Abstract
Multi-microorganisms mixed fermentation is the main characteristic of Chinese baijiu brewing. In this study, plate culture and high-throughput sequencing were used to investigate the changes in microbial community during baijiu brewing. The results revealed that fungi accounted for the largest number of culturable microorganisms, followed by bacteria and molds. The diversity of bacteria and fungi first increased and then decreased, before increasing slightly at the late stage. Most microbial species came from the environment or raw materials, and the microbial community changed greatly during fermentation; thus, the dominant microorganisms in the starter might not be the dominant organisms during fermentation. The bacterial composition changed in a certain pattern, and the core microbes were lactic acid bacteria and Acetobacter pasteurianus. The composition of each genus was relatively uniform at the early stage, while Lactobacillus spp. were dominant at the late stage. Among them, Lactobacillus sp., which ranked first, had low initial abundance, before reaching 63.7% at the end. Four fungi, Saccharomyces cerevisiae, Wickerhamomyces anomalus, Aspergillus sp. and Rhizopus oryzae, were dominant, accounting for 74.6%–93.6% of the species present, among which the first two accounted for 50.3%–81.2%. This study provides a basis for the mechanized production of Xiaoqu baijiu.
Introduction
Chinese baijiu is a traditional alcoholic beverage made from grains by natural fermentation [Citation1], and is one of the six major distilled liquors in the world. The most prominent feature of Chinese baijiu brewing is the use of a mixture of various microorganisms, which allows simultaneous saccharification and fermentation [Citation2]. Different types of baijiu are brewed using different types of microbial starter through different production processes. Currently, 12 types of baijiu with different styles and characteristics have been created. Among them, strong aroma, light aroma and sauce aroma are the mainstream aroma profiles. Light aroma baijiu has a long history, simple production process, high yield and short cycle; thus, it has become an important raw material for the production of medicinal and health-promoting liquors [Citation3].
Light-aroma Xiaoqu (XQ) liquors are formed by solid or semi-solid fermentation with sorghum as the main raw material and XQ (microbial starter in small balls or cubes) as the starter, followed by distillation and storage [Citation4]. Furthermore, in general, the microbial starter used in the production of Luzhou-flavor baijiu, Fen-flavor baijiu and Maotai-flavor baijiu is Daqu. The main microorganisms in Daqu are Aspergillus, Acetobacter, Lactobacillus and yeast, while they include Rhizopus, Mucor, and a small amount of yeast in XQ [Citation5–7]. XQ not only provides abundant enzymes for degrading the substrates and synthesizing flavor substances [Citation7, Citation8], but also provides a large volume of microorganisms and alcohol produced by their metabolism for baijiu fermentation [Citation9]. Among many natural microbial starters, only a few types of microorganisms can drive the fermentation process. Not only do they produce various flavor substances, but more importantly, they maintain the interactions among microorganisms [Citation10], which are known as the core microorganisms, and their composition and function determine the quality of the baijiu [Citation11].
Although Chinese baijiu production has a long history, traditional manual production is labor-intensive, costly, and inefficient [Citation12], and the open environment can easily cause problems, such as difficulty in controlling the fermentation temperature and susceptibility to contamination by undesirable microorganisms, resulting in variable quality of baijiu [Citation1]. Jing Brand Co., Ltd. is the largest producer of XQ baijiu in China, and the baijiu produced by it is a typical representative of XQ baijiu in China. The construction of its mechanized brewing base was completed in 2013, providing an annual output of 50,000 tons of baijiu. The mechanized production workshop changed the fermentation temperature from natural room temperature to a constant temperature of 21–23 °C all year round, and the raw materials no longer touch the ground or come into direct contact with humans.
