Abstract
To identify mechanisms of starch degradation in rice leaf sheaths at the post-heading stage, we investigated the function of OsBAM2 and OsBAM3, which encode plastid-targeted active β-amylase isoforms, in starch remobilization in leaf sheaths. The starch content in the second leaf sheaths below the flag leaf (the third leaf sheaths) peaked at the flag leaf emergence stage and gradually decreased until 15 days after heading. The mRNA levels of OsBAM2 and OsBAM3 in the third leaf sheaths increased from the flag leaf emergence stage to the heading stage when the starch content began to decrease. However, these mRNA levels did not always remain high during post-heading. Overexpression of OsBAM2 or OsBAM3 markedly repressed starch accumulation in the third leaf sheaths, showing that OsBAM2 and OsBAM3 function in starch degradation in rice leaf sheaths. In contrast, no significant differences in starch content in the third leaf sheaths were detected between knockdown plants of OsBAM2 or OsBAM3 and non-transgenic wild-type plants. Our results suggest that reduced expression of the individual genes, OsBAM2 or OsBAM3, does not result in excess accumulation of starch in the leaf sheaths, probably because of the complementary function of another gene or the action of other genes encoding starch-degrading .
Abbreviation:
- AGPase: ADP-glucose pyrophosphorylase
- BSA: bovine serum albumin
- EDTA: ethylenediamine tetraacetate
- GBSS: granule-bound starch synthase
- GWD: glucan, water dikinase
- KD: knockdown
- OE: overexpression
- PCR: polymerase chain reaction
- PWD: phosphoglucan, water dikinase
- RT-PCR: reverse transcriptase polymerase chain reaction
- SBE: starch branching enzyme
- SEstandard error
- SSS: soluble starch synthase
- WT: non-transgenic wild-type
Rice (Oryza sativa L.) leaf sheaths serve as temporary storage organs for photoassimilates. Starch is accumulated at a high level in leaf sheaths prior to the heading stage and remobilized for grain filling after heading (Perez et al., Citation1971). Approximately 30% of the final grain yield is derived from carbohydrates accumulated transitorily in the leaf sheaths and culms (Cock & Yoshida, Citation1972). Thus, identifying the mechanisms of starch accumulation at the pre-heading stage and starch remobilization at the post-heading stage in leaf sheaths is important for improving rice grain yield.
Physiological and biochemical studies have been performed to investigate the correlation between starch accumulation and starch-synthesizing enzymes in rice leaf sheaths (He et al., Citation2005; Hirano et al., Citation2005; Hirose et al., Citation1999, Citation2006; Perez et al., Citation1971; Watanabe et al., Citation1997). The activity of granule-bound starch synthase (GBSS) is positively correlated with the starch content of leaf sheaths (Perez et al., Citation1971). Watanabe et al. (Citation1997) suggested that starch branching enzyme (SBE) is primarily involved in the regulation of starch accumulation in the leaf sheaths. The starch content of leaf sheaths at the heading stage was lower in rice plants grown under a high-nitrogen fertilizer regime than that under a low-nitrogen fertilizer regime, which was accompanied by a decrease in SBE activity (Hirano et al., Citation2005). Hirose et al. (Citation2006) have shown that starch accumulation in leaf sheaths before the heading stage was accompanied by a transient rise in ADP-glucose pyrophosphorylase (AGPase), soluble starch synthase, GBSS, and SBE activities. They analyzed in detail the transcript levels of genes encoding these enzymes to develop and propose a profile of starch synthesis in leaf sheaths for comparison with that in endosperms. In this profile, AGPL1, AGPS1, SSI, SSIIb, SSIIIb, GBSSII, and BEIIa were expressed predominantly in leaf sheaths during the pre-heading stage (Hirose et al., Citation2006).
In contrast, the details of the enzymes involved in starch remobilization in leaf sheaths are largely unknown. Only a few reports have addressed the correlations between changes in starch content and starch-degrading enzymes in leaf sheaths during the post-heading stage. After heading, the activity of α-amylase was consistent with the degree of starch degradation in leaf sheaths (Ishimaru et al., Citation2004). Chen and Wang (Citation2008) reported that α-amylase 2A (RAmy2A) and β-amylase (BAM) genes (Accession Number; AK068968, OsBAM3) were highly expressed in leaf sheaths at the post-heading stage. Recently, Sugimura et al. (Citation2015) proposed that RAmy2A genes play an important role in leaf sheath starch degradation at the post-heading stage. These findings seem to support the involvement of α-amylase in starch degradation in leaf sheaths during the post-heading stage.
