Abstract
Sanghuangporus baumii, a macrofungus belonging to the Hymenochaetaceae family, has been widely used as a traditional medicine in Asia for centuries. Triterpenoids in S. baumii contribute to the pharmacological activities, however, their content is low in the fruiting body. Phosphomevalonate kinase (PMK) is an important rate-limiting enzyme in the triterpenoids synthesis pathway. In this study, a PMK gene (SbPMK) was cloned and analyzed. Sequence analysis showed that SbPMK has six exons and five introns. The 1,488 bp open reading frame encodes a 489 amino acid polypeptide with a predicted protein molecular weight of 52.47 kDa. The amino acid sequence of SbPMK shares highly similarity with that of PMK sequences from other species, especially in three functional motifs (MI, MII and MIII). The promoter of SbPMK contains typical cis-acting regulatory elements involved in MeJA responsiveness, light responsiveness, anaerobic induction, etc. Furthermore, recombinant SbPMK was overexpressed in Escherichia coli BL21, with a satisfactory solubility. During the fermentation culture of S. baumii, the biomass increased with fermentation and peaked on day 18 (4.80 g/L). The SbPMK expression and the triterpenoids content was peaked on day 6, and the triterpenoids content stabilized at 20.8 mg/g on day 18.
Supplemental data for this article is available online at https://doi.org/10.1080/13102818.2021.1938678 .
Introduction
Sanghuangporus baumii is a macrofungus that belongs to the Hymenochaetaceae family [Citation1, Citation2]. It has been used as a traditional medicine for treating various diseases in China, Japan and some other Asian countries for centuries [Citation3]. Triterpenoids are among the main bioactive components in S. baumii [Citation4–7], and they are responsible for diverse pharmacological activities, including anti-tumour, anti-oxidative and anti-inflammatory properties [Citation8–11]. Despite of these important pharmacological activities, the medical application of S. baumii triterpenoids is limited by the low yield of triterpenoids. Metabolic engineering is an effective way to increase the yield of triterpenoids in S. baumii. However, the lack of work on the biosynthetic pathway of S. baumii triterpenoids makes metabolic engineering difficult.
In eukaryotes, triterpenoids are synthesised via the mevalonate (MVA) pathway (), in which mevalonate is converted through a series of chemical reactions to isopentenyl pyrophosphate, the building block of terpenoids [Citation12–14]. This process requires the involvement of several enzymes, including phosphomevalonate kinase (PMK), an important rate-limiting enzyme. PMK catalyzes a reversible ATP-dependent phosphorylation, converting mevalonate 5-phosphate to mevalonate 5-diphosphate (). As a representative kinase in the MVA pathway, the structure and function of PMK has been widely studied in human [Citation15–17], bacteria [Citation18], yeast [Citation19] and plants [Citation20]. Studies on the phosphorylation mechanism of Streptococcus pneumonia PMK indicate that PMK anchors ATP through a conserved serine residue (Ser106), and another serine residue (Ser213) located on the α-helix is responsible for facilitating the proton transfer [Citation21]. In addition to the functional study of PMK, metabolic engineers found that the expression level of PMK was relatively low through the proteomics data, which may be a potential bottleneck in the MVA pathway. Hence, they significantly increased the expression levels of PMK and another protein, mevalonate kinase (MK), by optimizing the genes codon and using a stronger promoter, and increased the final diene titer by more than three times [Citation22]. These studies suggest that PMK is the key to improve MVA pathway products through metabolic engineering. However, the literature contains very few studies on S. baumii PMK [Citation23].
Figure 1. Sequence analysis of SbPMK and its promoter region. (A) Exon and intron sites of SbPMK. (B) Phylogenetic analysis of PMK from S. baumii and other species. (C) Functional elements of the SbPMK promoter.
