Abstract
Hepatitis C is a public health problem worldwide, affecting chronically about 71 million people, or 1% of the world population. The early detection of infection in clinical practice routine and blood donors is still important, being necessary for the development of new specific, sensitive, reliable tests that can improve the screening of HCV infection. Flow cytometry beads assay opens the possibility to detect multiple antigens, decrease cross-reactivity, increase in the sensitivity, with low sample volumes. Here, we describe proof-of-concept of a method for anti-Core and anti-NS5a detection using magnetic beads and flow cytometry. The immunoassay presented 93.33% sensitivity and 100% specificity for rCore magnetic beads (MgBs), and 93.33% sensitivity and 96.67% specificity for rctNS5a MgBs. The accuracy values for the tests using rCore and rctNS5a MgBs were 96.67% and 95%, respectively. The results of the ROC curve were 0.97 and 0.99 for rCore and rctNS5a MgBs, respectively. Therefore, we concluded that the method presented here for the detection of anti-HCV antibodies using magnetic beads analyzed by flow cytometry showed promising results, achieving satisfactory sensitivity and specificity values, with future potential to be an alternative method for screening anti-HCV antibodies in blood banks and blood centers.
Introduction
Hepatitis C is a public health problem worldwide, affecting chronically about 71 million people, or 1% of the world population. The mortality per year of people afflicted with chronic infection of hepatitis C virus (HCV), reaches approximately 399,000, being caused by severe liver diseases such as cirrhosis and hepatocellular carcinoma [Citation1]. Approximately 11.9 new cases per 100.000 are confirmed for the HCV in Brazil [Citation2].
The viral genome of HCV consists of ribonucleic acid of a single strand, with positive polarity composed of 9,600 nucleotides and a coding region with a long open reading frame that expresses a polyprotein with 3000 amino acids [Citation3]. Between the structural proteins expressed, the one called “Core” is highly conserved in all genotypes and forms the viral nucleocapsid [Citation4,Citation5]. Among the non-structural proteins is the “NS5a”, which is a protein that has conserved regions in different genotypes and contributes to viral replication, pathogenesis, modulation of cell signaling pathways and apoptosis of the host cell [Citation6–8].
According to the guidelines of the World Health Organization [Citation1], the diagnosis of HCV infection can be made by serologic and molecular assays. The serological screening tests detect antibodies against proteins of the virus, the enzymatic immunoassay (EIA) being commonly used in routine laboratory practice for the detection of anti-HCV antibodies in serum and plasma of patients [Citation9].
Although the diagnostic tests used for HCV detection present a significant evolution and its incidence has decreased enough in developed countries, a large portion of the world population have an undiagnosed asymptomatic infection and live with severe problems related to the liver. In addition, the screening tests give false-negative results in the window period in patients with low titers of antibody, or false-positive in some autoimmune diseases or infections by other pathogens [Citation10]. Thus, the early detection of infection in routine clinical practice and blood donors is still important, which calls for the development of new specific, sensitive, reliable tests that can improve the screening of HCV infection.
Single and multiplex immunoassays using flow cytometry and beads coupled with proteins or antibodies are a potent diagnostic methodology for the identification and confirmation of microbial antigens in biological samples in infectious diseases. The results of these studies have demonstrated significant gains in time, cost, sensitivity and specificity of the diagnostics, mainly in assays using different antigens at the same time [Citation11,Citation12].
Thus, we present here a proof-of-concept of an immunodiagnostic method to detect antibodies against Core and NS5a proteins of the hepatitis C virus using its recombinant forms coupled to functionalized magnetic microbeads associated to flow cytometry. This test may become an alternative for the detection of HCV infections, with reliable sensitivity and specificity.