A pathway of starch degradation in leaves has been well elucidated in Arabidopsis thaliana (L.) Heynh and potato (Zeeman et al., Citation2010). The starch degradation depends on the reversible phosphorylation of glucans at the surface of the starch granule (Zeeman et al., Citation2010). Two classes of enzymes, glucan, water dikinase (GWD) (Mikkelsen et al., Citation2004; Ritte et al., Citation2002, Citation2004; Yu et al., Citation2001) and phosphoglucan, water dikinase (PWD) (Baunsgaard et al., Citation2005; Kötting et al., Citation2005), are implicated in the glucan phosphorylation. Maltose is the major product of starch hydrolysis and is exported from chloroplasts at night (Chia et al., Citation2004; Fulton et al., Citation2008; Niittylä et al., Citation2004; Weise et al., Citation2004). The hydrolysis of α-1, 4-glucan chains is catalyzed primarily by β-amylases localized in chloroplasts (Fulton et al., Citation2008; Kaplan & Guy, Citation2005; Lao et al., Citation1999; Scheidig et al., Citation2002; Sparla et al., Citation2006). There are nine genes encoding β-amylase-like proteins in the A. thaliana genome. Total β-amylase activity was significantly reduced in the leaves of bam1 and bam3 mutants as compared with the wild-type (WT) plants (Fulton et al., Citation2008). The bam3 mutant accumulated excess starch in leaves and the bam1 bam3 double mutant had a more severe phenotype than the bam3 mutant (Fulton et al., Citation2008). Both BAM1 and BAM3 encode catalytically active, plastid-localized proteins (Kaplan & Guy, Citation2005; Sparla et al., Citation2006). These findings show that BAM3 is a β-amylase isoform that functions predominantly in starch degradation in A. thaliana leaves and that BAM1 also plays an important role in this process, at least in the absence of BAM3 (Fulton et al., Citation2008). In contrast, in rice, although two β-amylase isoforms, OsBAM2 and OsBAM3, have been reported to be plastid-targeted active enzymes (Hirano et al., Citation2011), it is still unclear whether OsBAM2 and OsBAM3 are actually involved in starch remobilization in rice leaves.
Our objectives in the present study were to elucidate the function of OsBAM2 and OsBAM3 in starch remobilization in rice leaf sheaths during the post-heading stage. We analyzed in detail the changes in OsBAM2 and OsBAM3 mRNA levels in leaf sheaths at the pre- and post-heading stages. In addition, we generated OsBAM2- and OsBAM3-overexpressing plants and OsBAM2- and OsBAM3-knockdown plants to evaluate the effect of the expression levels of OsBAM2 and OsBAM3 in starch remobilization in leaf sheaths, and investigated their phenotypes.
2. Materials and methods
2.1. Plant materials and sampling
For analyses of carbohydrate content, β-amylase activity and mRNA levels of OsBAM2 and OsBAM3, three rice seedlings (O. sativa L., cv. Nipponbare) per pot were transplanted to 1/5000 are Wagner pots (three plants per pot) containing 3.0-kg paddy soil on 5 June, 2014 and were grown outdoors at Meijo University. As a basal fertilizer, chemical fertilizer consisting of N (15%), P (15%), and K (10%) was applied at 2.0 g per pot. Nitrogen fertilizer in the form of ammonium sulfate (0.3-g N per pot) was also applied at the 10th leaf stage. The second leaf sheaths below the flag leaf (the third leaf sheaths) were harvested between 9:30 and 10:30 am at the second leaf emergence stage, the flag leaf emergence stage, the heading stage, and 5, 10, 15, 20, and 40 days after heading.
Overexpression and knockdown plants of OsBAM2 and OsBAM3 (T2 generation) were grown under a photoperiod of 13-h light (845 μmol photon m− 2 s−1)/11-h dark at 32/25 °C and approximately 60% relative humidity in a growth chamber. The third leaf sheaths were harvested between 9:30 and 10:30 am at the heading stage and 20 days after heading. All harvested samples were longitudinally divided into two equal parts, immediately frozen in liquid nitrogen, and stored at −80 °C.
2.2. Vector construction and transformation of rice
All primers used for vector construction are listed in Table . Full-length cDNAs of OsBAM2 and OsBAM3 were amplified from cDNA prepared from rice (cv. Nipponbare) leaf blades by polymerase chain reaction (PCR) using PrimeSTARTM HS DNA Polymerase (TaKaRa) and inserted immediately downstream of the maize Ubi-1 promoter of pAHC17 (Christensen et al., Citation1992). The resulting plasmids, pAHC17-BAM2, or pAHC17-BAM3, were digested with HindIII and ligated together with a SacI/HindIII adapter into the binary vector pBECKS2000.2 (McCormac et al., Citation1999) digested with SacI to generate the constructs for OsBAM2 and OsBAM3 overexpression.
Table 1. Gene-specific primers used in this study.
The inverted repeat (IR) regions triggering RNA-silencing of OsBAM2 and OsBAM3 were amplified by the method described above. The PCR products were cloned into the pENTR/D-TOPO cloning vector (Life Technologies) to generate entry vectors. The RNA-silencing vectors were produced by an LR Clonase reaction (Life Technologies) between each entry vector containing the IR region of OsBAM2 or OsBAM3 and the pANDA vector (Miki & Shimamoto, Citation2004; Miki et al., Citation2005).
Transgenic rice plants were generated by Agrobacterium tumefaciens-mediated transformation of rice (cv. Sasanishiki) calli derived from scutellum (Hiei et al., Citation1994). Plants (T0 generation) were regenerated from transformed calli selected with hygromycin B and transplanted to plastic pots containing nursery soil. The presence of the transgene was verified by PCR using genomic DNA extracted from the leaf blades of the resistant plants as the template.