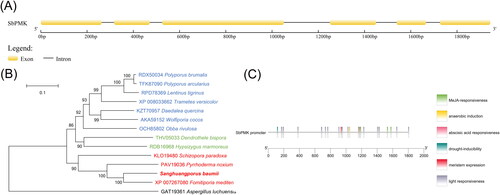
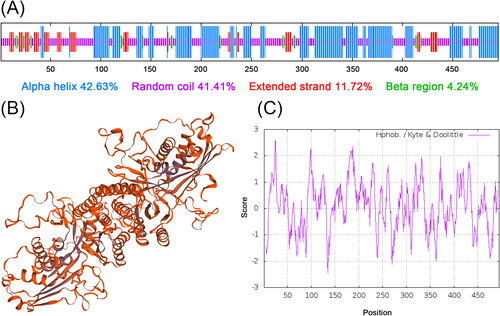
Figure 2. Analysis of the secondary structure and predicted three-dimensional structure of SbPMK. (A) Annotation of the secondary structure of SbPMK. (B) Predicted three-dimensional structure of SbPMK. (C) Hydrophobicity analysis of SbPMK.
In this study, in order to explore the function of PMK in S. baumii, we cloned and analysed the structure of the SbPMK cDNA, DNA and promoter for the first time, and successfully expressed SbPMK in Escherichia coli. These studies will provide the basis for codon-optimization of the PMK gene and cognate promoter for metabolic engineering. Fermentation is an efficient method for triterpenoids production [Citation24]. However, it remains unknown how the expression level of PMK affects the accumulation of S. baumii triterpenoids. Therefore, triterpenoids content and SbPMK transcript levels during fermentation of S. baumii were also measured.
Materials and methods
Strains and culture conditions
S. baumii strain was collected from Syringa reticulata at Yichun city, Heilongjiang Province, China, and identified based on internal transcribed spacer (ITS) sequence alignment in the National Center for Biotechnology Information (NCBI) database (GenBank accession number KP974834). Mycelia were cultured on potato-dextrose agar (PDA) plates to rejuvenate, then transferred to potato dextrose broth (PDB) medium and cultured in a constant temperature shaking incubator at 25 °C and 160 rpm to harvest mycelia. Escherichia coli DH5α and BL21 (DE3), both from TIANGEN (Beijing, China), were cultured on Luria-Bertani (LB) medium at 37 °C.
Nucleic acids isolation and cDNA preparation
Genomic DNA was extracted from the S. baumii mycelia using the cetyl trimethylammonium bromide (CTAB) method [Citation25]. Total RNAs were extracted using TRIzol reagent (TaKaRa, Dalian, China) according to the manufacturer’s instructions, then treated with RNase-free DNase I (TaKaRa). Subsequently, a PrimeScript II 1st Strand cDNA Synthesis Kit (TaKaRa) was used to synthesise first-strand complementary DNA (cDNA).
Cloning and bioinformatics analysis
According to the annotation in the transcriptome of S. baumii mycelia, 1 putative phosphomevalonate kinase gene was obtained. Combining the genomic data of S. baumii present in the NCBI database (GenBank accession number: LNZH00000000), primers PMK-F and PMK-R () were designed using Primer Premier 5.0 software (PREMIER Biosoft Co., Palo Alto, CA, USA). Using cDNA and Genomic DNA as a template, polymerase chain reaction (PCR) was performed to obtain the complete full-length cDNA and DNA sequence respectively. Specific primers were also designed to amplify the homologous promoter of SbPMK (promoter-F, promoter-R, ). PCR products were purified and sequenced.
Table 1. List of primers used in the study.