Subjects and methods
Recombinant protein expression and purification
Synthetic genes were designed for recombinant protein production. They were based on the GenBank accession ACJ37216.1 (gi|212675045, polyprotein [Hepatitis C virus subtype 1a, NCBI]). The genes were optimized for Escherichia coli using the software offered by the manufacturer IDT - Integrated DNA Technologies. The 594-bp core gene was synthesized, flanked with the restriction enzyme sites of XhoI/PstI, 5′ and 3′, respectively. The NS5a gene C- terminus (ctNS5a), comprising 692 bp and was constructed with restriction sites for BamHI/PstI enzymes. The amino-acid sequences were analyzed in the Protparam program, which predicted the molecular mass, GRAVY, among other parameters. The synthetic genes were excised, purified by gel extraction kit QIAquick (QIAGEN), according to the manufacturer's instructions. Subsequently, the fragments were ligated to the pRSET expression vector (Invitrogen), and competent cells of Escherichia coli, strain TOP 10, were transformed with this plasmid. Cloning of the insert was confirmed through enzymatic restriction and sequencing in an Abi a 3100 Genetic Analyzer (Applied Biosystems). E. coli, strain BL21 (DE3) pLysS, competent cells were transformed with the plasmid through electroporation. Expression of proteins was performed in 500 mL of LB medium, with 1 mmol/L IPTG (isopropyl β-D-1-thiogalactopyranoside), at 37 °C, under constant agitation, for three hours.
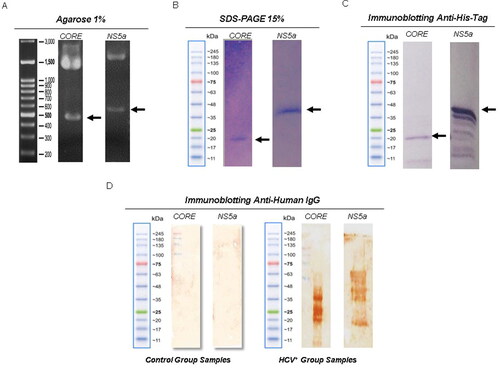
Recombinant proteins were purified using an NTA-Ni column (Qiagen®), following the manufacturer’s instructions. The proteins were separated by 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) () [Citation13]. The proteins were quantified using the Bradford method (Bio-Rad®). The presence of the polyhistidine tag in the recombinant antigen was confirmed by immunoblotting [Citation14]. For this, proteins were separated by 15% SDS-PAGE and transferred to a PVDF membrane using electrotransfer (Semidry system, Bio-rad, Hercules, USA) [Citation13]. Subsequently, the visualization was performed using a commercial WesternBreeze kit (Invitrogen), following the manufacturer's recommendations.
Figure 1. Confirmation and expression of recombinant proteins and analysis of reactivity against anti-HCV antibodies. (A) Confirmation by restriction enzyme of the correct insertion of recombinant genes. Lane 1, molecular weight marker (Jena Bioscience); Lane 2, recombinant protein CORE gene with ∼594 bp; Lane 3,recombinant protein gene ctNS5a (∼692 bp) to the expression vector pRSET A (∼2897 bp). (B) SDS PAGE (15%) of the purified CORE (rCORE) and ctNS5a (rctNS5a) recombinant proteins. (C) Western blot with anti-histidine tag against rCORE and rctNS5a. (D) Immunoblot using a pool of 10 HCV + sera against the rCORE (well 1) and against the rctNS5a (well 2). Immunoblot using a pool of HCV negative serum did not present any reactivity against the proteins.
