Abstract
Lavender (Lavandula angustifolia Mill.) is an essential oil-bearing and medicinal plant of great economic and social importance to Bulgaria, which is the world largest producer of lavender oil nowadays. Currently, the industrial cultivation of lavender in Bulgaria employs up to seven varieties established during the last century. Despite the growing industrial lavender cultivation, few molecular markers have been applied for characterization of lavender genetic resources and breeding. The present study employed sequence-related amplified polymorphism (SRAP) markers to characterize the genetic resources and genetic diversity within and between two groups of ten Bulgarian and five foreign lavender varieties and breeding lines. The data generated following analysis with 51 SRAP primer pairs showed amplification of a high number (4697) and ratio (77.2%) of polymorphic SRAP fragments resulting in relatively high value of the polymorphism information content, with an average of 0.27 ± 0.03 for the used primer pairs. Analysis of molecular variance of the SRAP data further revealed high level of genetic diversity (96.6% of the total variations) within the analyzed groups of Bulgarian and foreign varieties and rather low diversity (3.4% of total variations) between the two groups. The possibilities for further applications of SRAP markers in lavender breeding and cultivation are discussed.
Introduction
Lavender (Lavandula angustifolia Mill.) is an important oil-bearing plant belonging to the mint family (Lamiaceae), which is cultivated as medicinal and garden plant, and for essential oil production worldwide [Citation1]. Large and increasing volumes of lavender essential oil are applied in a wide range of home and personal care products, perfumery, aroma-therapy and alternative medicine [Citation2]. During the last decade Bulgaria has emerged as the world’s largest producer and exporter of lavender essential oil [Citation2, Citation3]. This was related to expansion of the lavender growing areas, including establishment of new lavender fields in the country regions with diverse soil and climatic conditions, which have been traditionally used for growing other crops. All these increased the demand for better characterization and utilization of the existing lavender genetic resources and initiation of a lavender breeding programme supported by marker-assisted selection for development of new varieties [Citation3].
So far, molecular genetic research related to Lavandula species has been focused mainly on cloning and characterization of terpene synthase genes involved in biosynthesis of the essential oil compounds [Citation4–9] and draft genome sequence [Citation10]. However, molecular marker development and application for characterization of the genetic resources and biodiversity of lavender and other Lavandula species has been restricted to a few reported studies. Curto et al. developed molecular markers from single-copy nuclear genes for phylogenetic analyses at the species level in the Lamiaceae family [Citation11] and Hind et al. were able to discriminate the cultivated L. latifolia from that of L. angustifolia and L. x intermedia using DNA barcoding [Citation12]. Hnia et al. reported characterization of the genetic diversity of Tunisian L. multifida populations using RAPD markers [Citation13]. Adal et al. developed the first set of SSR markers for Lavandula sp. based on sequences from the L. angustifolia and L. x intermedia EST databases [Citation14]. However, the efficiency of their application in the characterization of closely related sets of lavender genetic resources remains to be evaluated. Thus, in spite of the increased interest on lavender cultivation and essential oil production, the genetic diversity of the employed lavender genetic resources remains poorly studied and no reliable molecular marker methods are currently applied for testing the genetic authenticity and homogeneity of the industrially used lavender planting material.
Sequence-related amplified polymorphism (SRAP) markers developed by Li and Quiros [Citation15] have been widely applied in a number of genetic studies due to their simplicity, reliability, effectiveness and genome-wide coverage [Citation16, Citation17]. Although the results from applications of SRAP markers in taxonomic studies could be influenced by anthropogenic or other types of selective pressure [Citation17], during the last decade SRAP markers have been successfully applied in a number of studies on characterization of plant genetic resources and natural populations, including species from the Lamiaceae family outside the genus Lavandula [Citation18–23].
In this study, we report SRAP markers for characterization of the genetic diversity in a set of lavender genetic resources including seven Bulgarian industrially cultivated lavender essential oil varieties, three breeding essential oil lines and five lavender cultivars grown in other countries. The potential of SRAP markers in lavender breeding and cultivation are discussed.