2.3. Semiquantitative RT-PCR
Total RNA was purified from germinating seeds, leaf blades at the active tillering stage, leaf sheaths at the active tillering stage, leaf sheaths at 10–14 days after heading, and developing caryopses at 10–14 days after heading, following the procedures described by Hirano et al. (Citation2011). The purified total RNA (1 μg) was reverse-transcribed using SuperScriptTM III Reverse Transcriptase (Life Technologies) with 50 pmol of Oligo(dT)20 Primer (Life Technologies). After the reaction, the cDNA solution was treated with 2 U of Escherichia coli RNase H (Life Technologies) at 37 °C for 20 min. The mRNA levels of β-amylase genes were evaluated by semiquantitative RT-PCR using the gene-specific primers listed in Table . An aliquot of cDNA solution corresponding to 20 ng total RNA extracted from the third leaf sheaths was used as the template. The reaction was performed using PrimeSTARTM HS DNA polymerase (TaKaRa) under the following conditions: 24, 26, or 28 cycles of 10 s at 98 °C, 5 s at 55 °C, and 0.5 min at 72 °C. The amplified fragments were separated by electrophoresis on 1.5% (w/v) agarose gel.
2.4. Quantitative RT-PCR
Total RNA purification and cDNA synthesis were performed by the methods described above. The mRNA levels of OsBAM2 and OsBAM3 were evaluated by quantitative RT-PCR using the gene-specific primers listed in Table . An aliquot of cDNA solution corresponding to 20-ng total RNA extracted from the third leaf sheaths was used as the template. Quantitative RT-PCR was performed using a StepOnePlus Real-Time PCR System (Life Technologies) in a 20-μl reaction mixture containing Power SYBER Green PCR Master Mix (Life Technologies). A serial dilution series of plasmid DNA containing the partial cDNAs of the genes was prepared to construct a standard curve. The mRNA level estimated from the standard curve was standardized to the expression of OsEF1α1 (RAP-ID: Os03g0177400) (Hirano et al., Citation2011; Sugimura et al., Citation2015), which encodes the translation elongation factor 1A.
2.5. Determination of starch and sugar contents
Starch, sucrose, and maltose contents in the third leaf sheaths were determined by the method of Sugimura et al. (Citation2015).
2.6. Determination of β-amylase activity
The activity of β-amylase was determined by the method of Scheidig et al. (Citation2002) with modifications. Frozen leaf sheaths were extracted with 2 ml of ice-cold buffer containing 50 mM MOPS–KOH (pH 7.5), 20 mM MgCl2, 2 mM CaCl2, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.1% (w/v) bovine serum albumin (BSA), and 0.1% (v/v) β-mercaptoethanol and 2% (w/v) polyvinylpolypyrrolidone. The samples were centrifuged at 20,000× g for 10 min at 4 °C. For the determination of β-amylase activity, 50 μl of the supernatant was mixed with 200 μl of buffer containing 100 mM sodium acetate (pH 5.4), 2 mM EDTA, 0.1% (w/v) BSA, and 0.1% (v/v) β-mercaptoethanol. Assays were started by the addition of 50 μl of Betamyl-3® substrate solution (Megazyme) containing p-nitrophenyl-β-D-maltotrioside as a substrate and β-glucosidase as a coupling enzyme at 40 °C. After 15 min, the reaction was stopped by the addition of 1.5 ml of 1% (w/v) Trizma base. The absorbance of the reaction solutions was analyzed at 400 nm.
3. Results
3.1. Changes in carbohydrate content and β-amylase activity in the leaf sheaths
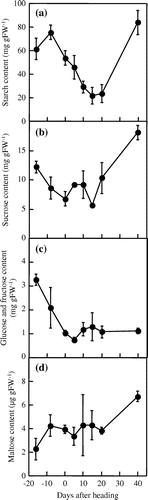
Starch content in the third leaf sheaths peaked at the flag leaf emergence stage and then gradually decreased until 15 days after heading (Figure (a)). Beginning at 20 days after heading, starch content rapidly increased until 40 days after heading. The sucrose contents in the third leaf sheaths gradually decreased from the second leaf emergence stage until the heading stage (Figure (b)). The sucrose contents rapidly increased from 15 days after heading until 40 days after heading. The glucose and fructose contents exhibited maximum levels at the second leaf emergence stage and gradually decreased until 5 days after heading (Figure (c)). The maltose content in the third leaf sheaths remained constant from the flag leaf emergence stage until 20 days after heading (Figure (d)).
Figure 1. Changes in carbohydrate contents in the third leaf sheaths. The third leaf sheaths were harvested at the second leaf emergence stage, the flag leaf emergence stage, the heading stage, and 5, 10, 15, 20, and 40 days after heading. (a) Starch, (b) sucrose, (c) glucose and fructose, and (d) maltose. Data represent means ± SE of three replications.