The open reading frame (ORF) was predicted with ORF Finder on the NCBI website (http://www.ncbi.nlm.nih.gov/orffinder/). The theoretic molecular weight, isoelectric point and absorption coefficient were predicted using ExPASy Proteomics Server (http://www.expasy.ch/tools/protparam.html). The protein domain structure was analysed using ExPASy PROSITE (http://www.expasy.ch/prosite/). Protein secondary structure was analysed with PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/). Transmembrane regions were predicted with TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). The 3 D structure was predicted using SWISS-MODEL online homologous modelling software (http://swissmodel.expasy.org). The promoter was analyzed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Phylogenetic analysis
Sequence similarity searches were performed using GenBank and the basic local alignment search tool (BLAST) algorithm (http://www.ncbi.nlm.nih.gov/blast) on the NCBI website, with the SbPMK sequence as a query. Homologous sequences were downloaded and aligned, and a neighbour-joining tree was constructed with MEGA 5.0 software [Citation3]. The sequence was aligned using DNAMAN (Version 7) (Lynnon Corporation, Quebec, Canada).
Heterologous expression of SbPMK in E. coli
The entire SbPMK ORF was amplified using primers PMK-B and PMK-X harbouring BamHI and XhoI restriction sites, respectively (). Afterwards, the target fragment was incorporated into the pET-32a vector (TaKaRa), and transferred into E. coli DH5α competent cells for plasmid propagation. After propagation, recombinant plasmid pET-32a-SbPMK was transferred into E. coli BL21, and positive clones were screened by PCR using the vector primers T7 and T7t (). Individual E. coli BL21 colonies containing pET-32a-SbPMK were transferred into 5 mL LB medium and cultured to an optical density at 600 nm (OD 600) of 0.5–0.8, at which point isopropyl b-d-1-thioga-lactopyranoside (IPTG) was added to a final concentration of 1 mmol/L, and culturing was continued for 10 h. Samples (1 mL) were harvested every two hours during induction, centrifuged at 12,000 g for 10 min, and cells were re-suspended in 100 μL of 4× protein buffer (Solarbio, Beijing, China). Cells were subsequently disrupted using a boiling water bath for 10 min, centrifuged at 12,000 g for 5 min, and 7.5 μL of supernatant was analysed by 12% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE).
Determination of biomass and triterpenoids
The strain was grown at 25 °C for 7 days in liquid PDB medium with shaking at 160 rpm. Fermentation was performed in PDB medium after inoculation with 5% (v/v) of the liquid culture. Mycelia were harvested at 3, 6, 9, 12, 15 and 18 days, followed by 45 °C of drying to a constant dry weight to obtain the biomass of mycelia. The dried mycelia were pulverized and extracted by 70% (v/v) ethanol (solid-liquid ratio 1:20) for 40 min at 60 °C. After removal of the mycelia by centrifugation, the supernatants were dried at 70 °C under vacuum conditions. Residues were added 4 mL 5% vanillin-acetic acid and 1 mL perchloric acid, incubated at 60 °C for 20 min, and cooled rapidly. Subsequently, 5 mL glacial acetic acid was added and incubated at room temperature for 15 min. The absorbance was detected at 546 nm in a spectrophotometer (Laspec, Shanghai, China). The content of triterpenoids was calculated according to a standard curve prepared by using ursolic acid [Citation24].
SbPMK expression during fermentation
Fermentation was performed in 500 mL flasks containing 250 mL of PDB after inoculation with 5% (v/v) of the liquid culture. Mycelia were harvested at 3, 6, 9, 12, 15 and 18 days, and frozen at −80 °C until RNA extraction. Total RNA was extracted using TRIzol reagent, then reverse-transcribed to cDNA using a PrimeScript TM RT reagent Kit (TaKaRa) according to the manufacturer’s instructions. Primers PMK-qF and PMK-qR were used to evaluate levels of the SbPMK transcript, and α-tubulin was used as a reference gene (). Next, real-time quantitative reverse transcription–PCR was performed using an Mx3000P Sequence Detection System (Agilent Technologies, Santa Clara, CA, USA). Mycelia harvested at 0 days served as a reference, and relative transcript levels were calculated according to the 2-ΔΔct method.
Data analysis
Quantitative data are presented as mean values from three independent experiments with standard deviation (±SD). Statistical analysis was performed using Tukey tests with STATGRAPHICS PLUS software (STATPOINT TECHNOLOGIES, INC., Virginia). Values were considered statistically significantly different at the p < 0.05 level.