Magnetic beads preparation
Purified proteins were coupled in BcMag carboxy-terminated magnetic beads (Bioclone Inc.). For this, 20 mg of magnetic microbeads were activated according to the manufacturer’s instructions. After the first wash with ultrapure water, coupling buffer (10 mmol/L potassium phosphate + 0.15 mol/L NaCl pH 5.5) was added and beads were incubated for 3 min at room temperature without stirring. Then, the supernatant was removed, and this process was repeated twice. Subsequently, 10 μg of purified proteins were put in contact with the activated beads, homogenized by agitation using a vortex for 3 min; 3 mmol/L of EDAC (N-Ethyl-N´-(3-dimethyl aminopropyl) carbodiimide hydrochloride) were added and this mixture was incubated at 4 °C for 24 h. Beads coupled with recombinant proteins were washed 2 times with 1 mL of the washing buffer (10 mmol/L Tris base + 0.15 mol/L NaCl + 0.1% BSA + 1 mmol/L EDTA+ 0.1% Sodium azide - pH 7.5). After washing, the solution containing HCV proteins coupled to magnetic beads was blocked with 500 μL of 1 mol/L glycine, pH 8.0, for 1-2 h. After the blocking period, the solution was washed twice with a washing buffer and stored at 4 °C until its use.
Ethics approval and consent to participate
This study was approved with written consent by the Ethics Committee Fundação Hospitalar de Hematologia e Hemoterapia do Amazonas with the approval number of CAAE: 49652815.8.0000.0009. Written informed consent was obtained from all the participants.
Sample collection
To evaluate the proposed test, the samples included in this study consist of patients with a confirmed diagnosis of chronic hepatitis C. Sample collection was made at Fundação de Medicina Tropical Dr. Heitor Vieira Dourado (FMT-HVD) with patients who agreed to participate in the study by signing the Term of Informed consent of the approved project. Both sexes were included, aged between 18 and 70 years old, with and without prior treatment or presence of fibrosis (F1-F2 and F3-F4). Indigenous patients were not included, or pregnant women or any patients who presented co-infection. The control group included candidates who attended at the blood donation at Fundação Hospitalar de Hematologia e Hemoterapia do Amazonas (FHEMOAM), of both sexes, with a minimum age of 18 years, resident in Manaus. The control group samples did not present reactivity to any infection evaluated in serological and molecular screening tests. Standardization of immunodiagnostic tests occurred with 30 analyzed HCV positive (HCV+) samples without any previous treatment: 21 samples from male patients with ages ranging from 29 to 59 years, and nine samples from HCV + female patients with ages ranging from 49 to 78 years.
Flow cytometry antibody detection using magnetic beads
For the detection of anti-Core and anti-ctNS5a, 1 μL of suspension of magnetic beads with coupled HCV proteins was added in 50 μL of samples in dilution of 1:200 with PBS (samples were treated previously with 10% E. coli BL21 (DE3) pLysS bacterial lysate for the blockade of any possible anti-E. coli antibodies present in human serum). This mixture was incubated with gentle agitation for 30 min, at room temperature, protected from the light. Then, the beads were washed with PBS-W (Phosphate Buffered Saline 1x + 0.5% Bovine Serum Albumin) and harvested at 0.3 g for 5 min. The supernatant was discarded and 1 μL of the secondary antibody (anti-IgG human Alexa Flour 488, Invitrogen) was added and homogenized and incubated for 30 min. Subsequently, magnetic beads were washed three times with PBS-W and read in the BD FACS Canto II flow cytometer. All procedures were done in a magnetic bead's separation rack. Only magnetic beads and beads coupled with proteins but without primary antibody were used as a negative control. Median fluorescence intensity (MFI) was obtained excluding the values acquired by magnetic beads without modifications in histogram graphic analysis using FlowJo software v.10.
Data analysis
FlowJo® software v.10 program was used for flow cytometry result analysis, calculating the median fluorescence intensity (MFI) and cutoff point, removing the normal bead fluorescence. The statistical and graphic analyses were done in GrahPadPrism® software v.7 program, the cutoff point being obtained by adding the average of the negative samples plus two times their standard deviation. For HCV positive samples, Mann–Whitney test was performed with p < 0.0001 in both beads/HCV-Core and beads/HCV-NS5A C-terminal tests. ROC curve test was used to determine the test sensitivity and specificity.