During baijiu fermentation, microorganisms play a pivotal role in flavor development and aroma profile. Studying the composition, diversity and variation patterns of microorganisms is of great significance in baijiu production. In our previous study, metagenomic and metatranscriptomic analyses were used to investigate the microbial communities in Xiaoqu baijiu brewing produced by different technologies, and 5 core microbes (Saccharomyces cerevisiae, Rhizopus delemar, Pichia kudriavzevii, Lactobacillus helveticus and R. oryzae) were found in the fermentation [Citation13]. However, because of the small sample size, there is currently no exact blueprint for the main sources of microorganisms, their changing patterns, or the composition of core microorganisms at every stage of fermentation of XQ baijiu produced by a mechanized process. In this study, the composition and changes of bacteria and fungi during the mechanized brewing process of XQ baijiu are expounded by using high-throughput sequencing and culturable techniques to provide a theoretical basis for baijiu production and process improvement.
Materials and methods
Experimental design and sampling
The fermented grain sample was obtained from the Fenglin Winery of the Jing Brand Co., Ltd. The fermentation period lasted for 15 days. On day 0 (d0; beginning of fermentation), 1 (d1), 3 (d3), 5 (d5), 7 (d7), 10 (d10) and 15 (d15; the end of fermentation), samples were collected via the five-point sampling method, i.e. five sub-samples were collected at different locations of the fermenter and mixed evenly into one sample. Each sample was divided into two parts, one of which was stored at −80 °C for DNA extraction, and the other was stored at 4 °C for the immediate detection of culturable microorganisms and physicochemical indices.
Physicochemical index determination
The water content was determined by the weighing method. Specifically, 20.00 g of sample was placed in a crucible to be dried in an oven at 105 °C for 3–5 h, and the water content was calculated according to the weight difference before and after drying. The reducing sugar content was determined by Fehling’s method [Citation14]. The process of starch detection was as follows: 5.00 g of sample was weighed and placed into a 250 mL conical flask, added to 100 ml of 5% HCl solution, bathed in boiling water for 30 min, quickly cooled, added to 20% NaOH for neutralization until the solution became slightly acidic, filtered, added to distilled water to a volume of 250 mL, and shaken well. Then, the same method as that for reducing sugar detection was applied. Acidity was determined by titration method with 0.1 mol/L NaOH solution [Citation15]. Detection of alcohol by volume; 100.00 g of fermented grains were added to 200 mL of water, from which 100 mL of solution was distilled with a 500 mL all-glass distiller and the alcohol content was measured with a DMA5000 densimeter.
Plate counting
First, 25.00 g of fermented grain sample was placed in a conical flask, added to 225 mL of distilled water, and shaken on a shaker at 120 r/min for 30 min. Then, 1 mL of the suspension was pipetted and diluted to an appropriate concentration successively by 10-times gradients. The bacteria were cultured in Luria − Bertani medium (LB) at 37 °C for two days [Citation16], the molds were cultured in Czapek Dox medium [Citation17] containing 100 μg/mL ampicillin at 30 °C for three days, and the yeasts were cultured in YPD medium (1% of yeast extract, 2% of peptone, and 2% of glucose) containing 100 μg/mL ampicillin at 30 °C for three days [Citation18], after which they were counted separately.
High throughput sequencing and analysis
Genomic DNA was extracted from the samples using reagent kits, and DNA quality was detected by 1% agarose gel electrophoresis. High-throughput sequencing was completed by Shanghai Majorbio Bio-Pharm Technology Co., Ltd., and the libraries were built according to Illumina’s standard process. The libraries of bacteria were built with primers targeting the V3-V4 region, namely, 341 F: 5’-CCTAYGGGRBGCASCAG-3’ and 806 R: 5’-GGACTACNNGGGTATCTAAT-3’, and those of fungi were built with primers targeting the ITS1 region, namely, ITS5-1737F: 5’-GGAAGTAAAAGTCGTAACAAGG-3’ and ITS2-2043R: 5’-GCTGCGTTCTTCATCGATGC-3’. PCR reaction system (50 µL); 25 µL of 2× Premix Taq, 3 µL of template (20 ng/µL), 1 µL of each primer F and R (10 μmol/L), and 20 µL of nuclease-free water. PCR reaction conditions; 94 °C for 5 min, 94 °C for 30 s, 52 °C for 30 s, and 72 °C for 30 s, for a total of 30 cycles; 72 °C for 10 min (PCR Amplifier: ABI GeneAmp® 9700; ABI, CA, USA). The constructed amplicon libraries were subjected to PE300 paired-end sequencing on the Illumina Hiseq3000 platform. The PE reads obtained by sequencing were spliced according to the overlap relationship among PE reads, and the sequence quality was controlled and filtered at the same time. The samples were distinguished according to the barcodes at the beginning and end of the sequences as well as the primer information to obtain the effective sequences, the sequence directions were corrected, and the optimized sequences were obtained using FLASH and Trimmomatic software.