Results
Gene sequence analysis
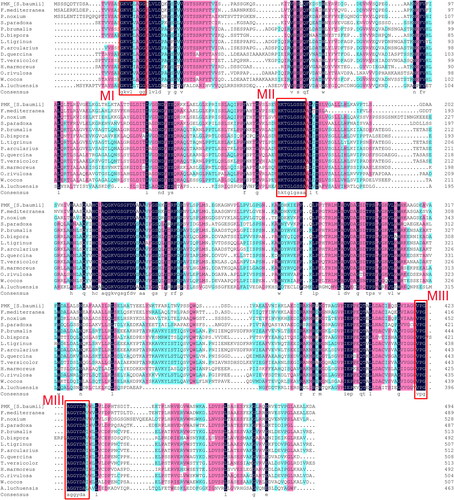
The gene fragment was cloned and sequenced. Protein BLAST search demonstrated that it encodes phosphomevalonate kinase, and belongs to the Isoprenoid_Biosyn_C1 Superfamily. Thus, it was designated as SbPMK. Comparison of the cDNA and DNA sequence revealed that the SbPMK gene contains six exons (nucleotides 1–263, 318–471, 526–1051, 1251–1450, 1540–1666, 1727–1944) and five introns (264–317, 472–525, 1052–1250, 1451–1539, 1667–1726; ). The 1,488 bp ORF of SbPMK encodes a 495 amino acid polypeptide (Supplemental Figure S1) and shares 76.55% percent identity with the phosphomevalonate kinase of Fomitiporia mediterranea (Sequence ID: XP_007267080.1). The SbPMK sequence was deposited in GenBank under accession number MT040050.
Phylogenetic tree () was constructed based on PMK sequences of 14 species () of which 13 species belonged to Basidiomycota, and one species (Aspergillus luchuensis) belonged to Ascomycota. The basidiomycetes of phylogenetic trees are divided into three branches, Agaricales, Polyporales and Hymenochaetales, corresponding to the blue, dark green and red parts in .
Table 2. Origins of homologous amino-acid sequences from other species.
Promoter analysis
The 1,806 bp SbPMK promoter contains typical eukaryotic promoter elements, including 20 CAAT-boxes (common cis-acting element in promoter and enhancer regions), 5 TATA boxes (with a core promoter element 30-bp upstream of the transcription start site), 8 G-boxes (cis-acting regulatory element involved in light responsiveness), 7 ABRE (cis-acting element involved in the abscisic acid responsiveness), 10 CGTCA-motif (cis-acting regulatory element involved in the MeJA-responsiveness), 1 ARE (cis-acting regulatory element essential for the anaerobic induction), 1 MBS (MYB binding site involved in drought-inducibility), 1 CAT-box (cis-acting regulatory element related to meristem expression) ().
Protein sequence analysis
The predicted molecular weight of the SbPMK protein is 52.47 kDa for the molecular formula C2352H3740N626O714S8. The theoretical isoelectric point of the protein is 5.77, with 52 negatively charged residues (Asp + Glu) and 43 positively charged residues (Arg + Lys). The instability index of the protein is 45.10, classifying the protein as stable. The SbPMK does not contain transmembrane region (Supplemental Figure S1) and signal peptides (Supplemental Figure S2), indicating that it is synthesized on free ribosomes and retained in the cytoplasmic matrix for catalysis, which is consistent with the current state of knowledge [Citation26].
Secondary structure analysis predicted that the SbPMK contains 42.63% alpha helix, 41.41% random coil, 11.72% extended strand and 4.24% beta region (). The predicted three-dimensional structure of SbPMK is shown in . The grand average of hydropathicity (GRAVY) of SbPMK is 0.072 (), without obvious hydrophobic and hydrophilic regions. Comparison of PMK amino acid sequences from 14 species () demonstrated that the sequences share high similarity, especially in the three functional motifs originally used to define the GHMP kinase superfamily (MI, MII and MIII, ). Of these, the most conserved MII has a Pro-X-X-Gly-Leu-X-Ser-Ser-Ala sequence that is folded into a phosphate-binding loop or P-loop for ATP binding [Citation21].
