Abstract
Bacterial leaf blight (BLB) and blast are among the most destructive diseases of rice. In order to develop new varieties that can resist various races of the pathogens Xanthomonas oryzae pv. oryzae (Xoo) and Magnaporthe oryzae, which cause these two diseases, respectively, it is important to identify suitable markers and a marker-aided selection strategy. Seventeen foreground microsatellites associated with resistance against BLB and blast were screened for polymorphism for possible adoption in a breeding program for resistant cultivar development. A total of 472 background simple sequence repeats (SSRs) were also screened for polymorphism in order to recover the recurrent parent genome. All 72 F1 progenies were also analysed with the 17 foreground and 472 background markers in comparison to the alleles of their parents. The markers were validated using 218 BC2F2 plants. The result revealed six tightly linked polymorphic foreground markers (Xa21FR, pTA248, Xa13prom, MP, RM21 and RM13) associated with resistance against Xoo pathotype P7.7 and two microsatellites (RM6836 and RM8225) associated with resistance against blast pathotype P7.2. A total of 79 polymorphic background markers were identified for recovering the recurrent parent genome. The result also showed that the markers had goodness of fit (blast resistance) to 1:2:1 ratio for single gene model in BC2F2 progenies. These genetically confirmed microsatellite markers were recommended for use in marker-assisted backcross breeding and other marker-assisted selection of BLB and blast resistance genes in order to develop varieties with durable and broad spectrum resistance against the two major rice diseases.
Introduction
Rice is a major cereal crop with high demand in the world. It is a vital staple food crop with high concentration of carbohydrate utilized by almost 50% of the world’s populace [Citation1]. However, significant yield decrease results from diseases such as bacterial leaf blight (BLB) caused by Xanthomonas oryzae pv. oryzae (Xoo).The disease causes yield reduction as a result of partial grain filling associated with constrained photosynthetic region of rice [Citation2].The most effective approach to prevent the disease involves development of resistant rice variety as use of chemical control agents is not an effective control measure. Magnaporthe oryzae is the causal agent for blast disease. The disease is notorious for rice destruction in Asia and other popular rice agro-ecologies [Citation3, Citation4]. Microsatellites which are also referred to as simple sequence repeat markers (SSRs) are tandemly arranged repeats of short DNA motifs (1–6 bp in length) that usually display differences in the number of repeats at a particular locus. Microsatellites are a useful source of DNA-based markers because they are capable of detecting high polymorphism and are abundant in nature [Citation5–7]. These markers are the most suitable for rice among other molecular markers available because they show co-dominance in inheritance, high reproducibility and polymorphism, ease of use and moderate price. Miah et al. [Citation8] reported that over 20,000 microsatellites were available in rice and are well distributed throughout the genome of rice. It is very vital to study the genetic underpinnings of the microsatellite markers and their distribution across the 12 rice chromosomes as a fundamental basis for developing a resistant rice variety against BLB and blast. Therefore, this work was conducted to genetically analyze and select the suitable SSR markers associated with resistance against BLB and blast diseases of rice for use in developing a new resistant rice variety.
Materials and methods
Genetic materials
A total of 72 F1 plants were developed from a cross between Putra-1 and IRBB60 rice varieties at Ladag 10, Institute of Tropical Agriculture and Food Security (ITAFoS), UPM from September 2017 to January 2018, under glass house conditions. The F1 seeds were later used to develop BC1F1, BC2F1 and BC2F2 populations between February 2018 and July 2019 and evaluated in field conditions. A cross made between MR219 × Pongsu Seribu 1 [Citation9, Citation10] at ITAFoS led to the development of Putra-1 rice variety, while IRBB60 is an International Rice Research Institute (IRRI) variety. Putra-1 has a characteristic high yield and resistance to blast with inherent Piz, Pi2 and Pi9 R-genes but is susceptible to BLB infection. The IRBB60 has BLB resistance genes, namely, Xa4, xa5, xa13 and Xa21. Both IRBB60 and Putra-1 possessed broad spectrum resistance against BLB and blast diseases due to their genetic background. The blast resistance genes are all in dominant condition, while bacterial leaf blight resistance genes were of dominant and recessive conditions.