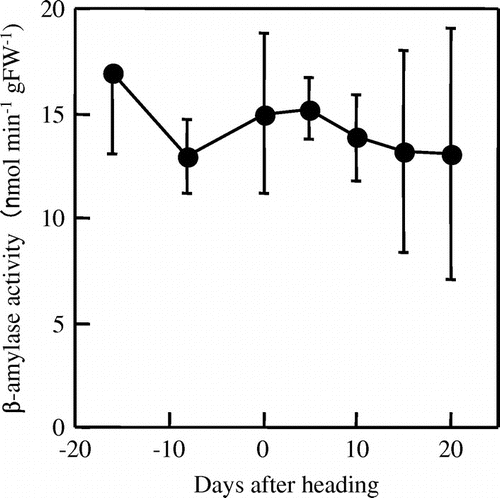
The β-amylase activity in the third leaf sheaths remained substantially constant throughout the experimental period (Figure ).
3.2. Expression analysis of β-amylase genes
Of the nine genes predicted to encode β-amylase isoforms annotated in the rice genome, OsBAM1 (RAP-ID: Os07g0543300) and OsBAM7 (RAP-ID: Os07g0667100) were expressed specifically in germinating seeds and developing caryopses, respectively (Hirano et al., Citation2011). Accordingly, we initially analyzed the transcript levels of β-amylase genes other than OsBAM1 and OsBAM7 in several organs. The transcript levels of OsBAM2 and OsBAM3 were highest in leaf sheaths after heading (Figure (a)). OsBAM2 and OsBAM3 were also highly expressed in leaf blades. OsBAM4 was clearly expressed in leaf blades and leaf sheaths after heading. OsBAM5 was expressed in leaf blades, leaf sheaths after heading and developing caryopses. The expression of OsBAM6 was detected specifically in leaf blades. OsBAM8 and OsBAM9 were constitutively expressed in all organs analyzed. OsBAM9 was strongly expressed in leaf sheaths after heading.
Figure 3. Expression analyses of β-amylase genes. (a) Semiquantitative RT-PCR of OsBAM2, OsBAM3, OsBAM4, OsBAM5, OsBAM6, OsBAM8, and OsBAM9 in various rice organs. Leaf blade and leaf sheath before heading were harvested at the active tilling stage, and leaf sheath after heading and developing caryopsis were harvested at 10–14 days after heading. OsEF1α1 encoding translation elongation factor 1A was used as the internal standard. (b) Changes in transcript levels of OsBAM2 and OsBAM3 in the third leaf sheaths by quantitative RT-PCR. The third leaf sheaths were harvested at the second leaf emergence stage, the flag leaf emergence stage, the heading stage, and 5, 10, 15 and 20 days after heading. Values were standardized to the transcript level of OsEF1α1 and represent means ± SE of three replications.
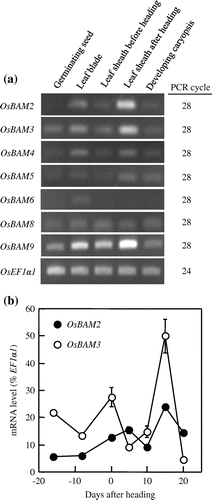
The mRNA levels of OsBAM2 and OsBAM3 were analyzed in detail in the leaf sheaths during the pre- and post-heading stages. The mRNA level of OsBAM2 gradually increased from the flag leaf emergence stage until 5 days after heading (Figure (b)). Subsequently, the mRNA levels peaked at 15 days after heading and then decreased following 15 days after heading. The mRNA level of OsBAM3 decreased from heading stage to 5 days after heading. However, the level rapidly increased starting 10 days after heading, reached a maximum at 15 days after heading. Thereafter, the level decreased markedly from 15 to 20 days after heading.
3.3. Phenotypic analysis of overexpression and knockdown lines of OsBAM2 and OsBAM3
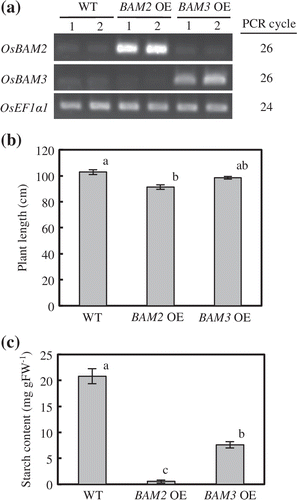
We generated overexpression lines of OsBAM2 (BAM2 OE) and OsBAM3 (BAM3 OE) to determine whether the overexpression of OsBAM2 and OsBAM3 in fact reduces starch content in rice leaves. The mRNA levels of OsBAM2 and OsBAM3 markedly increased in the third leaf sheaths of BAM2 OE and the BAM3 OE plants, respectively (Figure (a)). The length of BAM2 OE plants was significantly shorter than that of non-transgenic WT plants (Figure (b)). BAM3 OE plants exhibited a slightly dwarf phenotype. Starch content in the third leaf sheaths was markedly lower in BAM2 OE and BAM3 OE plants than in WT plants at the heading stage (Figure (c)). The starch content of BAM3 OE plants was significantly higher than that of BAM2 OE plants.