Heterologous expression of SbPMK in E. coli
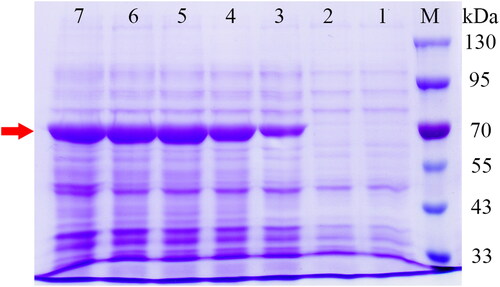
E. coli BL21 was selected as a host to stably overexpress SbPMK. A clear protein band of the expected molecular weight (52.47 kDa + tag protein 22.68 kDa) was observed on the 12% SDS-PAGE gel (), while no target protein was found in the control cells carrying empty plasmids. The results showed that SbPMK was successfully overexpressed in E. coli cells and the recombinant protein had satisfactory solubility.
Figure 4. Heterologous expression of SbPMK in E. coli following IPTG induction (1 mmol/L). The bands indicated by the red arrow are target proteins. Lane M, protein molecular weight marker (Solarbio, Beijing, China); Lane 1, blank control group: E. coli harbouring empty vector; Lane 2–7, E. coli harbouring pET-32a-SbPMK induced by IPTG at 0 h, 2 h, 4 h, 6 h, 8 h, 10 h.
Biomass during fermentation
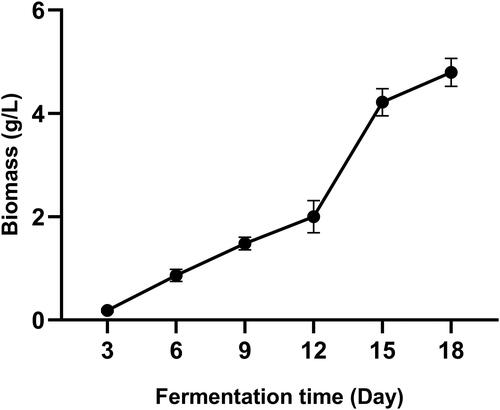
The graph in displays the kinetics of mycelial growth during fermentation using PDB medium. The biomass increased slowly in the first 12 days, during which time the mycelia remained in a lag phase. After day 12, the mycelia entered a period of rapid growth, and the biomass doubled from day 12 to day 15. After day 15, the biomass kept increasing and reached the highest level of 4.80 g/L dry weight (DW) on day 18.
SbPMK expression and triterpenoids content during fermentation
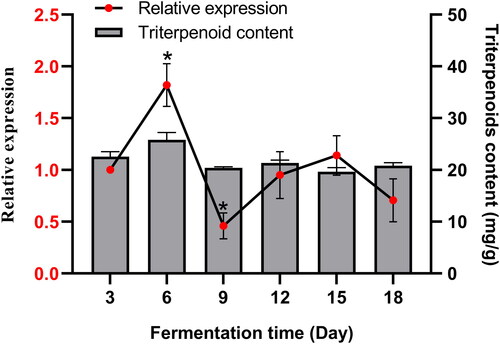
To determine the expression pattern of SbPMK during S. baumii fermentation, qRT-PCR was performed. Mycelia were harvested at different fermentation stages (3, 6, 9, 12, 15 and 18 days) for differential expression analysis. The α-tubulin gene served as a reference. The graph in displays the expression pattern of SbPMK during S. baumii fermentation. The SbPMK gene peaked on day 6 (1.82-fold higher than that on day 3), and decreased to the lowest expression on day 9 (0.46-fold). Bars in display triterpenoids content in unit mycelia during fermentation. The highest triterpenoids content was 27.48 mg/g (DW, dry weight) on day 6, and the lowest reached 19.63 mg/g (DW) on day 15.