Xoo inoculum preparation and inoculation
Pure culture of the most virulent Xoo pathotype P7.7 was collected from MARDI and sub-cultured at the Bacteriology laboratory of ITAFoS UPM. Five milliliters of distilled water were poured in each culture plate and colonies of bacteria were suspended into it. The inoculums’ concentration was adjusted to 108 cfu/mL [Citation4]. All isolate suspensions were bulked and homogenized. Wet conditions suitable for development of disease were established by spraying distilled water on the rice leaves. Upon inoculation, sterilized scissors were dipped into the prepared inoculums and used to cut five leaves approximately 5 cm from the tip. Disease data were taken after 14 days of inoculation to verify the degree of pathogenicity using the standard evaluation system (SES). The disease incidence was determined by calculating the blight lesion degree (BLD), which is the proportion, or percentage, of infected leaves per total number of observed plants. On the other hand, disease severity was determined by measuring the lesion length and width, which were used to calculate the percentage of disease leaf area or the total area of infected leaves.
Inoculation with Magnaporthe oryzae pathoype P7.2
The two parents and BC2F2 plants were inoculated with the most virulent Magnaporthe oryzae pathoype P7.2 isolated at Malaysia Rice research center MARDI. The plants were screened phenotypically for resistance to the pathogen. Twenty-one day-old rice plants were inoculated by spraying spore suspension diluted to a concentration of 1.5 × 105 conidia/mL on the plants. Humid environment was maintained at 90% by covering the inoculated rice plants with plastic bags and this encouraged disease development. Disease data for blast was taken after a period of nine days. Incidence of the disease was determined by calculating the blast lesion degrees (BLDs) on the plant. The disease leaf area (DLA) was calculated from 0 − 100 percent, while the blast lesion type (BLT) was also scored following the standard evaluation system of IRRI [Citation11].
Genomic DNA extraction and genetic analysis of microsatellites
Young and fresh rice leaves were collected from 3-week-old individual plants (before inoculation) and their gDNAs were extracted using the CTAB method [Citation12]. A total of 17 foreground markers were screened and their polymorphisms between the two parents (Putra-1 x IRBB60) were genetically analysed, as well as their position and location on the rice chromosome. Sets of primers comprising 472 microsatellites spread across the 12 rice chromosomes which were already mapped by Akagi et al. [Citation13], Temnykh et al. [Citation5] and McCouch et al. [Citation14] were selected from gramene marker database (www.gramene.com) for background selection. The two parental varieties were used to screen for polymorphism of SSR markers. Genotype data was obtained by analyzing DNA with SSR markers using 15-μL polymerase chain reaction (PCR) mixtures. After initial denaturation for 5 min at 94 °C, each cycle comprised 1 min denaturation at 94 °C, 1 min annealing at 55 °C and 2 min extension at 72 °C with a final extension for 5 min at 72 °C at the end of 35 cycles (Thermocycler, T100TM, Bio-Rad, London, UK). Electrophoresis was run for 45 min and the bands obtained were read with the aid of Molecular Imager.
Marker genotyping and statistical analysis
The whole 72 F1 progenies were scored with the 17 foreground and 472 background markers in comparison to the alleles of their parents. The selected markers were validated using BC2F2 plants. The markers were screened using 3% MetaphorTM agarose gel. The allele size of each progeny was compared with the allelic size of its parents for the particular marker with a clear polymorphism. A DNA ladder was added in the first loading and used for confirming the observed allelic sizes in the parents and progeny. Bands were scored as A, B or H representing homozygous alleles (single bands) similar to the recurrent parent, homozygous alleles (single bands) similar to the donor parent and heterozygous alleles (double bands) with genes of both parents, respectively. Descriptive statistics were done using SAS 9.4 software. The BC2F2 population segregation data were analysed using a chi-square test (χ2). An analysis for goodness of fit to the expected ratio of 1:2:1 was calculated using SAS software version 9.4.