Materials and methods
Plant material
The lavender genotypes used in the present study included (a) the seven Bulgarian essential oil varieties cultivated currently: ‘Yubileina’, ‘Hemus’, ‘Hebar’, ‘Raya’, ‘Sevtopolis’, ‘Druzhba’ and ‘Karlovo’; (b) three Bulgarian superior essential oil breeding lines: ‘D1’, ‘S1’, ‘Y1’ and (c) five lavender varieties grown in other countries: ‘Blue ice’, ‘Hidcote’, ‘Mellisa Lilac’, ‘Munstead’ and ‘Rosea’. The Bulgarian varieties and breeding lines were maintained in the genetic resources collections of the Institute of Rose and Aromatic Plants, Agricultural Academy, Kazanlak, Bulgaria, and ZP Janeta Staneva Nursery, Kazanlak, Bulgaria. The rest of the lavender varieties were purchased from Peter Beales Roses Ltd., Norwich, UK. Leaves from single lavender bushes were collected, immediately frozen in liquid nitrogen and stored at –80 °C until DNA extraction.
DNA extraction
Lavender leaves were ground into fine powder using liquid nitrogen frozen stainless steel jars and TissueLyser II (QIAGEN GmbH), at 30 Hz for 2 min. Genomic DNA was extracted using a CTAB protocol [Citation24]. The purified total DNA was quantified spectrophotometrically using Nanodrop 2000 (Thermo Scientific), diluted to 25 ng/μL and stored at –20 °C until use.
PCR amplification and fragment analysis
The SRAP primers used in the study were designed according to Li and Quiros [Citation15] and are shown in . The forward ‘ME’ primers were 5′ end labeled with FAM dye. PCR reactions were carried out in a volume of 25 μL containing: 2 μL of DNA template (25 ng/μL), 0.1 μmol/L of each forward and reverse primer, 12.5 μL of MyTaq HS DNA polymerase (Bioline Ltd., UK). Samples were PCR amplified using the following thermal profile: 5 min at 94 °C; 3× cycles of 1 min at 94 °C, 1 min at 35 °C and 1 min at 72 °C; 35× cycles of 1 min at 94 °C, 1 min at 50 °C and 1 min at 72 °C and a final elongation step of 3 min at 72 °C. PCR products were diluted tenfold. For electrophoresis, 1 μL of the diluted PCR amplified product was mixed with 0.5 μL of the GeneScan 600 LIZ Size Standard and 9 μL of Hi-Di Formamide (Thermo Fisher Scientific Inc.). The mixture was denatured and analyzed on an Applied Biosystems 3130 Genetic Analyzer using 3130 POP-7 Polymer and 36 cm capillary array (Thermo Fisher Scientific Inc.).
Table 1. Sequences of SRAP primers used in the analysis.
Data analysis
SRAP fragment analysis was performed with the GeneMapper Analysis Software v4.0 (Thermo Fisher Scientific Inc.). The threshold for peaks assignation was set to 350 relative fluorescence units. Fragment analysis was carried out for fragment sizes in the range of 50–950 bp. All clearly distinguished SRAP fragments were scored as (1) present or (0) absent, and the obtained data were used for construction of a SRAP data matrix. The polymorphism information content (PIC) values were calculated using the ‘GDdom’ online tool [Citation25]. Phylogenetic analysis and analysis of molecular variance (AMOVA) analysis were performed with the FAMD software [Citation26].
