein antigen. The biopanning epitopes may not be identical to the original antigenic structure but mimic the antigenic properties. Thus, phage display peptide libraries have been widely used to study the intermolecular interactions, antigenicity [Citation23] or enzymatic activity [Citation24]. Beghetto et al. [Citation25] screened the epitope of M. pneumoniae by phage display technology and confirmed that P1 adhesin was the main immunogen of M. pneumoniae. And, its immunogenic dominant region was mainly located at amino acids 1125–1131 and 1382–1394. Interestingly, the homologous segment of the 12 peptides in this study was located at 1266–1272 of P1 protein, which further confirms that the C-terminus of the P1 protein is an important epitope region [Citation26, Citation27].
The representative phages could specifically bind to positive serum
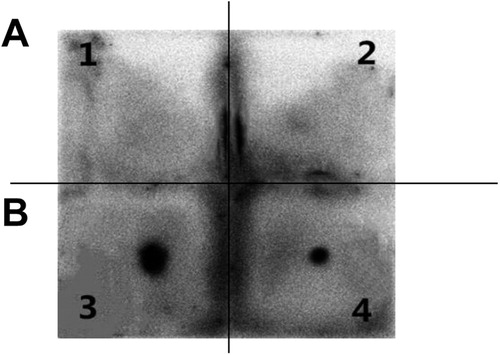
Dot blotting was performed to detect whether representative phage 1 with highest occurrence frequency could specifically bind to purified P1' antibodies. As shown in , there were clear spots for the representative phage 1 and positive control. However, there was no obvious spot for the wild-type phage and negative serum control. These results further demonstrate that the corresponding polypeptide displayed by representative phage 1 can specifically bind to the anti-P1' polyclonal antibody, which suggests that this epitope may be a dominant epitope of the P1 protein.
Figure 4. Dot immuno-binding assay of the representative phages. A1, Wild-type vcsM13 phage control + purified anti-P1' polyclonal antibody. A2, Representative phage 1 + M. pneumoniae negative serum control. B3, Purified anti-P1' polyclonal antibody (positive control). B4, Representative phage 1+ purified anti-P1' polyclonal antibody.
Synthetic peptides have good serological reactivity
To evaluate the serological diagnostic value of exogenous polypeptides displayed by representative phage 1, we tested the reactivity between the synthesised peptide and different M. pneumoniae positive sera and negative sera by indirect ELISA. As shown in , the OD450 values of the synthetic polypeptide to the positive serum were significantly higher than the negative serum and BSA control groups (p < 0.05). In addition, the sensitivity of peptide 1 was 81.87%, the specificity was 95%, and the AUC value was 0.942 (). These results indicated that the synthetic peptide 1 has good application value for the diagnosis of M. pneumoniae infection. Compared with heptapeptide 1 (TVNFKLY) and heptapeptide 2 (LPQRLRT) screened in our previous study [Citation13], the sensitivity of peptide 1 screened in this study was lower than heptapeptide 1 but higher than heptapeptide 2, and the specificity of peptide 1, heptapeptide 1 and heptapeptide 2 were all higher than 90%. Larralde and Petrik [Citation5] screened 14 mimic epitopes of Hepatitis E Virus capsid protein from a phage display 12-mer random peptide library and used them for the serological diagnosis of Hepatitis E virus with high specificity. Our previous study also confirmed that the mimic epitopes of Mycobacterium tuberculosis screened from the phage display random 12-mer peptide library had high sensitivity and specificity for the sero-diagnosis of tuberculosis [Citation14].
Conclusions
We expressed and purified the M. pneumoniae P1' protein. The peptide of HLQMRLTKLRMP was identified as the dominant mimotope from the phage display random 12-mer peptide library using the polyclonal antibody against M. pneumoniae P1' protein as the target molecule. This epitope had good sensitivity and specificity in response to M. pneumoniae-positive sera and may be used for serological diagnosis of M. pneumoniae infection. Our next study will expand the amount of serum samples and test the reactivity of the other three epitopes displayed by representative phages 2–4 with positive M. pneumoniae sera. By linking these polypeptides together, we will further examine the value of these recombinant polypeptides in the serological diagnosis of M. pneumoniae infection.
Funding
This work was supported by the [Program of Department of science and technology of Hunan Province] under Grant [2017SK51105], [Natural Science foundation of Hunan Province] under Grant [2019JJ3509], [Health and Family Planning Commission of Hunan province] under Grant [B20180621], [Chenzhou Municipal Bureau of Science and Technology] under Grant [czkj2016045], and the [Project of Chenzhou First People's Hospital] under Grant [N2014-001].
Disclosure statement
All authors declare no financial competing interests. All authors declare no non-financial competing interests.
References
- Parrott GL, Kinjo T, Fujita J. A compendium for Mycoplasma pneumoniae. Front Microbiol. 2016;7:513.
- Lee WJ, Huang EY, Tsai CM, et al. Role of serum Mycoplasma pneumoniae IgA, IgM, and IgG in the diagnosis of Mycoplasma pneumoniae-related pneumonia in school-age children and adolescents. Clin Vaccine Immunol. 2017; 24:pii: e00471–16.