Results and discussion
Pathogens trait variation on inoculated plants
The trait variations (means of donor and recipient parents as well as the BC2F2 population) of pathotype P7.7 and P7.2 for BLB and blast pathogens were investigated. For BLB, the average leaf blight severity scores recorded in Putra-1 were 54.76 and 3.82 for BLD and BLT, respectively, with 56.56% DLA. No infection was found in IRBB60 in any disease severity parameters. The result obtained on the challenging of BC2F2 population with pathotype P7.7 of Xoo indicated a score of 8.11 and 0.64 for BLD and BLT, respectively, with 2.20% DLA. The BC2F2 population showed resistance response against BLB infection. The present findings were in agreement with Chukwu et al. [Citation10], who found both IRBB60 and BC2F2 populations to be resistant to Xoo pathotype P7.7 in their studies that involved marker-assisted introgression of multiple resistance genes against bacterial leaf blight and blast diseases in Putra-1 rice variety. In the case of blast disease, Putra-1 showed no symptoms of blast. The average leaf blast severity scores recorded in IRBB60 were 28.13 and 3.38 for BLD and BLT, respectively, with 47.89% DLA. The result also showed that the BC2F2 population challenged with pathotype P7.2 of Magnarporthe oryzae scored 8.33 and 0.98 with regard to BLD and BLT, respectively, while the DLA obtained was 0.48%. This result indicated that the BC2F2 population showed resistance response against blast infection. Ashkani et al. [Citation4] and Miah et al. [Citation9] both reported resistance response against the blast pathogen (Magnaporthe oryzae) in their selected populations.
Foreground SSR markers
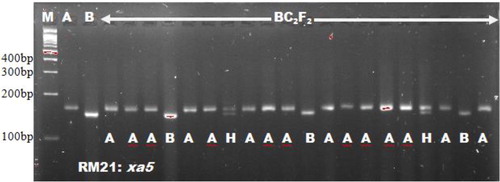
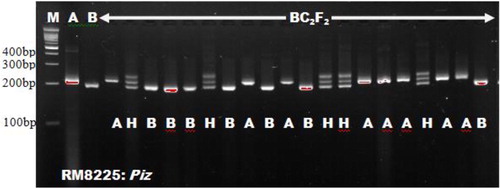
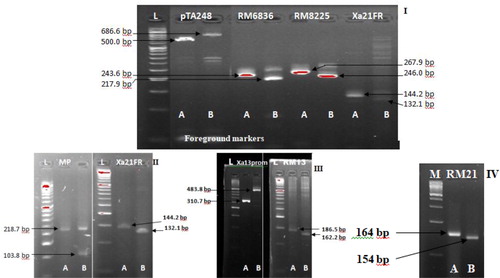
With the exception of RG556, the other foreground markers screened for polymorphism with respect to Xoo resistance genes displayed adequate amplification given the PCR and thermal cycling adopted. Polymorphic/heterogeneous bands were obtained for the two parents; Putra-1 × IRBB60, in six markers that were linked to Xoo resistance genes. The markers were RM13, MP, Xa21FR, Xa13prom, RM21 and pTA248. The amplified products for RM13 were 186.5 × 162.2 bp, MP 218.7 × 103.8 bp, Xa21FR 144.2 × 132.1 bp, Xa13prom 310.7 × 483.8 bp, RM21 164.0 × 154.0 bp and pTA248 731.6 × 686.6 bp fragments, for Putra-1 × IRBB60, respectively. The two foreground markers; RM8225 and RM6836, screened for polymorphism with respect to blast resistance genes were found to be polymorphic. The product amplification obtained for RM8225 were 267.9 × 246.0 bp fragments for blast resistant parent (Putra-1 allele) and blast susceptible parent (IRBB60 allele), respectively (). Similarly, the product amplifications for RM6836 were 243.6 × 217.9 bp. A total of eight foreground markers were confirmed polymorphic from this study which involved the two parental combinations (Putra-1 × IRBB60). Other researchers who found the markers to be polymorphic in their own parental combinations included Deshmukh et al. [Citation15], Dokku et al. [Citation16], Ji et al. [Citation17] and Singh et al. [Citation18].
Figure 1. Genetic analysis of foreground markers and their target genes.