Results and discussion
HCV genotype type 1 presented high prevalence
Among the analyzed samples, the most frequent HCV genotype was type 1 (70%), followed by type 3 (20%), type 2 (6.66%) and a sample with type 1 and type 2 (3.33%), superinfection with the presence of two genotypes.
The HCV genotype distribution observed in our study corroborates to the prevalence found in Brazil with the highest frequency of genotype 1 followed by genotype 3 and genotype 2 [Citation15] Araújo et al. [Citation16] in their study characterizing hepatitis C virus genotypes of patients in the state of Amazonas, also obtained results with higher prevalence of genotype 1 followed by genotypes 3 and 2.
Recombinant proteins were successfully obtained from soluble E. coli fraction
Recombinant his-tagged HCV Core and NS5a were successfully expressed and purified using E. coli host and nickel column purification system. In our experiments, it was possible to obtain around 5 mg of rCore and 8 mg of NS5a per liter of culture using the described method. Proteins of molecular weight lower than the expected were detected, and probably correspond to c-terminal cleavages of recombinant proteins during the expression process.
E. coli lysate reduces non-specific reactivity in magnetic bead assay
MFI observed in HCV + samples was higher when bacterial lysate adsorption was used. Our test was standardized using serum samples at 1:200 dilutions plus 10% of E. coli lysate. We noted that although the gene sequence of the recombinant ctNS5a (rctNS5a) and Core (rCore) were obtained from genotype 1a, antibodies of genotypes 2 and 3 infection samples could recognize the produced proteins coupled to magnetic beads (). Analysis of negative and positive HCV samples for both recombinant proteins with magnetic beads (MgBs) showed statistically significant results (p < 0.0001) (), MFI from rCore-MgBs being higher than rctNS5a-MgBs.
Figure 2. Comparisons between the concentrations of primary antibodies with or without the presence of bacterial lysate. (A) Pools of ten HCV negative samples and pools of 10 HCV + samples diluted at concentrations of 1:50, 1:200 and 1:800, without the addition of bacterial lysate proteins. (B) Analysis of negative and positive samples at 1:50 1:200 and 1:800 dilutions adsorbed with 10% of bacterial lysate. (C) Comparison of HCV + pools with and without the addition of bacterial protein lysate.
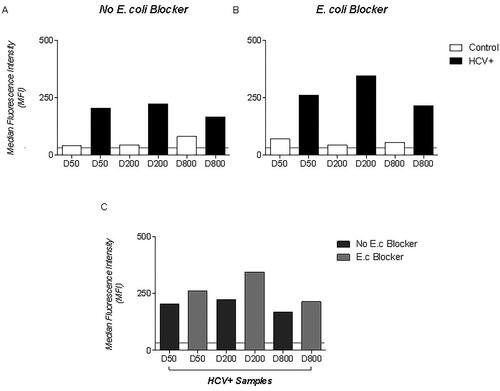
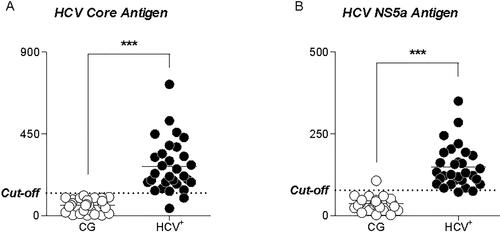
Figure 3. Comparison of MFI between 30 HCV + samples and 30 negative HCV samples using beads coupled with rCORE (A) and rctNs5a (B).
Note: Panel (A) shows the highest recognition of anti-Core HCV antibodies present in HCV + samples. No recognition of the control samples getting all below the cutoff. Panel (B) We observed increased recognition of Anti-ctNS5a antibodies in HCV + samples compared to control.