Figure 4. Phenotypic analyses of overexpression plants of OsBAM2 and OsBAM3. (a) Semiquantitative RT-PCR of OsBAM2 and OsBAM3 in the third leaf sheaths at the heading stage. OsEF1α1 was used as internal standard. (b) Plant lengths at the heading stage. Data represent means ± SE of five replications. Data followed by different letters represent significant differences at the 5% level by Tukey’s test. (c) Starch contents in the third leaf sheaths at the heading stage. Data represent means ± SE of three replications. Data followed by different letters represent significant differences at the 5% level by Tukey’s test.
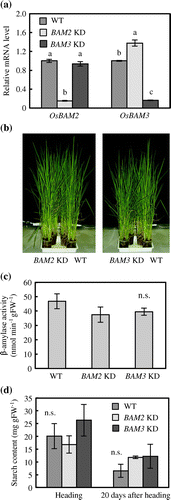
Knockdown lines of OsBAM2 (BAM2 KD) and OsBAM3 (BAM3 KD) were generated by the RNA interference method to evaluate the effect of the repressed expression of OsBAM2 or OsBAM3 on the starch content in leaf sheaths. The mRNA level of OsBAM2 was significantly lower in the third leaf sheaths of BAM2 KD plants than in those of WT and BAM3 KD plants (Figure (a)). Similarly, the third leaf sheaths of BAM3 KD plants exhibited drastically lower levels of OsBAM3 mRNA than those of WT plants and BAM2 KD plants. Few differences in the growth habits were observed between knockdown and WT plants (Figure (b)). No significant differences in panicle weight and yield components among the three plant lines were observed (data not shown). Although β-amylase activities in the third leaf sheaths tended to be lower in BAM2 KD and BAM3 KD plants than in WT plants, no significant differences were detected among the three plant lines (Figure (c)). BAM2 KD and BAM3 KD plants accumulated starch in the third leaf sheaths at levels similar to those of WT plants at the heading stage (Figure (d)). Although, at 20 days after heading, the starch contents of BAM2 KD and BAM3 KD plants tended to be slightly higher than that of WT plants, there were no significant differences among the three plant lines.
Figure 5. Phenotypic analyses of knockdown plants of OsBAM2 and OsBAM3. (a) Quantitative RT-PCR of OsBAM2 and OsBAM3 in the third leaf sheaths at the heading stage. OsEF1α1 was used as the internal standard. Data are relative to the transcript level of wild-type (WT) plants and represent means ± SE of three replications. Data followed by different letters represent significant difference at the 5% level by Tukey’s test. (b) Photographs of knockdown plants of OsBAM2 and OsBAM3 at booting stage. (c) β-Amylase activity in the third leaf sheaths at the heading stage. Data represent means ± SE of three replications. n.s., no significant difference at the 5% level by Tukey’s test. (d) Starch contents in the third leaf sheaths at the heading and 20 days after heading. Data represent means ± SE of three replications. n.s., no significant difference at the 5% level by Tukey’s test.
4. Discussion
In rice leaf sheaths, starch is accumulated at a high level prior to the heading stage and remobilized for grain filling during the post-heading stage (Perez et al., Citation1971). In the present study, starch content in the third leaf sheaths peaked at the flag leaf emergence stage and then gradually decreased (Figure (a)), suggesting that starch degradation in the third leaf sheaths was activated to provide carbohydrate to the ripening grains after the flag leaf emergence stage. The starch content was reduced to the lowest levels at 15 days after heading and then rapidly increased from 20 days after heading. These results suggest that starch remobilization was almost complete by 20 days after heading. We accordingly evaluated the β-amylase activity and the mRNA levels of β-amylase genes in the third leaf sheaths until 20 days after heading.
Perez et al. (Citation1971) analyzed β-amylase activity in leaf sheaths harvested at 6, 8, 10, 11 (booting stage), 12 (flowering stage), and 14 weeks after transplanting, but there is little information concerning continuous changes in β-amylase activity in leaf sheaths from the heading to the grain filling stage. In the present study, we analyzed in detail the β-amylase activity in the third leaf sheaths at the pre- and post-heading stages. The activity remained substantially constant from the third leaf emergence stage to 20 days after heading (Figure ). Thus, no significant correlation between β-amylase activity and the amount of starch loss in the third leaf sheaths during the post-heading stage was detected. In A. thaliana, BAM5, an active β-amylase protein localized in the cytoplasm, may account for up to 80% (Caspar et al., Citation1989; Lin et al., Citation1988) or over 70% (Monroe et al., Citation2014) of total β-amylase activity in leaves. Thus, analysis using a bam5 mutant allowed the evaluation of the contribution of each plastid-localized β-amylase isoform to total β-amylase activity (Monroe et al., Citation2014). In rice, the existence of a cytosolic β-amylase isoform that actively functions in leaves is still undetermined. Identification of a cytoplasm-localized β-amylase protein and analysis of its activity may be needed to evaluate the correlations between changes in starch content and β-amylase activity in the leaf sheaths.