Discussion
Triterpenes are a class of secondary metabolites with medicinal value, and their biosynthetic pathways have important clinical, pharmaceutical and biotechnology significance. Phosphomevalonate kinase is a key kinase in triterpenoids biosynthesis pathway. In this study, a PMK gene was cloned and analyzed from medicinal fungi S. baumii. Bioinformatics analysis identified SbPMK as a phosphomevalonate kinase that encoding a 495 amino acid polypeptide. Phylogenetic analysis revealed high similarity between SbPMK and the PMK sequences from other species, especially species belonging to Hymenochaetales. SbPMK share the closest relative with the PMK from the highly similar species Fomitiporia mediterranea, and the furthest relative with the PMK in A. luchuensis.
In addition to the common cis-acting elements, the homologous promoter of SbPMK also contains cis-acting regulatory elements involved in MeJA-responsiveness, etc. A large number of stress and hormone-related cis-elements were also observed in the promoter region of GHMP (Galactokinase, Homoserine kinase, Mevalonate kinase and Phosphomevalonate kinase) superfamily in wheat [Citation27] and Arabidopsis [Citation28]. These regulatory elements control the expression of genes in the MVA pathway in response to various hormones and stressors. In our previous studies, similar cis-regulatory elements were also found in the homologous promoters of the key enzyme genes of S. baumii’s MVA pathway, acetyl-CoA C-acetyltransferase synthase gene [Citation29] and farnesyl pyrophosphate synthase gene [Citation30]. Moreover, under the stimulation of MeJA, the content of triterpenoids in the mycelia of S. baumii increased by 4 times [Citation31], which may be related to the MeJA-responsiveness elements in the promoters. Similarly, Ren et al. also reported that MeJA could induce the biosynthesis of ganoderic acid, a type of triterpenoid, through the transcriptional induction of genes in Ganoderma lucidum, which is consistent with our study [Citation32].
The MVA pathway is an important conduit for the production of a wide array of bioactive substances. Heterologous expression of the MVA pathway in E. coli or yeast is used commercially to produce drugs, chemicals and fuels, including terpenoids [Citation33–35]. The recombinant protein obtained in this experiment can be used to explore the kinetics of SbPMK, which is critical to optimize the pathway for high flux.
Submerged fermentation is an efficient method to produce biomass and bioactive metabolites. The medicinal mushrooms mycelia possess a number of medicinal properties such as anti-tumour, immunomodulatory, antigenotoxic, anti-inflammatory [Citation36]. A number of triterpenoids were extracted and identified from the mycelia of S. sanghuang, a similar species of S. baumii [Citation37]. Therefore, it is necessary to study the growth kinetics of mycelia in the fermentation process of S. baumii. During the submerged fermentation of S. baumii, the triterpenoids content fluctuated around 20 mg/g, indicating that the triterpenoids did not accumulate in the fermentation process. The transcription level of SbPMK also decreased after the peak on day 6 and remained at a lower level throughout the fermentation period. The similar trends of triterpenoids content and gene expression levels indicated that the low triterpenoids content was related to the low expression of SbPMK. In metabolic engineering, the low expression of PMK is considered to be a bottleneck in the pathway, limiting terpene production. Codon-optimization of the PMK gene and expression from a stronger promoter can significantly increase the kinase levels and corresponding terpene production [Citation22].