There was also a decrease in non-specific reactivity after the use of 10% of E. coli BL21 DE3 pLysS bacterial lysate in the serum samples. Other authors reported the use of bacterial protein lysate for the improvement of MFI in positive samples and 1:200 dilution offers a greater recognition of HCV antibodies [Citation17,Citation18]. Crestani et al. [Citation17] added bacterial lysate in samples at 3% final concentration and observed an improvement in their bead assay specificity using recombinant proteins. It is important to point out that many recombinant proteins are produced in E. coli and even after many purification steps, it is possible to present impurities. The positive effect of the addition of bacterial lysate to the sample could be explained by the blocking of human serum anti-E. coli antibodies from bead interaction.
Magnetic bead assay specifically detect HCV positive samples
MFIs of HCV + samples were higher when compared to those of negative samples (<125MFI), indicating specific reactivity against rCore-MgBs and rctNS5a-MgBs (). However, two positive samples presented inconclusive results, since they showed MFIs below the cutoff (MFI = 78), indicating the absence of HCV Core antibodies in these samples. It is essential to point out that these same samples exhibited reactive antibodies to rctNS5a MgBs, showing that not all HCV infections presented a humoral response to all viral proteins. We also observed that a negative sample presented reactive antibodies against rctNS5a-MgBs with its MFI above the cutoff, representing a possible cross-reactivity against this protein.
The test showed excellent accuracy (96.7% and 95.0%) for rCore and rctNS5a mgBs (). Furthermore, the immunoassay presented 93.3% sensitivity for both proteins, with 100% and 96.7% specificity for rCore-MgBs and rctNS5a-MgBs, respectively. Negative predictive value (NPV) of 93.8% and 93.6%, and positive predictive value (PPV) of 100% and 96.7%, indicating a high probability of the disease presence when the test is positive. Finally, the results of the ROC curve were 0.97 and 0.99 for rCore () and rctNS5a MgBs (), respectively.
Figure 4. ROC curve. Analysis of positive and negative HCV samples by flow cytometry using beads coupled with Core (A) and ctNS5a (B) recombinant protein, presenting areas of 0.97 and 0.99, respectively.
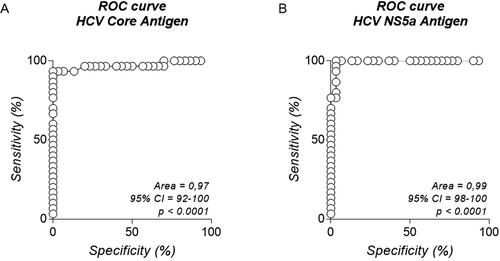
Table 1. Sensitivity and specificity parameters of tests.
The high sensitivity and specificity obtained in the assay presented here demonstrated that the test developed here is useful for detection of anti-HCV antibodies, allowing detection in dilutions as low as 1:200 and corroborating with literature. Core protein has a high recognition of reactive antibodies using different immunoassays. Fonseca et al. [Citation18] observed a sensitivity of 55.9% and specificity of 100%, PPV 100%, and NPV 69.4% in their standardized assay using commercial Luminex beads with NS5 protein. When they analyzed individually, there was an improvement when tested in a multiplex format. McHugh et al. [Citation19] also reported a high performance in their test using microsphere-coupled Core and NS3 and cytometry reading, reaching sensitivity five times higher than the ELISA test. These results demonstrate the enormous potential of using microbeads and flow cytometry in screening for anti-HCV antibodies.
The use of magnetic beads brought advantages in the execution of the test since the entire process of concentration of the beads was carried out in a magnetic beads separation rack, excluding the need for centrifugation, thus preventing the compaction of impurities with the sample. Another point that should be highlighted is the practicality in immobilizing proteins to these beads and the possibility of standardizing the method to use minimal concentrations of the antigen. Meng et al. [Citation20] also described high sensitivity and specificity in the identification of Staphylococcus aureus using the same approach used here, using minimal concentrations of capture protein in the beads. It is worth noting that the antigens and antibodies used in immunoassays are one of the most expensive elements in commercial kits. Therefore, reducing its use to very low quantities leads to a reduction in the analysis’ cost. Ondigo et al. [Citation21] also applied the flow cytometry beads assay in their work to detect multiple antigens of Plasmodium falciparum, and observed a decrease in cross-reactivity, reduction of the antigen concentration used, an increase in the sensitivity of the test, allowing the use of sample dilutions 5 times lower than those used in other tests. They also described a decrease in the time of diagnosis and an increase in the range of detection of antigens.