Note: A = Putra-1, B = IRBB60, (I) pTA248(Xa21), RM6836(Pi9, Pi2, Piz), RM8225(Piz), Xa21FR(Xa21); (II) MP(Xa4), Xa21FR(Xa21); (III) Xa13prom(xa13), RM13(xa5); (IV) RM21(xa5). L: 50bp DNA ladder, M: 100bp DNA ladder
The use of microsatellites in selecting for the Xoo resistance gene xa5 and blast R-genes corresponded with Singh et al. [Citation18], who adopted RM13 for selecting xa5 gene. Miah et al. [Citation9], Hasan et al. [Citation19] and Ashkani et al. [Citation4] all used microsatellite markers for selecting the blast resistance genes. The microsatellites RM8225 and RM6836 linked to blast resistance genes were found to be polymorphic in the works of Ashkani et al. [Citation4]. They reported 221 bp and 240 bp fragments for the resistant donor parent allele in RM8225 and RM6836, respectively. These findings followed the result of our present study. Out of the eight polymorphic foreground markers, MP, pTA248, xa13prom and Xa21FR were functional markers. These markers have been reported by Ma et al. [Citation20], Ronald et al. [Citation21], Sujatha et al. [Citation22] and Deshmukh et al. [Citation15], respectively. Functional markers were emphasized in some Xoo resistance genes such as Xa4, xa13 and Xa21 due to the benefits of functional markers over microsatellites. Functional markers could reduce the linkage drag close to the gene of interest [Citation23, Citation24]. Bharani et al. [Citation25] and Perumalsamy et al. [Citation26] both made use of functional makers in their studies. Except the eight polymorphic markers obtained, the other foreground microsatellite markers were found to be monomorphic between the two parents studied and were not associated with their resistance against bacterial leaf blight and blast diseases. A similar report on monomorphism was reported by Rajpurohit et al. [Citation27] for RG136 marker linked to xa13. Monomorphic markers cannot detect heterozygous alleles and as such are not useful in selection. The results in show the marker information with respect to the linked genes for both blast and Xoo resistance. and show the foreground marker analysis of BC2F2 plants using RM21 linked to Xoo resistance gene xa5 and RM8225 linked to blast resistance gene Piz.
Table 1. Polymorphic tightly linked markers for Blast (Pi) and BLB (Xa) resistance genes.
Background SSR marker
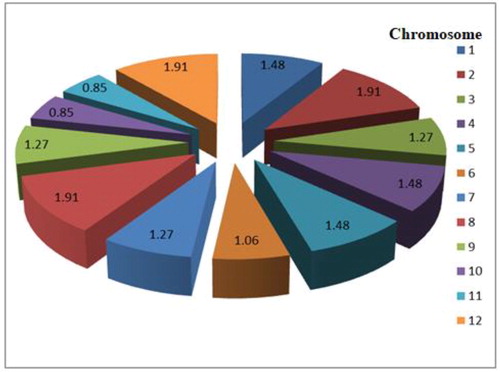
The results obtained from the background marker screening indicated that 79 out of the 472 screened were found to be polymorphic between the two contrasting parents. The polymorphism level recorded from the 472 markers was 16.74% distributed among the 12 rice chromosomes. Chromosomes 2, 8 and 12 each recorded nine polymorphic markers which were the highest per chromosome, while the minimum obtained was four polymorphic markers and this was observed in chromosomes 10 and 11 each (). Lau et al. [Citation28] reported similar results corresponding with our current findings. shows the percentage polymorphism of 472 screened background SSR markers.
Table 2. Percentage polymorphism of screened background markers.
Suitable marker selection is vital in a marker-assisted backcross breeding for selection of target genes while avoiding linkage drag [Citation29–32]. It also facilitates the recurrent parent genome recovery. During the background marker analysis, selection was made preferably on the primers with more repeat motifs – hepta, tetra, tri and di-nucleotides [Citation33, Citation34]. This was done because the relative frequencies of the nucleotide repeat motifs of the selected markers were compared on chromosome-wise basis. The 16.74% background marker polymorphism obtained from a list of 472 SSR markers screened in this study was similar to the report of Linh et al. [Citation33]. They obtained 18.7% marker polymorphism from a list of 477 microsatellites in a cross that involved BT7 × FL478. Huyen et al. [Citation35] reported 12.6% marker polymorphism. Khanh et al. [Citation36] found the sum of 15.1%, while Basavaraj et al. [Citation37] reported 17.74% polymorphic microsatellites in their various parental combinations. These results all validated our present findings on the suitability and selection of microsatellites for resistance against bacterial leaf blight and blast diseases.
Conclusions
The functional and microsatellite markers that were found to be polymorphic between the two parents studied and were also linked to the target genes could be useful for foreground selection of target genes in a marker-assisted backcross breeding program. The 79 polymorphic background markers obtained in this study could be useful in recovering the recurrent parent genome at every stage of marker-assisted backcross breeding. The genetic analysis of selected microsatellites in BC2F2 population showed that these markers were associated with BLB and blast resistance genes following the genotypic and phenotypic trait variation. The deployment of major rice resistance genes would reduce selection pressure and as such prevent the evolution of resistance in pathogen races as a result of broad spectrum resistance. Adopting this approach would assist breeders to increase research activities in rice crop by enabling genotypic selection rather than phenotypic selection alone. The microsatellites reported here provide a useful tool to breeders and geneticists for marker-assisted selection of bacterial leaf blight and blast resistance genes.