Results and discussion
A total of 51 SRAP primer pairs were selected after preliminary testing of all possible 100 EM1/EM10 and ME1/ME10 primer combinations (). The primer pairs were selected for their ability to prime PCR amplification from DNA of lavender var. Druzhba and after assessment of the SRAP peaks quality in the obtained electropherograms. The selected primer pairs were further used for PCR amplifications of DNA isolated from seven Bulgarian essential oil lavender varieties, three breeding lines and five foreign lavender varieties. The results from the comparative analysis of the amplified SRAP fragments across all studies genotypes are summarized in . The tested 51 primer pairs amplified a total of 6088 different SRAP fragments with sizes between 50 and 950 bp, represented by well-resolved peaks on the electropherograms. Of them, 4697 fragments showed polymorphisms after SRAP analysis of the studied genotypes. The data analysis further showed that the number of the amplified SRAP fragments varied significantly among the tested primer pairs and studied genotypes, ranging between 38 and 220 fragments per primer pair (). Accordingly, 29%–97% of the amplified fragments appeared to be polymorphic in the studied genotypes pool, as the average percentage of polymorphic fragments was 74.3 ± 13.3. The average number of the polymorphic SRAP fragments per one genotype also varied significantly depending on the primer pair, ranging from 6.3 ± 1.6 polymorphic fragments per genotype for primer pair ЕМ7/МЕ9 to 52.6 ± 13.3 polymorphic fragments for primer pair ЕМ2/МЕ10. The average number of polymorphic SRAP fragments amplified by one primer pair per one genotype for the entire study was 25.5 ± 12.8. The calculated PIC values were relatively high for all primer pairs, varying from 0.22 to 0.39, with an average of 0.27 ± 0.03 for the study. These data and the calculated PIC values demonstrated a relatively high level of information of the SRAP markers, with a large number of loci producing unique fingerprints.
Table 2. Summary of SRAP fragments data following SRAP analysis of all studied genotypes.
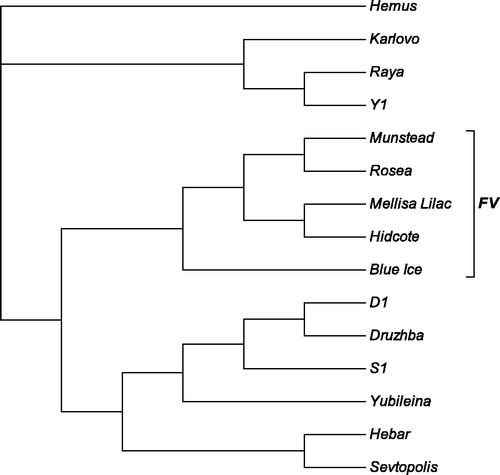
The obtained SRAP data were further used to determine the genetic relatedness among the studied lavender genotypes. The genetic similarity estimates based on the Jaccard similarity coefficients ranged from 0.22 to 0.395 for the tested genotypes and the overall similarity for the tested genotypes was 0.30 ± 0.04. The constructed phylogenic tree shows that all foreign varieties form a distinct cluster and were embedded among the clusters formed by the Bulgarian essential oil varieties and breeding lines suggesting relatively higher level of genetic diversity within the Bulgarian varieties (). The AМOVA results of the SRAP data for the two groups of Bulgarian essential oil and foreign varieties provided further support for this. The AМOVA revealed that only 3.4% of the overall SRAP variations in the study are related to differences between the two groups of Bulgarian essential oil and foreign lavender varieties, as the remaining 96.6% of the variations are due to the genetic diversity within the studied groups. This supports our earlier results from the analysis of flower volatiles composition, showing high level of variation in volatiles related to the essential oil of Bulgarian lavender varieties [Citation27]. Although the present study involves a restricted number of lavender varieties and breeding lines, the obtained results suggest that the level of genetic diversity of Bulgarian essential oil varieties and breeding lines is relatively high and the genetic resources available in the country (analyzed here and an additional set of breeding lines) could be used as a basis for further lavender breeding without an immediate need for expansion of the genetic resources pool.
Figure 1. Neighbour-joining phylogenetic tree constructed from SRAP data following analysis of Bulgarian industrially cultivated lavender essential oil varieties and breeding lines, and foreign lavender cultivars. The names of the analyzed varieties and breeding lines are shown on the right. The cluster formed by foreign varieties is designated with the abbreviation ‘FV’.