In the rice genome, there are nine genes predicted to encode β-amylase-like proteins (Hirano et al., Citation2011; Saika et al., Citation2005). Of these genes, at least OsBAM2 and OsBAM3 encode plastid-targeted active isoforms (Hirano et al., Citation2011). The finding that the overexpression of OsBAM2 or OsBAM3 markedly reduced starch accumulation in the third leaf sheaths at the heading stage (Figure (c)) shows that OsBAM2 and OsBAM3 can in fact function in the degradation of starch stored transiently in plastid in vivo. The degree of starch loss was more pronounced in BAM2 OE than in BAM3 OE plants (Figure (c)). This phenomenon may result from the differences in expression levels between the overexpressed OsBAM2 and OsBAM3 or the kinetic properties between OsBAM2 and OsBAM3 isoforms. A rice mutant lacking OsAGPL1 encoding a large subunit of ADP-glucose pyrophosphorylase, which had markedly reduced starch content in the stem, displayed the shorter plant length than WT plants (Okamura et al., Citation2013). In the present study, the plant length of BAM2 OE, which accumulated much less starch in the leaf sheaths, was significantly shorter than that of WT plants, similarly to the mutant defective in OsAGPL1 (Figure (b)). These results suggest that the repression of starch accumulation in the leaf sheaths affects the plant growth via the abnormal supply of carbohydrate.
The mRNA levels of OsBAM2 and OsBAM3 were higher in leaf sheaths after heading than in those before heading and in leaf blades (Figure (a)). In addition, the mRNA levels of both OsBAM2 and OsBAM3 increased from the flag leaf emergence stage to the heading stage when the starch content began to decrease in the third leaf sheaths (Figure (b)). These results raise the possibility that OsBAM2 and OsBAM3 play a role in starch degradation in the leaf sheaths. However, the mRNA level of OsBAM2 decreased from 5 to 10 days after heading and that of OsBAM3 rapidly decreased from the heading to 5 days after heading. Subsequently, the mRNA levels of both OsBAM2 and OsBAM3 reached a maximum at 15 days after heading. Chen and Wang (Citation2008) showed that the β-amylase gene (cDNA accession number: AK068968, designated as OsBAM3 in this study) was highly expressed in leaf sheaths coincidentally with a decrease in starch content during the post-heading stage. However, our results indicate that the mRNA levels of OsBAM2 and OsBAM3 did not always remain high from the heading stage to 15 days after heading, when the starch content gradually decreased in the third leaf sheaths. In A. thaliana, expression of BAM5 is induced by sugar (Mita et al., Citation1995). Similarly in rice leaves, it is possible that the expression of OsBAM2 and OsBAM3 may be tightly regulated in response to changes in carbon status in cells. Such a mechanism may at least in part cause the drastic changes in the mRNA levels of OsBAM2 and OsBAM3 in the third leaf sheaths during the post-heading stage.
Although the starch contents in the third leaf sheaths tended to be higher in both BAM2 KD and BAM3 KD than in WT plants at 20 days after heading, no significant differences were detected among their starch contents (Figure (d)). These results indicate that the knockdown of the individual genes, OsBAM2 or OsBAM3, did not lead to the hyperaccumulation of starch in the leaf sheaths. In fact, the β-amylase activities in the third leaf sheaths were not significantly lower in BAM2 KD and BAM3 KD than in WT plants at the heading stage (Figure (c)). In A. thaliana, a bam3 mutant deficient for BAM3, one of the β-amylase isoforms targeted to plastids, accumulated an excess of starch in leaves, whereas the bam1 mutant deficient for plastid-targeted BAM1 showed normal accumulation of starch (Fulton et al., Citation2008). Monroe et al. (Citation2014) reported that only the bam3 mutant among the single β-amylase mutants (bam1, bam2, bam3, and bam6) accumulated an excess of starch in leaves of five-week-old plants, but in eight-week-old plants all mutants displayed a starch-excess phenotype. Moreover, the bam1 bam3 (Fulton et al., Citation2008), the bam2 bam3, and the bam3 bam6 double mutants (Monroe et al., Citation2014) displayed a more extreme phenotype for leaf starch accumulation than the bam3 mutant. Phenotypic analysis of double knockdown of OsBAM2 and OsBAM3 may reveal the contribution of these genes to starch degradation in leaf sheaths.
Asatsuma et al. (Citation2005) showed that α-amylase I-1 encoded by RAmy1A plays a role in starch degradation in rice leaves. The expression level of RAmy2A, encoding one of the α-amylase isoforms, increased significantly in rice leaf sheaths at the post-heading stage (Chen & Wang, Citation2008; Sugimura et al., Citation2015). The disruption of OsGWD1 encoding α-glucan water dikinase displayed an excess-starch phenotype in rice leaves (Hirose et al., Citation2013). In the present study, OsBAM4, OsBAM5, OsBAM8, and OsBAM9 were distinctly expressed in leaf sheaths after heading (Figure (a)), raising the possibility that these β-amylase genes may contribute to starch degradation in leaf sheaths during the post-heading stage. These results suggest the involvement of several other genes as well as OsBAM2 and OsBAM3 in starch degradation in leaf sheaths, perhaps accounting for the absence of hyperaccumulation of starch in the leaf sheaths of BAM2 KD and BAM3 KD.