Operational taxonomic unit (OTU) cluster analysis was carried out using Usearch software. Statistical analysis of biological information was carried out for OTUs with similarity levels greater than 99%, and each OTU represented a species [Citation19]. Based on OTU cluster analysis and species comparison results at different classification levels, the diversity index was analyzed based on OTUs using BIO-DAP (Fundy National Park, Canada) software. The V3-V4 regions of 16S rDNA genes of bacteria were compared using the Greengenes database for bacteria and the RDP Classifier. The ITS1 regions of fungi were compared using the UNITE database for fungi and the Basic Local Alignment Search Tool (BLAST). Using QIIME platform and KRONA software, species annotation and abundance analysis were carried out.
Results and discussion
Changes in the physical and chemical index
In baijiu brewing, temperature, water content and acidity are usually considered as key environmental variables, which not only jointly reflect the fermentation process with indexes, such as reducing sugar and starch [Citation20], but are also closely related to the activities of microorganisms and play an important role in ensuring the quality of the base liquor [Citation12, Citation21]. In the brewing process, the temperature first rose rapidly and reached the maximum on day 5, and then slowly decreased. The acidity was stable for the first 5 days, gradually increased, and was the highest at the end (). At the late stage of baijiu fermentation, Lactobacilli and other microorganisms generally produce a large amount of acidic substances [Citation22]. Appropriate acidity is beneficial to the gelatinization and saccharification of fermented grains and can also suppress the growth of harmful microorganisms and contribute to the taste of baijiu [Citation20].
Figure 1. Physical and chemical index, and culturable microorganisms in fermented grains. Temperature and acidity (A); water, starch, reducing sugar and alcohol (B); culturable microorganisms determined by plate counting method (C); n = 3.
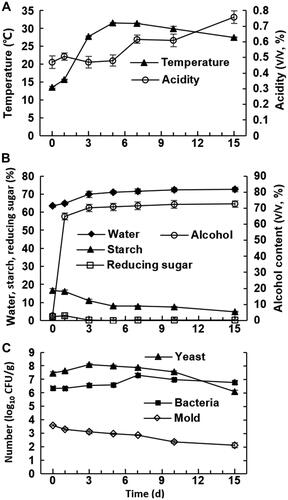
As shown in , the water content increased slowly at first and remained unaltered after the day 5. Alcohol was mainly produced on day 1, and the alcohol by volume increased rapidly from 3.13 to 64.83 on day 1, reached 70.20 on day 3, and then remained stable. When the concentration of alcohol or acid was relatively high, the growth of yeast was inhibited [Citation23]. Starch is the material basis for alcohol fermentation. The content of starch gradually decreased during fermentation, especially rapidly from day 1 to 3, and the growth of yeast was very active at this stage. Furthermore, the reducing sugar content basically stabilized to zero on day 3 of fermentation.
Changes in the numbers of culturable microorganisms
The yeast levels increased rapidly in the first 3 days, reaching the maximum of 1.26 × 108 CFU/g at day 3 (day 0, 2.9 × 107 CFU/g), and then decreased slowly. The bacteria levels increased slowly in the first 5 days, increased rapidly from d5 to d7, and then decreased slowly after reaching the maximum of 2.0 × 107 CFU/g at day 7. At the beginning, the bacteria levels were higher than those of yeasts, but at the end, the opposite trend was observed. The mold levels decreased continuously, with 3.9 × 103 CFU/g found at the start of fermentation and 1.3 × 102 CFU/g at the end of fermentation ().