- Li W, Liu Y, Zhao Y, et al. Rapid diagnosis of Mycoplasma pneumoniae in children with pneumonia by an immuno-chromatographic antigen assay. Sci Rep. 2015;5:15539.
- Loens K, Goossens H, Ieven M. Acute respiratory infection due to Mycoplasma pneumoniae: current status of diagnostic methods. Eur J Clin Microbiol Infect Dis. 2010;29:1055–1069.
- Larralde O, Petrik J. Phage-displayed peptides that mimic epitopes of hepatitis E virus capsid. Med Microbiol Immunol. 2017;206:301–309.
- Moise L, Gutierrez A, Kibria F, et al. An integrated toolkit for the selection and optimization of antigens and the design of epitope-driven vaccines. Hum Vaccin Immunother. 2015;11:2312–2321.
- Chen X, Dreskin SC. Application of phage peptide display technology for the study of food allergen epitopes. Mol Nutr Food Res. 2017;61:1600568.
- Lebreton A, Simon N, Moreau V, et al. Computer-predicted peptides that mimic discontinuous epitopes on the A2 domain of factor VIII. Haemophilia. 2015;21:e193–e201.
- Nakane D, Adan-Kubo J, Kenri T, et al. Isolation and characterization of P1 adhesin, a leg protein of the gliding bacterium Mycoplasma pneumoniae. J Bacteriol. 2011;193:715–722.
- Chourasia BK, Chaudhry R, Malhotra P. Delineation of immunodominant and cytadherence segment(s) of Mycoplasma pneumoniae P1 gene. BMC Microbiol. 2014;14:108.
- Rastawicki W, Rokosz N, Gierczyński R, et al. Use of recombinant P1 protein of Mycoplasma pneumoniae for the serodiagnosis of mycoplasmosis. Med Dosw Mikrobiol. 2012;64:229–237.
- Tekewe A, Connors NK, Middelberg AP, et al. Design strategies to address the effect of hydrophobic epitope on stability and in vitro assembly of modular virus-like particle. Protein Sci. 2016;25:1507–1516.
- Shi W, Zhao L, Li S, et al. Serological diagnosis of Mycoplasma pneumoniae infection by using the mimic epitopes. World J Microbiol Biotechnol. 2018;34:82.
- Zeng Y, Liu L, He J, et al. Screening and identification of the mimic epitope of the adhesion protein of Mycoplasma genitalium. Can J Microbiol. 2012;58:898–908.
- Wang L, Deng X, Liu H, et al. The mimic epitopes of Mycobacterium tuberculosis screened by phage display peptide library have serodiagnostic potential for tuberculosis. Pathog Dis. 2016;74:pii: ftw091.
- Das SC, Morales RAV, Seow J, et al. Lipid interactions modulate the structural and antigenic properties of the C-terminal domain of the malaria antigen merozoite surface protein 2. Febs J. 2017;284:2649–2662.
- Han Y, Guo W, Su B, et al. High-level expression of soluble recombinant proteins in Escherichia coli using an HE-maltotriose-binding protein fusion tag. Protein Expr Purif. 2018;142:25–31.
- Rueda F, Gasser B, Sánchez-Chardi A, et al. Functional inclusion bodies produced in the yeast Pichia pastoris. Microb Cell Fact. 2016;15:166.
- Liu W, Liu G, Zhou H, et al. Computer prediction of paratope on antithrombotic antibody 10B12 and epitope on platelet glycoprotein VI via molecular dynamics simulation. Biomed Eng Online. 2016;15:152.
- Guanhua X, Ling C, Luoping W, et al. Evaluation of P1 adhesin epitopes for the serodiagnosis of Mycoplasma pneumoniae infections. FEMS Microbiol Lett. 2013;340:86–92.
- Zhang F, Wang M, Qiu Z, et al. Identification and characterization of strychnine-binding peptides using phage-display screening. Protein Pept Lett. 2017;24:626–632.
- Pinschewer DD. Virally vectored vaccine delivery: medical needs, mechanisms, advantages and challenges. Swiss Med Wkly. 2017;147:w14465.
- Portes LDS, Kioshima ES, de Camargo ZP, et al. Subtractive phage display selection for screening and identification of peptide sequences with potential use in serodiagnosis of paracoccidioidomycosis caused by Paracoccidioides brasiliensis. Lett Appl Microbiol. 2017;65:346–353.
- Wei X, Liu Q, Gao Y, et al. Two epitopes responsible for the catalytic activity of heme oxygenase-1 identified by phage display. FEBS Open Bio. 2017;7:719–726.
- Beghetto E, Paolis FD, Montagnani F, et al. Discovery of new Mycoplasma pneumoniae antigens by use of a whole-genome lambda display library. Microbes Infect. 2009;11:66–73.
- Chen C, Yong Q, Jun G, et al. Designing, expression and immunological characterization of a chimeric protein of Mycoplasma pneumoniae. Protein. PPL. 2016;23:592–596.
- Meng YL, Wang WM, Lv DD, et al. The effect of Platycodin D on the expression of cytoadherence proteins P1 and P30 in Mycoplasma pneumoniae models. Environ Toxicol Pharmacol. 2017;49:188–193.