Unlike other immunoassays such as ELISA and immunoblot, immunoassays using flow cytometers apply lasers that recognize only the soluble particles marked and of pre-established size. Recent studies have demonstrated its potential use in new applications [Citation22,Citation23]. This methodology presents a minimum interference of unmarked particles, thus allowing reduction of the washing steps, handling and testing time [Citation24,Citation25]. Commercial enzymatic immunoassays take about 2 h to be executed. In our study, the use of flow cytometry, in association with magnetic beads, this time was reduced to 1 h and 30 min, mainly due to the quick separation of the beads during all procedures and the fast reading process of the equipment. The technique described here could allow the reduction in test cost, sample volume, time for result and can open the possibility for multiple tests [Citation18,Citation26]. It can also be applied to analyze the treatment response of patients according to anti-NS5a titers [Citation27] and anti-core titers to distinguish acute and chronic HCV infection [Citation28].
Conclusions
This study demonstrated that the method presented here for the detection of anti-HCV antibodies using magnetic beads analyzed by flow cytometry showed promising results, achieving satisfactory sensitivity and specificity values, promising to be in the future an alternative method for screening anti-HCV antibodies in blood banks and blood centers. However, more studies must be made, applying the same methodology to other proteins of HCV and evaluating it in a multiplex format with principal structural and non-structural proteins of the Hepatitis C virus. Future work will also expand the number of samples and analyze the cross-reactivity with other epidemic diseases in the region.
Disclosure statment
All authors declare that they have no competing interests regarding this manuscript.
Ethics approval and consent to participate
This study was approved with written consent by the Ethics Committee Amazonas Hospital Foundation of Hematology and Hemotherapy (HEMOAM), with the approval number of CAAE: 49652815.8.0000.0009, and written informed con-sent was obtained from all the participants
Additional information
Funding
References
- World Health Organization. Global hepatitis report 2017. World Health Organisation, 2017.
- BRASIL. Ministério da Saúde. Hepatites virais - Boletim Epidemiológico. Ministério da Saúde. 2018;49(31):1–69.
- Choo QL, Kuo G, Weiner AJ, et al. Isolation of a cdna clone derived from a blood-borne non-A, non-B viral hepatitis genome . Science. 1989;244(4902):359–362.
- Gawlik K, Science M, Jolla L, et al. HCV core protein and virus assembly: what we know without structurescess. Immunologic Research. 2014;60(1):1–10.
- Bukh J, Purcell RH, Miller RH. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc Natl Acad Sci USA. 1994;91(17):8239–8243. (August 1994):
- Gale MJ, Korth MJ, Tang NM, et al. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230(2):217–227.
- Macdonald A, Crowder K, Street A, et al. The hepatitis C virus NS5A protein binds to members of the Src family of tyrosine kinases and regulates kinase activity. J Gen Virol. 2004;85(Pt 3):721–729.
- Gu M, Charles RM. Structures of hepatitis C virus nonstructural proteins required for replicase assembly and function. Curr Opin Virol. 2013;3(2):129–136.
- Alborino F, Burighel A, Tiller FW, et al. Multicenter evaluation of a fully automated third-generation anti-HCV antibody screening test with excellent sensitivity and specificity. Med Microbiol Immunol. 2011;200(2):77–83.
- Moorman AC, Drobenuic J, Kamili S. Prevalence of false-positive hepatitis C antibody results, National Health and Nutrition Examination Study (NHANES) 2007-2012. J Clin Virol. 2017;89:1–4.