Acknowledgments
The authors are grateful to the technical staff and technologists in the molecular laboratory of Institute of Tropical Agriculture and Food Security, Universiti Putra Malaysia, especially Mrs. Nooraishah Abd Aziz for the technical supports while the work lasted. All colleagues from various universities whose research output were useful are well appreciated.
Disclosure statement
The authors declare no conflict of interest.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article. The supplementary materials and other raw data generated at Molecular Laboratory of the Institute of Tropical Agriculture and Food Security (ITAFoS), Universiti Putra Malaysia, are available from the corresponding author (MYR – [email protected]) on request.
Additional information
Funding
References
- Oladosu Y, Rafii MY, Magaji U, et al. Genotypic and phenotypic relationship among yield components in rice under tropical conditions. Biomed Res Int. 2018; 2018:1–10.
- Pradhan SK, Nayak DK, Mohanty S, et al. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety. Rice (N. Y.). 2015; 8(1):51.
- Akos IS, Yusop MR, Ismail MR, et al. A review on gene pyramiding of agronomic, biotic and abiotic traits in rice variety development. Int J Appl Biol. 2019; 3(2):65–96.
- Ashkani S, Rafii MY, Rusli I, et al. SSRs for marker-assisted selection for blast resistance in rice (Oryza sativa L.). Plant Mol Biol Rep. 2012;30(1):79–86.
- Temnykh S, Cartinhour S, Park W, et al. Mapping and genome organization of microsatellite sequence in rice (Oryza sativa L.). Theor Appl Genet. 2000;100(5):697–712.
- Chukwu SC, Rafii MY, Ramlee SI, et al. Bacterial leaf blight resistance in rice: a review of conventional breeding to molecular approach. Mol Biol Rep. 2019; 46(1):1519–1532.
- Chukwu SC, Rafii MY, Ramlee SI, et al. Marker-assisted selection and gene pyramiding for resistance to bacterial leaf blight disease of rice (Oryza sativa L.). Biotechnol Biotechnol Equip. 2019; 2:1–16.
- Miah G, Rafii M, Ismail M, et al. A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int J Mol Sci. 2013; 14(11):22499–22528.
- Miah G, Rafii MY, Ismail MR, et al. Marker-assisted introgression of broad-spectrum blast resistance genes into the cultivated MR219 rice variety. J Sci Food Agric. 2017; 97(9):2810–2818.
- Chukwu SC, Rafii MY, Ramlee SI, et al. Marker-assisted introgression of multiple resistance genes confers broad spectrum resistance against bacterial leaf blight and blast diseases in Putra-1 rice variety. Agronomy. 2020; 10(1):42.
- IRRI. Standard evaluation system for rice. 4th ed. Manila, the Philippines: International Rice Research Institute; 1996.
- Ashkani S, Rafii MY, Rahim HA, et al. Mapping of the quantitative trait locus (QTL) conferring partial resistance to rice leaf blast disease. Biotechnol Lett. 2013; 35(5):799–810.
- Akagi H, Yokozeki Y, Inagaki A, et al. Microsatellite DNA markers for rice chromosomes. Theor Appl Genet. 1996; 93(7):1071–1077.
- McCouch SR, Teytelman L, Xu Y, et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 2002;9(6):199–207.
- Deshmukh UC, Verma RK, Saxena RR, et al. Marker assisted selection for bacterial leaf blight resistance in segregating populations of Karma Mahsuri. Vegetos- An Inter J Plnt Res. 2017; 30(1):55–58.
- Dokku P, Das KM, Rao GJN. Genetic enhancement of host plant-resistance of the Lalat cultivar of rice against bacterial blight employing marker-assisted selection. Biotechnol Lett. 2013; 35(8):1339–1348.
- Ji ZJ, Yang SD, Zeng YX, et al. Pyramiding blast, bacterial blight and brown planthopper resistance genes in rice restorer lines. J Integr Agric. 2016; 15(7):1432–1440.