The results of the present study clearly demonstrate that the SRAP markers are a powerful tool for studying genetic diversity in lavender. SRAP markers offer a non-specific genome wide coverage which could well complement the use of other markers related to single or small number of loci, e.g. SSR and SNP markers. The reported SRAP markers could be successfully applied in different areas of lavender research and cultivation. The SRAP analysis of genetic diversity could be readily applied for (re-)evaluation of the full set of lavender genetic resources including a number of breeding lines developed during the last five decades. The large number of polymorphic SRAP fragments/markers described in the study suggests they could be used as affordable and efficient tool for development of the first lavender genetic map and initiation of breeding programmes based on marker-assisted selection. One of the current problems related to the market of lavender planting material for industrial cultivation is the lack of efficient routine methods for quality control. The planting of homogenous vegetatively propagated lavender planting material is essential for the high yield and quality of the produced essential oil [Citation3]. The high number of polymorphic SRAP fragments generated by a single primer pair, accounting to an average of 25.5 ± 12.8 (), suggests that SRAP markers could offer an affordable method for routine quality testing of the origin and homogeneity of the vegetatively propagated planting material used for establishment of new plantations. Indeed, the analysis of vegetatively propagated lavender plantlets using one or few SRAP primers pairs could be sufficient to confirm or reject their declared origin via comparison of their SRAP pattern with those of a reference sample from the corresponding lavender variety. Our preliminary study involving SRAP analysis with three primer pairs of 24 vegetatively propagated plants from var. Hemus shows that all of them display one and the same pattern of SRAP fragments confirming the reproducibility of the results from SRAP marker application, which is mandatory for development of a routine testing procedure.
Conclusions
The overall results from the present study demonstrate that SRAP markers are a powerful and efficient tool to study the lavender genetic diversity and exhibit high potential for a wide range for further applications in lavender breeding and cultivation including: characterization of lavender genetic resources pool, characterization of lavender segregating populations and genetic mapping studies and marker-assisted selection. Additionally, the tested SRAP markers could be used as a basis for development of an affordable and routine procedure for quality testing of vegetatively propagated lavender planting material. The SRAP data analysis demonstrates a relatively high level of genetic diversity within the tested group of Bulgarian industrially cultivated lavender essential oil varieties and breeding lines, suggesting the currently available genetic resources pool is a sufficient basis for initiation of a long-term lavender breeding programme.
Disclosure statement
The authors report no conflict of interest
Additional information
Funding
References
- Lis-Balchin M, editor. Lavender: the genus Lavandula. London: Taylor and Francis; 2002.
- Wells R, Truong F, Adal AM, et al. Lavandula essential oils: a current review of applications in medicinal, food, and cosmetic industries of lavender. Nat Prod Commun. 2018;13(10):1403–1417.
- Stanev S, Zagorcheva T, Atanassov I. Lavender cultivation in Bulgaria – 21st century developments, breeding challenges and opportunities. Bulg J Agric Sci. 2016;22(4):584–590.
- Adal AM, Sarker LS, Lemke AD, et al. Isolation and functional characterization of a methyl jasmonate-responsive 3-carene synthase from Lavandula x intermedia. Plant Mol Biol. 2017;93(6):641–657.
- Adal AM, Sarker LS, Malli RP, et al. RNA-Seq in the discovery of a sparsely expressed scent-determining monoterpene synthase in lavender (Lavandula). Planta. 2019;249(1):271–290.
- Demissie ZA, Erland LA, Rheault MR, et al. The Biosynthetic origin of irregular monoterpenes in Lavandula Isolation and biochemical characterization of a novel cis-prenyl diphosphate synthase gene, lavandulyl diphosphate synthase. J Biol Chem. 2013;288(9):6333–6341.
- Guitton Y, Nicolè F, Moja S, et al. Differential accumulation of volatile terpene and terpene synthase mRNAs during lavender (Lavandula angustifolia and L. x intermedia) inflorescence development. Physiol Plant. 2010;138(2):150–163.