- Binder SR. Autoantibody detection using multiplex technologies. Lupus. 2006;15(7):412–421.
- Ernst D, Bolton G, Recktenwald D, et al. Bead-based flow cytometric assays: a multiplex assay platform with applications in diagnostic microbiology. In: Advanced Techniques in Diagnostic Microbiology [Internet]. Boston, MA: Springer US; 2006. p. 427–443. Available from:
- Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: a laboratory manual [Internet]. Cold Spring Harbor Laboratory Press; 1982. p. 545. (A Manual for genetic engineering). Available from: http://books.google.com.br/books?id=HZRqAAAAMAAJ
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. 1979 [ classical article]. Biotechnology. 1992;24(9):145–149. http://www.ncbi.nlm.nih.gov/pubmed/1422008
- Campiotto S, Pinho JRR, Carrilho FJ, et al. Geographic distribution of hepatitis C virus genotypes in Brazil. Braz J Med Biol Res. 2005;38(1):41–49.
- Araújo AR, de Almeida CM, Fraporti L, et al. Characterization of hepatitis C virus in chronic hepatitis patients: genotypes in the State of Amazonas, Brazil. Rev Soc Bras Med Trop. 2011;44(5):638–640.
- Crestani S, Leitolis A, Lima LFO, et al. Enhanced target-specific signal detection using an Escherichia coli lysate in multiplex microbead immunoassays with E. coli-derived recombinant antigens. J Immunol Methods. 2016;435:17–26.
- Fonseca BPF, Marques CFS, Nascimento LD, et al. Development of a multiplex bead-based assay for detection of hepatitis C virus. Clin Vaccine Immunol. 2011;18(5):802–806.
- McHugh TM, Viele MK, Chase ES, et al. The sensitive detection and quantitation of antibody to HCV by using a microsphere-based immunoassay and flow cytometry. Cytometry. 1997;29(2):106–112.
- Meng X, Yang G, Li F, et al. Sensitive detection of staphylococcus aureus with vancomycin-conjugated magnetic beads as enrichment carriers combined with flow cytometry. ACS Appl Mater Interfaces. 2017;9(25):21464–21472.
- Ondigo BN, Park GS, Gose SO, et al. Standardization and validation of a cytometric bead assay to assess antibodies to multiple Plasmodium falciparum recombinant antigens. Malar J. 2012;11(1):427.
- Irumva V, Waihenya R, Mwangi AW, et al. Evaluation of the TbgI 2 and TbgI 17 Tandem Repeat Antigens as Potential Antigens for the Diagnosis of Trypanosoma brucei rhodesiense. OJCD. 2019;09(04):152–163.
- Wang H, Cong F, Guan J, et al. Establishment of xMAP for the simultaneous detection of antibodies to Newcastle disease virus and avian influenza virus. Poultry Science. 2019;98(3):1494–1499.
- Duensing TD, Watson SR. Antibody screening using high-throughput flow cytometry. Cold Spring Harb Protoc. 2018;2018(1):pdb.top093773–12.
- Edwards BS, Oprea T, Prossnitz ER, et al. Flow cytometry for high-throughput, high-content screening. Curr Opin Chem Biol. 2004;8(4):392–398. https://linkinghub.elsevier.com/retrieve/pii/S1367593104000833
- Kellar KL, Iannone MA. Multiplexed microsphere-based flow cytometric assays. Exp Hematol. 2002;30(11):1227–1237.
- Desombere I, Vlierberghe H, Van Weiland O, et al. Serum levels of anti-NS4a and anti-NS5a predict treatment response of patients with chronic hepatitis C. J Med Virol. 2007;79(6):701–713. (February):
- Nikolaeva LI, Blokhina NP, Tsurikova NN, et al. Virus-specific antibody titres in different phases of hepatitis C virus infection. J Viral Hepat. 2002;9(6):429–437.