The results of plate counting in this study indicated that yeast proliferated rapidly in from day 1 to 3, reaching the highest level, then gradually decreased. Bacteria grew slowly in between day 1 and 5 because the temperature at this stage was low and unsuitable for bacterial growth. On day 5, the fermentation temperature was above 30 °C, and some alcohol-tolerant bacteria multiplied. At different stages, the types of microorganisms changed in certain patterns. At the late stage, constant-temperature fermentation can be modified to variable-temperature fermentation to optimize the fermentation conditions.
Microbial community based on high-throughput sequencing
In baijiu brewing, bacteria are mainly used to produce aroma components and their precursors and are an important source of the unique flavor of light-aroma XQ baijiu [Citation24]. As shown in , at the phylum level, Firmicutes and Proteobacteria were predominant, and the number of bacteria belonging to other phyla was very small. At the genus level, there were 15 main genera, including Lactobacillus, Acetobacter, Weissella, Bacillus, Lactococcus, and Gluconobacter. At the early stage of day 1 to 3, the composition of each genus was relatively uniform. During the brewing process, Lactobacillus showed an upward trend, with an absolute predominance from day 5, and its abundance was over 50%. showed the top 30 most abundant species, collectively accounting for approximately more than 90% of all species present, among which Lactobacillus species accounted for the highest proportion with 10 species. The no. 1 Lactobacillus sp. had a low initial abundance but reached 63.7% by the end.
Figure 2. Microbial community in fermented grains. Bacteria at phylum (A) and genus (B) levels. Fungi at phylum (C) and genus (D) levels; * refers to the next taxon was unclassified; n = 3.
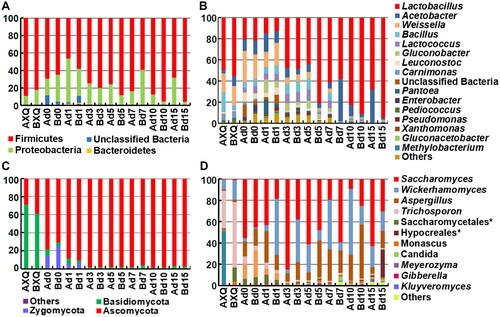
Table 1. Relative abundance of bacterial species based on high throughput sequencing (top 30, %).
According to the previous study, it was most likely the Lactobacillus helveticus [Citation13]. Other Lactobacillus species present include Weissella paramesenteroides, Lactococcus lactis, Lactococcus piscium, Leuconostoc pseudomesenteroides and Leuconostoc lactis. Acetobacter pasteurianus ranked second, with an abundance of 0.62% at the beginning and 16.6% at the end. In the fermentation process, the bacterial community changed dramatically and the top 10 species in XQ were not dominant at the end of fermentation.
In this study, the results showed the core bacteria were Lactobacillus sp., A pasteurianus, W. paramesenteroides, Gluconobacter sp., and Bacillus sp. (). It was different from Daqu baijiu production, in which Bacillus dominated the process of brewing [Citation2, Citation9]. Lactobacillus sp. play a dominant role in the fermentation process of light-aroma baijiu [Citation6, Citation25]. They can produce lactic acid and acetic acid via metabolism to maintain an acidic environment and reduce the diversity of microorganisms, thereby promoting fermentation of baijiu [Citation26]. The lactic acid produced is conducive to the formation of ethyl lactate, which is an important basic substance for the formation of aroma components in baijiu [Citation27]. The sour flavor substances in baijiu are mainly lactic acid and acetic acid. A. pasteurianus can convert ethanol to acetic acid, while Gluconobacter oxidizes ethanol into acetic acid. However, the difference is that Gluconobacter appears in sugar-rich environments, so its abundance is relatively high at the early stage of fermentation, while A. pasteurianus mostly appears in alcohol-rich environments, thus, has high abundance at the late stage of fermentation. W. paramesenteroides exists in soy sauce, pickles, sausages and other fermented foods, and plays an important role in the synthesis of organic acids, esters and short chain fatty acids in food [Citation28]. Bacillus mainly exists in distiller’s yeast, and its abundance is relatively high when the temperature is high in the fermentation process. It enhances the secretion of amylase [Citation29], provides amino acids, organic acids and other compounds, and can interact with other microorganisms to form representative aroma components [Citation10, Citation30].