- Jullien F, Moja S, Bony A, et al. Isolation and functional characterization of a τ-cadinol synthase, a new sesquiterpene synthase from Lavandula angustifolia. Plant Mol Biol. 2014;84(1–2):227–241.
- Sarker LS, Demissie ZA, Mahmoud SS. Cloning of a sesquiterpene synthase from Lavandula x intermedia glandular trichomes. Planta. 2013;238(5):983–989.
- Malli RP, Adal AM, Sarker LS, et al. De novo sequencing of the Lavandula angustifolia genome reveals highly duplicated and optimized features for essential oil production. Planta. 2019;249(1):251–256.
- Curto MA, Puppo P, Ferreira D, et al. Development of phylogenetic markers from single-copy nuclear genes for multi locus, species level analyses in the mint family (Lamiaceae). Mol Phylogenet Evol. 2012;63(3):758–767.
- Hind KR, Adal AM, Upson TM, et al. An assessment of plant DNA barcodes for the identification of cultivated Lavandula (Lamiaceae) taxa. Biocatal Agric Biotechnol. 2018;16:459–466.
- Hnia C, Yosr Z, Mohamed B. Genetic diversity of natural Tunisian Lavandula multifida L.(Lamiaceae) populations assessed by allozymes and random amplification of polymorphic DNA (RAPD). Afr J Biotechnol. 2013;12(7):648–657.
- Adal AM, Demissie ZA, Mahmoud SS. Identification, validation and cross-species transferability of novel Lavandula EST-SSRs. Planta. 2015;241(4):987–1004.
- Li G, Quiros CF. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet. 2001;103(2–3):455–461.
- Aneja B, Yadav NR, Chawla V, et al. Sequence-related amplified polymorphism (SRAP) molecular marker system and its applications in crop improvement. Mol Breeding. 2012;30(4):1635–1648.
- Robarts DW, Wolfe AD. Sequence-related amplified polymorphism (SRAP) markers: a potential resource for studies in plant molecular biology. Appl Plant Sci. 2014;2(7):1400017.
- Aghaei Z, Talebi M, Rahimmalek M. Assessment of genetic diversity within and among sage (Salvia) species using SRAP markers. Plant Genet Resour. 2017;15(3):279–282.
- Chen S-Y, Dai T-X, Chang Y-T, et al. Genetic diversity among Ocimum species based on ISSR, RAPD and SRAP markers. Aust J Crop Sci. 2013;7(10):1463–1471.
- Javan ZS, Rahmani F, Heidari R. Assessment of genetic variation of genus Salvia by RAPD and ISSR markers. Aust J Crop Sci. 2012;6(6):1068–1073.
- Saebnazar A, Rahmani F. Genetic variation among Salvia species based on sequence-related amplified polymorphism (SRAP) Marker. J Plant Physiol Breeding. 2013;3(1):71–78.
- Talebi M, Rahimmalek M, Norouzi M. Genetic diversity of Thymus daenensis subsp. daenensis using SRAP markers. Biologia. 2015;70(4):453–459.
- Taşcıoğlu T, Sadıkoğlu N, Doğanlar S, et al. Molecular genetic diversity in the Origanum genus: EST-SSR and SRAP marker analyses of the 22 species in eight sections that naturally occur in Turkey. Ind Crops Prod. 2018;123:746–761.
- Murray M, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucl Acids Res. 1980;8(19):4321–4326.
- Abuzayed M, El-Dabba N, Frary A, et al. GDdom: an online tool for calculation of dominant marker gene diversity. Biochem Genet. 2017;55(2):155–157.
- Schlueter PM, Harris SA. Analysis of multilocus fingerprinting data sets containing missing data. Mol Ecol Notes. 2006;6(2):569–572.
- Zagorcheva T, Rusanov K, Stanev S, et al. A simple procedure for comparative GC-MS analysis of lavender (Lavandula angustifolia Mill.) flower volatile composition. IOSR J Pharm Biol Sci. 2016;11(4):9–14.