- Singh AK, Dharmraj E, Nayak R, et al. Identification of bacterial leaf blight resistance genes in wild rice of eastern India. Turk J Bot. 2015; 39(6):1060–1066.
- Hasan MM, Yusop MR, Ismail MR, et al. Performance of yield and yield contributing characteristics of BC2F3 population with addition of blast resistant gene. Ciênc Agrotec. 2015; 39(5):463–476.
- Ma BJ, Wang WM, Zhao B, et al. Studies of PCR marker for the rice bacterial blight resistance gene Xa-4. Hereditas. 1999; 21:9–12.
- Ronald PC, Albano B, Tabien R, et al. Genetic and physical analysis of the rice bacterial blight disease resistance locus, Xa21. Mol Gen Genet. 1992; 236(1):113–120.
- Sujatha K, Natarajkumar P, Laha GS, et al. Inheritance of bacterial blight resistance in the rice cultivar Ajaya and high-resolution mapping of a major QTL associated with resistance. Genet Res (Camb). 2011; 93(6):397–408.
- Gupta PK, Kumar J, Mir RR, et al. Marker-assisted selection as a component of conventional plant breeding. Plant Breed Rev. 2010 [cited 2020 Jan 26]; 33:145.
- Bagge M, Lübberstedt T. Functional markers in wheat: technical and economic aspects. Mol Breed. 2008; 22(3):319–328.
- Bharani M, Nagarajan P, Rabindran R, et al. Bacterial leaf blight resistance genes (Xa21, xa13 and xa5) pyramiding through molecular marker assisted selection into rice cultivars. Arch Phytopathol Plant Prot. 2010; 43(10):1032–1043.
- Perumalsamy S, Bharani M, Sudha M, et al. Functional marker-assisted selection for bacterial leaf blight resistance genes in rice (Oryza sativa L.). Plant Breed. 2010; 129(4):400–406.
- Rajpurohit D, Kumar R, Kumar M, et al. Pyramiding of two bacterial blight resistance and a semi dwarfing gene in Type 3 Basmati using marker-assisted selection. Euphytica. 2011;178(1):111–126.
- Lau WC, Rafii MY, Ismail MR, et al. Review of functional markers for improving cooking, eating, and the nutritional qualities of rice. Front Plant Sci. 2015; 6:832–2020.
- Oladosu Y, Rafii MY, Samuel C, et al. Drought resistance in rice from conventional to molecular breeding: a review. IJMS. 2019; 20(14):3519. http://dx.doi.org/10.3390/ijms20143519
- Oladosu Y, Rafii MY, Fatai A, et al. Submergence tolerance in rice: Review of mechanism, breeding and, future prospects. Sustainability. 2020; 12(4):1632. http://dx.doi.org/10.3390/su12041632
- Muhammad I, Rafii MY, Ramlee SI, et al. Exploration of Bambara Groundnut (Vigna subterranean (L) Verdc), an underutilized crop, to aid global food security: varietal improvement, genetic diversity and processing. Agronomy. 2020; 10(6):766.
- Yusop MR, Sukaimi J, Amiruddin MD, et al. Genetic improvement of oil palm through recurrent selection. In: Ithnin M, Kushairi A, editors. The oil palm genome. Vol. 4. Cham: Springer; 2020. p. 35–46.
- Linh LH, Linh TH, Xuan TD, et al. Molecular breeding to improve salt tolerance of rice (Oryza sativa L.) in the Red River Delta of Vietnam. Int J Plant Genomics. 2012; 2012:949038–942020.
- Olalekan KK, Rafii MY, Salleh AM, et al. Analysis of recurrent parent genome recovery in marker-assisted backcross breeding programme in Watermelon. Int J Sci Technol Res. 2019; 8(08):945–955.
- Huyen LT, Cuc LM, Ismail AM, et al. Introgression the salinity tolerance QTLs Saltol into AS996, the elite rice variety of Vietnam. AJPS. 2012; 03(07):981–987.
- Khanh TD, Linh TH, Xuan TD. Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into the Vietnamese elite rice variety. J Plant Breed Crop Sci. 2013; 5(2):26–33.
- Basavaraj SH, Singh VK, Singh A, et al. Marker-assisted improvement of bacterial blight resistance in parental lines of Pusa RH10, a superfine grain aromatic rice hybrid. Mol Breeding. 2010; 26(2):293–305.