Funding
This work was supported in part by JSPS KAKENHI [grant number 24580027] and [grant number 26292009].
Disclosure statement
No potential conflict of interest was reported by the authors.
Acknowledgments
We thank Dr. Ko Shimamoto (Nara Institute of Science and Technology, Japan) for providing the pANDA vector.
References
- Asatsuma, S., Sawada, C., Itoh, K., Okito, M., Kitajima, A., & Mitsui, T. (2005). Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant and Cell Physiology, 46, 858–869.10.1093/pcp/pci091
- Baunsgaard, L., Lütken, H., Mikkelsen, R., Glaring, M. A., Pham, T. T., & Blennow, A. (2005). A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated α-glucans and is involved in starch degradation in Arabidopsis. The Plant Journal, 41, 595–605.10.1111/tpj.2005.41.issue-4
- Caspar, T., Lin, T. -P., Monroe, J., Bernhard, W., Spilatro, S., Preiss, J., & Somerville, C. (1989). Altered regulation of β-amylase activity in mutants of Arabidopsis with lesions in starch metabolism. Proceedings of the National Academy of Sciences USA, 86, 5830–5833.10.1073/pnas.86.15.5830
- Chen, H.-J., & Wang, S.-J. (2008). Molecular regulation of sink-source transition in rice leaf sheaths during the heading period. Acta Physiologiae Plantarum, 30, 639–649.10.1007/s11738-008-0160-8
- Chia, T., Thorneycroft, D., Chapple, A., Messerli, G., Chen, J., Zeeman, S. C., … Smith, A. M. (2004). A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. The Plant Journal, 37, 853–863.10.1111/tpj.2004.37.issue-6
- Christensen, A. H., Sharrock, R. A., & Quail, P. H. (1992). Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Molecular Biology, 18, 675–689.10.1007/BF00020010
- Cock, J. H., & Yoshida, S. (1972). Accumulation of 14C-labelled carbohydrate before flowering and its subsequent redistribution and respiration in the rice plant. Japanese Journal of Crop Science, 41, 226–234.10.1626/jcs.41.226
- Fulton, D. C., Stettler, M., Mettler, T., Vaughan, C. K., Li, J., Francisco, P., … Zeeman, S. C. (2008). β-amylase4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. The Plant Cell, 20, 1040–1058.10.1105/tpc.107.056507
- He, H. Y., Koike, M., Ishimaru, T., Ohsugi, R., & Yamagishi, T. (2005). Temporal and spatial variations of carbohydrate content in rice leaf sheath and their varietal differences. Plant Production Science, 8, 546–552.
- Hiei, Y., Ohta, S., Komari, T., & Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal, 6, 271–282.10.1046/j.1365-313X.1994.6020271.x
- Hirano, T., Saito, Y., Ushimaru, H., & Michiyama, H. (2005). The effect of the amount of nitrogen fertilizer on starch metabolism in leaf sheath of japonica and indica rice varieties during the heading period. Plant Production Science, 8, 122–130.10.1626/pps.8.122
- Hirano, T., Takahashi, Y., Fukayama, H., & Michiyama, H. (2011). Identification of two plastid-targeted β-amylases in rice. Plant Production Science, 14, 318–324.10.1626/pps.14.318
- Hirose, T., Aoki, N., Harada, Y., Okumura, M., Hashida, Y., Ohsugi, R., … Terao, T. (2013). Disruption of a rice gene for α-glucan water dikinase, OsGWD1, leads to hyperaccumulation of starch in leaves but exhibits limited effects on growth. Frontiers in Plant Science, 4, 147.
- Hirose, T., Endler, A., & Ohsugi, R. (1999). Gene expression of enzymes for starch and sucrose metabolism and transport in leaf sheaths of rice (Oryza sativa L.) during the heading period in relation to the sink to source transition. Plant Production Science, 2, 178–183.10.1626/pps.2.178
- Hirose, T., Ohdan, T., Nakamura, Y., & Terao, T. (2006). Expression profiling of genes related to starch synthesis in rice leaf sheaths during the heading period. Physiologia Plantarum, 128, 425–435.10.1111/ppl.2006.128.issue-3
- Ishimaru, K., Kosone, M., Sasaki, H., & Kashiwagi, T. (2004). Leaf contents differ depending on the position in a rice leaf sheath during sink-source transition. Plant Physiology and Biochemistry, 42, 855–860.10.1016/j.plaphy.2004.10.008
- Kaplan, F., & Guy, C. L. (2005). RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. The Plant Journal, 44, 730–743.10.1111/tpj.2005.44.issue-5
- Kötting, O., Pusch, K., Tiessen, A., Geigenberger, P., Steup, M., & Ritte, G. (2005). Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiology, 137, 242–252.10.1104/pp.104.055954
- Lao, N. T., Schoneveld, O., Mould, R. M., Hibberd, J. M., Gray, J. C., & Kavanagh, T. A. (1999). An Arabidopsis gene encoding a chloroplast-targeted β-amylase. The Plant Journal, 20, 519–527.10.1046/j.1365-313X.1999.00625.x
- Lin, T. -P., Spilatro, S. R., & Preiss, J. (1988). Subcellular localization and characterization of amylases in Arabidopsis leaf. Plant Physiology, 86, 251–259.10.1104/pp.86.1.251
- McCormac, A. C., Elliott, M. C., & Chen, D.-F. (1999). pBECK2000: A novel plasmid series for the facile creation of complex binary vectors, which incorporates ‘clean-gene’ facilities. Molecular and General Genetics, 261, 226–235.