At the phylum level, Basidiomycota, Ascomycota and Zygomycota were found. At the genus level, nine genera, including Saccharomyces, Wickerhamomyces, Aspergillus, Trichosporon, and Monascus, as well as two fungi of unknown genera were present. The proportion of the top 20 at the species level was generally over 97% (). The first two species were Saccharomyces cerevisiae and Wickerhamomyces anomalus, with a proportion of 50.3–81.2%, followed by two types of molds, Aspergillus sp. and Rhizopus oryzae. The proportions of these four fungi were between 74.6% and 93.6%, indicating that several types of microorganisms play a major role in baijiu brewing. Trichosporon sp. ranked first among the yeasts in XQ but were not dominant in the fermentation process.
Table 2. Relative abundance of fungal species based on high throughput sequencing (top 20, %).
The results showed that the core fungi were S. cerevisiae, W. anomalus, Aspergillus sp., R. oryzae, etc. It was a little different from the previous study which showed that the 4 core fungi were S. cerevisiae, Rhizopus delemar, Pichia kudriavzevii, and R. oryzae [Citation13]. Fungal communities mainly consisted of species of Saccharomycetaceae and Rhizopus [Citation6]. S. cerevisiae is the most important microorganism in the process of alcohol fermentation, and is involved in the formation of alcohol and flavor substances [Citation31–33]. W. anomalus is an aroma-producing yeast that can produce a large amount of ethyl acetate in the fermentation process [Citation34]. S. cerevisiae has important contributions to the fermentation rate and ethanol production, but generally cannot directly convert starch to glucose [Citation35]. Molds can produce hydrolases, such as amylase, protease and lipase, which are used for the saccharification of starch and decomposition of macromolecular substances. These enzymes also provide essential flavor compounds and enhance the formation of the aroma [Citation36]. Aspergillus produces proteolytic enzymes and other lyases, which promote the saccharification of starch, hydrolysis of proteins, and formation of flavonoids. Rhizopus is the main saccharifying microorganism in the process of baijiu brewing. For example, the abundance of R. oryzae could reach 88% at the beginning of Sichuan Xiaoqu baijiu brewing [Citation6]. Rhizopus widely exists in the starters and fermented grains of various liquors and can rapidly utilize various raw materials to produce glycerol, lactic acid, amylase and protease, among other hydrolases and volatile compounds [Citation29, Citation37].
Baijiu is produced by multi-microbes mixed fermentation. The synergy among populations is closely related to the quality of baijiu [Citation1, Citation28], and the uniformity of the community structure ensures its stability [Citation38]. Lactobacillus sp. and S. cerevisiae widely coexist during natural fermentation. Lactobacillus spp. promote the growth of brewing yeasts and facilitate their metabolism, resulting in the production of acids, alcohols, esters and other flavor substances [Citation39]. Some substances produced by the metabolism of S. cerevisiae can inhibit the synthesis of ethyl acetate by A. pasteurianus, and this inhibition is eliminated when there are live yeast in the fermentation system [Citation40]. The growth of S. cerevisiae and W. anomalus influence one another, and this co-culturing produces more varieties and higher contents of acetic ester, ethyl ester, higher alcohols, aldehydes and ketones [Citation41].
Sample relationship
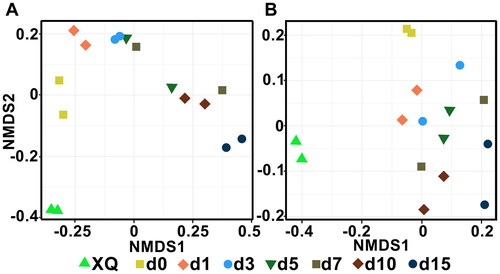
Non-metric multidimensional scaling (NMDS) analysis showed that the reproducibility between two replicate samples of bacteria composition was good, especially at d0, d1, d3, d10 and d15 (). The bacterial composition changed in a certain pattern, with close relationships among samples at d0, d1 and d3, as well as at d3, d5, d7 and d10. The composition of the fungi also had good reproducibility, but the change pattern was not significant compared with the composition of the bacteria.