Conclusions
In this study, a phosphomevalonate kinase gene and promoter were cloned and analyzed from medicinal fungi S. baumii. The SbPMK ORF encodes a 489 amino acid polypeptide and shares highly conserved functional domains with its orthologs in other species. The promoter of SbPMK contains cis-acting regulatory elements involved in MeJA-responsiveness, abscisic acid responsiveness, light responsiveness and anaerobic induction. These regulatory elements control the expression of genes in response to various hormonal and environmental stimuli. The SbPMK was synthesized in E. coli, which is helpful for studying the enzyme kinetics. During the fermentation of the mycelia of S. baumii, the expression of SbPMK and the content of triterpenoids were at a low level. It will be the next step to improve triterpenoids content through overexpression of SbPMK in S. baumii.
Data availability
All data that support the findings reported in this study are available from the corresponding author upon reasonable request.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental data
Supplemental data can be accessed at:
Additional information
Funding
References
- Han JG, Hyun MW, Kim CS, et al. Species identity of Phellinus linteus (sanghuang) extensively used as a medicinal mushroom in Korea. J Microbiol. 2016;54(4):290–295.
- Han JG, Oh J, Jo JW, et al. The complete mitochondrial genome of Sanghuangporus sanghuang (Hymenochaetaceae, Basidiomycota). Mitochondrial DNA B Resour. 2018;3(1):456–457.
- Wu SH, Dai YC, Hattori T, et al. Species clarification for the medicinally valuable ‘sanghuang’ mushroom. Bot Stud. 2012;53(1):135–149.
- Ge Q, Mao JW, Zhang AQ, et al. Purification, chemical characterization, and antioxidant activity of a polysaccharide from the fruiting bodies of sanghuang mushroom (Phellinus baumii Pilat). Food Sci Biotechnol. 2013;22(2):301–307.
- Lin WC, Deng JS, Huang SS, et al. Anti-Inflammatory Activity of Sanghuangporus sanghuang Mycelium. Int J Mol Sci. 2017;18(2):347.
- Lin WC, Deng JS, Huang SS, et al. Evaluation of antioxidant, anti-inflammatory and anti-proliferative activities of ethanol extracts from different varieties of Sanghuang species. RSC Adv. 2017;7(13):7780–7788.
- Liu K, Xiao X, Wang JL, et al. Polyphenolic composition and antioxidant, antiproliferative, and antimicrobial activities of mushroom Inonotus sanghuang. Lwt-Food Sci Technol. 2017;82:154–161.
- Liu C, Zhao C, Pan HH, et al. Chemical constituents from Inonotus obliquus and their biological activities. J Nat Prod. 2014;77(1):35–41.
- Kim YJ, Park J, Min BS, et al. Chemical constituents from the sclerotia of Inonotus obliquus. J Korean Soc Appl Bi. 2011;54(2):287–294.
- Handa N, Yamada T, Tanaka R. An unusual lanostane-type triterpenoid, spiroinonotsuoxodiol, and other triterpenoids from Inonotus obliquus. Phytochemistry. 2010;71(14-15):1774–1779.
- Muszyńska B, Grzywacz-Kisielewska A, Kała K, et al. Anti-inflammatory properties of edible mushrooms: A review. Food Chem. 2018;243:373–381.
- Landolfo S, Zara G, Zara S, et al. Oleic acid and ergosterol supplementation mitigates oxidative stress in wine strains of Saccharomyces cerevisiae. Int J Food Microbiol. 2010;141(3):229–235.
- Ajikumar PK, Xiao WH, Tyo KEJ, et al. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science. 2010;330(6000):70–74.
- Muntendam R, Melillo E, Ryden A, et al. Perspectives and limits of engineering the isoprenoid metabolism in heterologous hosts. Appl Microbiol Biotechnol. 2009;84(6):1003–1019.
- Herdendorf TJ, Miziorko HM. Phosphomevalonate kinase: Functional investigation of the recombinant human enzyme. Biochemistry. 2006;45(10):3235–3242.
- Herdendorf TJ, Miziorko HM. Functional evaluation of conserved basic residues in human phosphomevalonate kinase. Biochemistry. 2007;46(42):11780–11788.