- Miki, D., Itoh, R., & Shimamoto, K. (2005). RNA silencing of single and multiple members in a gene family of rice. Plant Physiology, 138, 1903–1913.10.1104/pp.105.063933
- Miki, D., & Shimamoto, K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant and Cell Physiology, 45, 490–495.10.1093/pcp/pch048
- Mikkelsen, R., Baunsgaard, L., & Blennow, A. (2004). Functional characterization of α-glucan, water dikinase, the starch phosphorylating enzyme. Biochemical Journal, 377, 525–532.10.1042/bj20030999
- Mita, S., Suzuki-Fujii, K., & Nakamura, K. (1995). Sugar-inducible expression of a gene for β-amylase in Arabidopsis thaliana. Plant Physiology, 107, 895–904.10.1104/pp.107.3.895
- Monroe, J. D., Storm, A. R., Badley, E. M., Lehman, M. D., Platt, S. M., Saunders, L. K., … Torres, C. E. (2014). β-amylase1 and β-amylase3 are plastidic starch hydrolases in Arabidopsis that seem to be adapted for different thermal, pH, and stress conditions. Plant Physiology, 166, 1748–1763.10.1104/pp.114.246421
- Niittylä, T., Messerli, G., Trevisan, M., Chen, J., Smith, A. M., & Zeemen, S. C. (2004). A previously unknown maltose transporter essential for starch degradation in leaves. Science, 303, 87–89.10.1126/science.1091811
- Okamura, M., Hirose, T., Hashida, Y., Yamagishi, T., Ohsugi, R., & Aoki, N. (2013). Starch reduction in rice stems due to a lack of OsAGPL1 or OsAPL3 decreases grain yield under low irradiance during ripening and modifies plant architecture. Functional Plant Biology, 40, 1137–1146.
- Perez, C. M., Palmiano, E. P., Baun, L. C., & Juliano, B. O. (1971). Starch metabolism in the leaf sheaths and culm of rice. Plant Physiology, 47, 404–408.10.1104/pp.47.3.404
- Ritte, G., Lloyd, J. R., Eckermann, N., Rottmann, A., Kossmann, J., & Steup, M. (2002). The starch-related R1 protein is an α-glucan, water dikinase. Proceedings of the National Academy of Sciences USA, 99, 7166–7171.10.1073/pnas.062053099
- Ritte, G., Scharf, A., Eckermann, N., Haebel, S., & Steup, M. (2004). Phosphorylation of transitory starch is increased during degradation. Plant Physiology, 135, 2068–2077.10.1104/pp.104.041301
- Saika, H., Nakazono, M., Ikeda, A., Yamaguchi, J., Masaki, S., Kanekatsu, M., & Nemoto, K. (2005). A transposon-induced spontaneous mutation results in low β-amylase content in rice. Plant Science, 169, 239–244.10.1016/j.plantsci.2005.03.022
- Scheidig, A., Fröhlich, A., Schulze, S., Lloyd, J. R., & Kossmann, J. (2002). Downregulation of a chloroplast-targeted β-amylase leads to a starch-excess phenotype in leaves. The Plant Journal, 30, 581–591.10.1046/j.1365-313X.2002.01317.x
- Sparla, F., Costa, A., Schiavo, F. L., Pupillo, P., & Trost, P. (2006). Redox regulation of a novel plastid-targeted β-amylase of Arabidopsis. Plant Physiology, 141, 840–850.10.1104/pp.106.079186
- Sugimura, Y., Michiyama, H., & Hirano, T. (2015). Involvement of α-amylase genes in starch degradation in rice leaf sheaths at the post-heading stage. Plant Production Science, 18, 277–283.10.1626/pps.18.277
- Watanabe, Y., Nakamura, Y., & Ishii, R. (1997). Relationship between starch accumulation and activities of the related enzymes in the leaf sheath as a temporary sink organ in rice (Oryza sativa). Australian Journal of Plant Physiology, 24, 563–569.10.1071/PP96107
- Weise, S. E., Weber, A. P. M., & Sharkey, T. D. (2004). Maltose is the major form of carbon exported from the chloroplast at night. Planta, 218, 474–482.10.1007/s00425-003-1128-y
- Yu, T. S., Kofler, H., Häusler, R. E., Hille, D., Flügge, U. I., Zeeman, S. C., … Weber, A. (2001). The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. The Plant Cell, 13, 1907–1918.10.1105/tpc.13.8.1907
- Zeeman, S. C., Kossmann, J., & Smith, A. M. (2010). Starch: Its metabolism, evolution, and biotechnological modification in plants. Annual Review of Plant Biology, 61, 209–234.10.1146/annurev-arplant-042809-112301