Figure 3. NMDS ranking of microbial community in fermented grains. Bacteria (A) and fungi (B). A closer distance between points indicates higher similarity; n = 2.
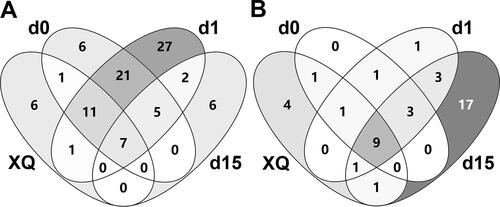
Chinese baijiu is a traditional fermented food, and much attention has been paid to the sources, composition and functions of microorganisms in its brewing process. Analysis of XQ and samples obtained on d0, d1 and d15 showed that there were 26 species of bacteria in the XQ sample, 51 in the d0 sample (19 from XQ), 74 in the d1 sample, and 20 in the d15 sample (). In addition, 7 species of bacteria were found in all four samples. A total of 16 fungal species were found in the XQ sample, 15 in the d0 sample (11 species from XQ), 19 in the d1 sample and 34 in the d15 sample. Additionally, 7 species of fungi were found in all four samples. The Venn diagrams revealed that most species came from the environment or raw materials (). These results indicate that the microorganisms not only came from the microbial starter, but also from the environment and raw materials.
Diversity index
As shown in , when the threshold was 0.01 to 0.05%, the number of OTUs was greatly reduced. The diversity index of d0 at the beginning of fermentation was higher than that of XQ. During the fermentation process, the bacterial diversity index first rose to the highest value on d1, then decreased to the minimum on d10, and finally increased slightly at the late stage. The diversity of fungi showed more complex variation than bacteria-first increasing to the maximum on d1, then rapidly decreasing until d3, remaining constant until d5, and finally increasing slightly.
Table 3. Microbial diversity index based on OTUs of high-throughput sequencing.
The initial fermented grains were rich in nutrition, inhibition by alcohol was minor, and various microorganisms grew rapidly, and therefore, the diversity increased rapidly. When the inhibition of alcohol occurred, the growth of many microorganisms was inhibited and diversity decreased. The diversity increased slightly after day 10, and many alcohol-tolerant microorganisms began to grow. The changes in environment and nutrition during brewing explain the changes in community species diversity [Citation42]. The diversity index reflects the changes in the microbial community, as confirmed in the present study.
Conclusions
In this study, traditional culture method and modern high throughput sequencing technology were used to investigate the changes in microbial community at every stage of baijiu brewing. The results revealed that most species of microorganisms originated from the environment or raw materials, and the dominant microorganisms in Xiaoqu are not necessarily dominant during the fermentation process. The diversity of bacteria and fungi increased initially, then decreased, and again increased slightly at the late stage of brewing process. The core species of bacteria were Lactobacillus spp., A. pasteurianus, W. paramesenteroides and L. lactis. The core species of fungi were S. cerevisiae, W. anomalus, Aspergillus sp. and R. oryzae. The microbial community changed greatly, with yeasts dominating the early stage and bacteria dominating the late stage. Thus, a temperature control process by stage could be adopted. This study provided fundamental understanding of the microbial communities in the mechanized production of light-aroma Xiaoqu baijiu.
Disclosure statement
The authors declare there are no competing interests. QY, SC, LZ are affiliated with Jing Brand Co., Ltd.
Data availability statement
All data generated is included in this present study, and the data are available on Mendeley:http://dx.doi.org/10.17632/mds4wpc5rn.1. Moreover, the sequencing data has been submitted to the NCBI website with a BioProject number of PRJNA699760.
Additional information
Funding
References
- Huang M, Huo J, Wu J, et al. Structural characterization of a tetrapeptide from Sesame flavor-type Baijiu and its interactions with aroma compounds. Food Res Int. 2019;119:733–740.
- He G, Dong Y, Huang J, et al. Alteration of microbial community for improving flavor character of Daqu by inoculation with Bacillus velezensis and Bacillus subtilis. LWT-Food Sci Technol. 2019;111:1–8.