- Hogenboom S, Tuyp JJM, Espeel M, et al. Phosphomevalonate kinase is a cytosolic protein in humans. J Lipid Res. 2004;45(4):697–705.
- McClory J, Hui CG, Zhang J, et al. The phosphorylation mechanism of mevalonate diphosphate decarboxylase: a QM/MM study. Org Biomol Chem. 2020;18(3):518–529.
- Garcia DE, Keasling JD. Kinetics of phosphomevalonate kinase from Saccharomyces cerevisiae. PLoS One. 2014;9(1):e87112.
- Song QL, Meng XX, Liao YL, et al. Transcriptome-guided gene isolation, characterization and expression analysis of a phosphomevalonate kinase gene (GbPMK) from Ginkgo biloba. Int J Agric Biol. 2018;20(5):1080–1088.
- Huang ML, Wei KX, Li X, et al. Phosphorylation mechanism of phosphomevalonate kinase: Implications for rational engineering of isoprenoid biosynthetic pathway enzymes. J Phys Chem B. 2016;120(41):10714–10722.
- Redding-Johanson AM, Batth TS, Chan R, et al. Targeted proteomics for metabolic pathway optimization: Application to terpene production. Metab Eng. 2011;13(2):194–203.
- Shao Y, Guo HW, Zhang JP, et al. The genome of the medicinal macrofungus Sanghuang provides insights into the synthesis of diverse secondary metabolites. Front Microbiol. 2020;10: 3035.
- Wu MD, Cheng MJ, Chen YL, et al. Secondary metabolites from the fermented whole broth of fungal strain Sanghuangporus sanghuang. Chem Nat Compd. 2019;55(1):36–40.
- Allen GC, Flores-Vergara MA, Krasnyanski S, et al. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 2006;1(5):2320–2325.
- Herdendorf TJ, Miziorko HM. Characterization & functional investigation of human phosphomevalonate kinase. Faseb J. 2006;20(4):A474–A474.
- N, Thakur Flowerika, Singh, PK, Kaur, K, et al. Genome-wide identification and analysis of GHMP kinase gene superfamily in bread wheat (Triticum aestivum L.). Plant Mol Biol Rep. 2020;39:455–470.
- Xiao WJ, Chang HP, Zhou P, et al. Genome-wide identification, classification and expression analysis of GHMP genes family in Arabidopsis thaliana. Plant Syst Evol. 2015;301(8):2125–2140.
- Wang X, Wang S, Xu X, et al. Molecular cloning, characterization, and heterologous expression of an acetyl-CoA acetyl transferase gene from Sanghuangporus baumii. Protein Expr Purif. 2020;170:105592.
- Wang XT, Sun TT, Sun J, et al. Molecular cloning, characterisation, and heterologous expression of farnesyl diphosphate synthase from Sanghuangporus baumii. Mol Biotechnol. 2020;62(2):132–141.
- Sun TT, Zou L, Zhang LF, et al. Methyl jasmonate induces triterpenoid biosynthesis in Inonotus baumii. Biotechnol Biotec Eq. 2017;31(2):312–317.
- Ren A, Qin L, Shi L, et al. Methyl jasmonate induces ganoderic acid biosynthesis in the basidiomycetous fungus Ganoderma lucidum. Bioresour Technol. 2010;101(17):6785–6790.
- Martin VJJ, Pitera DJ, Withers ST, et al. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21(7):796–802.
- Ro DK, Paradise EM, Ouellet M, et al. Keasling, Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943.
- Chang MCY, Eachus RA, Trieu W, et al. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol. 2007;3(5):274–277.
- Ulziijargal E, Mau JL. Nutrient compositions of culinary-medicinal mushroom fruiting bodies and mycelia. Int J Med Mushr. 2011;13(4):343–349.
- Cai CS, Ma JX, Han CR, et al. Extraction and antioxidant activity of total triterpenoids in the mycelium of a medicinal fungus, Sanghuangporus sanghuang. Sci Rep. 2019;9(1)7418.