- Dong W, Guo R, Sun X, et al. Assessment of phthalate ester residues and distribution patterns in Baijiu raw materials and Baijiu. Food Chem. 2019;283:508–516.
- Liu H, Sun B. Effect of fermentation processing on the flavor of Baijiu. J Agric Food Chem. 2018;66(22):5425–5432.
- Zhao QS, Yang JG, Zhang KZ, et al. Lactic acid bacteria in the brewing of traditional Daqu liquor. J Inst Brew. 2020;126(1):14–23.
- Wang M-Y, Zhao Q-S, Yang J-G. Analysis of the microbial community structure during brewing of Sichuan Xiaoqu Baijiu. J Am Soc Brew Chem. 2019;77(3):210–219.
- Xiao C, Lu Z, Zhang X, et al. Bio-heat is a key environmental driver shaping the microbial community of medium-temperature Daqu. Appl Environ Microb. 2017;83(23):e01550–17.
- Wang B, Wu Q, Xu Y, et al. Synergistic effect of multiple saccharifying enzymes on alcoholic fermentation for Chinese Baijiu production. Appl Environ Microb. 2020; 86(8):e00013–20.
- Wang P, Wu Q, Jiang X, et al. Bacillus licheniformis affects the microbial community and metabolic profile in the spontaneous fermentation of Daqu starter for Chinese liquor making. Int J Food Microbiol. 2017;250:59–67.
- Wang S, Wu Q, Nie Y, et al. Construction of synthetic microbiota for reproducible flavor compound metabolism in Chinese light-aroma-type liquor produced by solid-state fermentation. Appl Environ Microb. 2019;85(10):e03090–18.
- Song Z, Du H, Zhang Y, et al. Unraveling core functional microbiota in traditional solid-state fermentation by high-throughput amplicons and metatranscriptomics sequencing. Front Microbiol. 2017;8:1–14.
- Tang J, Chen S, Lin B, et al. Effects of microorganisms and environmental factors on changes in aroma composition in Fen-flavor Xiaoqu Baijiu. Food Ferment Ind. 2019;45(17):40–47.
- Hu Y, Yang Q, Chen D, et al. Study on microbial communities and higher alcohol formations in the fermentation of Chinese Xiaoqu Baijiu produced by traditional and new mechanical technologies. Food Res Int. 2021;140:109876.
- Penghui L, Lihong Z, Xiaowei D, et al. Dynamic analysis of physicochemical and biochemical indices and microbial communities of light-flavor daqu during storage. J Am Soc Brew Chem. 2019;77(4):287–294.
- Zheng N, Jiang S, He Y, et al. Production of low-alcohol Huangjiu with improved acidity and reduced levels of higher alcohols by fermentation with scarless ALD6 overexpression yeast. Food Chem. 2020;321:126691.
- Li M, Zhu L, Lin D. Toxicity of ZnO nanoparticles to Escherichia coli: mechanism and the influence of medium components. Environ Sci Technol. 2011;45(5):1977–1983.
- Abildgren M, Lund F, Thrane U, et al. Czapek‐Dox agar containing iprodione and dicloran as a selective medium for the isolation of Fusarium species. Lett Appl Microbiol. 1987;5(4):83–86.
- Martins D, English AM. Catalase activity is stimulated by H(2)O(2) in rich culture medium and is required for H(2)O(2) resistance and adaptation in yeast. Redox Biol. 2014;2:308–313.
- Daniel M, Cj C, Justin K, et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. GigaScience. 2012;1(1):2047–217X.
- Li P, Lin W, Liu X, et al. Environmental factors affecting microbiota dynamics during traditional solid-state fermentation of Chinese Daqu starter. Front Microbiol. 2016;7:1237.
- Wang H, Gao Y, Fan Q, et al. Characterization and comparison of microbial community of different typical Chinese liquor Daqus by PCR-DGGE. Lett Appl Microbiol. 2011;53(2):134–140.
- Mou R, Mao J, Meng X, et al. Analysis of fungi diversity and volatile flavor compounds in Chinese rice wine fermentation process. Food Sci Biotechnol. 2016;35(3):303–309.
- Liu P, Xiong X, Wang S, et al. Population dynamics and metabolite analysis of yeasts involved in a Chinese miscellaneous-flavor liquor fermentation. Ann Microbiol. 2017;67(8):553–565.
- Huang Y, Yi Z, Jin Y, et al. New microbial resource: microbial diversity, function and dynamics in Chinese liquor starter. Sci Rep. 2017;7(1):1–14.
- Chen L, Li Y, Jin L, et al. Analyzing bacterial community in pit mud of Yibin Baijiu in China using high throughput sequencing. PeerJ. 2020;8:e9122.
- Hu Y, Dun Y, Li S, et al. Changes in microbial community during fermentation of high-temperature Daqu used in the production of Chinese 'Baiyunbian' liquor. J Inst Brew. 2017;123(4):594–599.
- Liang C, Du H, Xu Y. The succession of procaryotic microbial community and the flavor components in the storage process of Daqu. Microbiol China. 2017;44(2):384–393.
- Wang M, Yang J, Zhao Q, et al. Research progress on flavor compounds and microorganisms of Maotai flavor Baijiu. J Food Sci. 2019;84(1):6–18.
- Wang C, Shi D, Gong G. Microorganisms in Daqu: a starter culture of Chinese Maotai-flavor liquor. World J Microbiol Biotechnol. 2008;24(10):2183–2190.
- Zhang Q, Yuan Y, Luo W, et al. Characterization of prokaryotic community diversity in new and aged pit muds from Chinese Luzhou-flavor liquor distillery. FSTR. 2017;23(2):213–220.
- Wang J, Zhong Q, Yang Y, et al. Comparison of bacterial diversity between two traditional starters and the round-koji-maker starter for traditional Cantonese chi-flavor liquor brewing. Front Microbiol. 2018;9:1053.
- Lin J, Wu Q, Xu Y. Dynamic profile of yeast community associated with urea metabolism in Chinese light-aroma liquor fermentation. Microbiol China. 2017;44(11):2522–2529.
- Duan G, Liu Y, Lv H, et al. Optimization of “Zaoheibao” wine fermentation process and analysis of aroma substances. Biotechnol Biotec Eq. 2020;34(1):1056–1064.
- Wu Q, Kong Y, Xu Y. Flavor profile of Chinese liquor is altered by interactions of intrinsic and extrinsic microbes. Appl Environ Microbiol. 2016;82(2):422–430.
- Ma R, Sui L, Zhang J, et al. Polyphasic characterization of yeasts and lactic acid bacteria metabolic contribution in semi-solid fermentation of Chinese Baijiu (traditional fermented alcoholic drink): towards the design of a tailored starter culture. Microorganisms. 2019;7(5):147.
- Liu J, Chen J, Fan Y, et al. Biochemical characterisation and dominance of different hydrolases in different types of Daqu - a Chinese industrial fermentation starter. J Sci Food Agric. 2018;98(1):113–121.
- Yin X, Yoshizaki Y, Ikenaga M, et al. Manufactural impact of the solid-state saccharification process in rice-flavor Baijiu production. J Biosci Bioeng. 2020;129(3):315–321.
- Pang X, Han B, Huang X, et al. Effect of the environment microbiota on the flavour of light-flavour Baijiu during spontaneous fermentation. Sci Rep. 2018;8(1):3396.
- Fang C, Du H, Zheng X, et al. Solid-state fermented Chinese alcoholic beverage (baijiu) and ethanol resulted in distinct metabolic and microbiome responses. Faseb J. 2019;33(6):7274–7288.
- Wu X, Li Z, Zhou S. Interaction of Saccharomyces cerevisiae and Acetobacter pasteurianus in liquor fermentation. Mod Food Sci Technol. 2017;33(12):61–67
- Ye M, Yue T, Yuan Y. Effects of sequential mixed cultures of Wickerhamomyces anomalus and Saccharomyces cerevisiae on apple cider fermentation. FEMS Yeast Res. 2014;14(6):873–882.
- Liu M, Tang Y, Zhang X, et al. Structural and functional changes in prokaryotic communities in artificial pit mud during Chinese Baijiu production. mSystems. 2020;5(2):e00829–19.