Abstract
Simultaneous perturbation stochastic approximation (SPSA) is a gradient-based optimization method which has become popular since the 1990s. In contrast with standard numerical procedures, this method requires only a few cost function evaluations to obtain gradient information, and can therefore be advantageously applied when identifying a large number of unknown model parameters, as for instance in neural network models or first-principles models. In this paper, a first-order SPSA algorithm is introduced, which makes use of adaptive gain sequences, gradient smoothing and a step rejection procedure to enhance convergence and stability. The algorithm performance is illustrated with the estimation of the most-likely kinetic parameters and initial conditions of a bioprocess model describing the evolution of batch animal cell cultures.
1. Introduction
Process modelling requires the estimation of several unknown parameters from noisy measurement data. In this connection, gradient-based optimization methods are popular tools for minimizing least-squares or maximum-likelihood cost functions – depending on the assumptions on the measurement noise – measuring the deviation between the model and real system outputs.
Several techniques for computing the gradient of the cost function are available, including finite difference approximations or direct differentiation, which involves the numerical solution of sensitivity equations.
In the above-mentioned techniques, the computational expense required to estimate the current gradient direction is directly proportional to the number of unknown model parameters, which becomes an issue for models involving a large number of parameters. This is typically the case in neural network (NN) modelling, but can also occur in other circumstances, such as the estimation of parameters and initial conditions in first-principles models. Moreover, the derivation of sensitivity equations requires analytic manipulation of the model equation, which is time consuming and prone to errors (note that an important stream of current research and development is dedicated to the automatic generation of such equations using symbolic manipulation).
In contrast to standard finite differences which approximate the gradient by varying all the parameters one at a time, the simultaneous perturbation (SP) approximation of the gradient proposed by Spall Citation1 makes use of a very efficient technique based on a simultaneous (random) perturbation in all the parameters. Hence, one gradient evaluation requires only two evaluations of the cost function. This approach has first been applied to gradient estimation in a first-order stochastic approximation (SA) algorithm Citation1, and more recently to Hessian estimation in an accelerated second-order SPSA algorithm Citation2.
Efficiency, simplicity of implementation and very modest computational costs make first-order SPSA (1SPSA) particularly attractive, even though it suffers from the classical drawback of first-order algorithms, i.e. a slowing down in the convergence as an optimum is approached. Unfortunately, this phenomenon is even more pronounced in the case of SP techniques since the gradient information is more delicate to ‘extract’ from the – usually rather ‘flat’ – neighbourhood of the optimum.
In this study, a variant of the original first-order algorithm is considered which makes use of adaptive gain sequences, gradient smoothing and a step rejection procedure, to enhance convergence and stability. To demonstrate the algorithm efficiency and versatility, attention is focused on the estimation of kinetic parameters (and initial conditions) in a macroscopic model of animal cell cultures. To this end, a maximum-likelihood cost function based on the experimental measurements of biomass, glucose, glutamine and lactate concentrations is minimized.
This paper is organized as follows. Section 2 introduces the basic principle of the first-order SPSA algorithm used throughout this study. In section 3, the algorithm is applied to the maximum-likelihood estimation of kinetic parameters and initial conditions of a bioprocess model from experimental measurements of several macroscopic component concentrations. Direct and cross-validation results demonstrate the good model agreement. Finally, section 4 is devoted to some concluding remarks.
2 A brief overview of SPSA
Consider the problem of minimizing a, possibly noisy, objective function J(θ) with respect to a vector θ of unknown parameters.
1SPSA is given by the following core recursion for the parameter vector θCitation1
In its original formulation Citation1, 1SPSA makes use of decaying gain sequences {ak } and {ck } in the form
In order to enhance convergence and stability, the use of an adaptive gain sequence for parameter updating is considered, i.e.
In addition to gain attenuation when the value of the criterion becomes worse, ‘blocking’ mechanisms Citation2 are also applied, i.e. the current step is rejected and, starting from the previous parameter estimate, a new step is accomplished (with a new gradient evaluation and a reduced updating gain). The parameter β in (4) represents the permissible increase in the criterion, before step rejection and gain attenuation occur.
A constant gain sequence ck = c could be used for gradient approximation, the value of c being selected so as to overcome the influence of (numerical or experimental) noise. In the optimum neighbourhood, a decaying sequence in the form (3) is required to evaluate the gradient with enough accuracy and avoid an amplification of the ‘slowing down’ effect mentioned in the introduction.
Finally, a gradient smoothing (GS) procedure is implemented, i.e. gradient approximations are averaged across iterations in the following way
The use of these numerical artifices, i.e. adaptive gain sequences, step rejection procedure and gradient smoothing, significantly improves the effective practical performance of the algorithm, which, in the following, is denoted ‘adaptive 1SP-GS’.
Inequality constraints can also be taken into account by a projection algorithm introduced in Sadegh Citation3: the current parameter estimate is projected onto a closed set included in the admissible region in such a way that no function evaluation is required outside this latter region. In this study, bound constraints (e.g. positivity constraints) are handled in this way.
3 Modelling batch CHO cell cultures
In this experimental study, rare and asynchronous measurements of biomass (X), glucose (G), glutamine (Gln) and lactate (L) concentrations are available. The measurement equation is given by
Data are collected from seven batch experiments corresponding to different initial glucose and glutamine concentrations. Five of these experiments are used for parameter estimation, the two remaining ones being used for cross-validation tests.
The parameter estimation procedure involves the minimization of a Gauss-Markov criterion
The model equations are derived from mass balances and are given by
The pseudo-stoichiometry can be estimated independently of the reaction kinetics by using a state transformation originally introduced in Bastin and Dochain Citation4, which is based on the following ideas.
If rank K = M, where M is the number of independent reactions, then there always exists a partition
Accordingly, the partition of ξ is given by
Using this solution, an auxiliary vector z∈ℜ N − M can be defined as
Based on
Equationequations (12) and Equation(13)
, a maximum-likelihood estimation Ĉ of the matrix C can be computed Citation5, and estimations [Kcirc]a
and [Kcirc]b
of the matrices Ka
and Kb
can be deduced from Equationequation (11)
. A systematic procedure for selecting the most-likely C-identifiable reaction scheme has been proposed in Bogaerts and Vande Wouwer Citation6.
This procedure can be applied to our case study with ξ
a
= [X G]
T
, ξ
b
= [G ln L]
T
, and dz(t)/dt = 0 (from
Equationequation (13) with D = 0). Then, the following macroscopic reaction scheme is derived.
Growth:
The estimated pseudo-stoichiometry matrix is then given by
The growth rate ϕg
and the maintenance rate ϕm
appearing in
Equationequations (14) and Equation(15)
are described by a general kinetic model structure proposed in Bogaerts and Hanus Citation5.
The eight kinetic coefficients in
Equationequations (17) and Equation(18)
and 20 initial concentrations appearing in Equationequations (19)
– Equation
Equation
Equation(22)
for the five batch experiments under consideration (initial concentrations are corrupted by measurement noise and their most-likely values have to be identified) are estimated by minimizing the cost function (7) with the adaptive 1SP-GS algorithm.
The tuning parameters in
Equationequations (1) – Equation
Equation
Equation
Equation(5)
are selected as follows: c = 10−6, γ = 1.5 (a decaying sequence ck
is used for gradient evaluation), a
0 = 10−8, η = 1.01, μ = 0.9, β = 0 (no relative increase in the criterion is allowed), ρ0 = 0.99. Starting with the measured initial concentrations (which are affected by measurement errors) and an initial guess for the kinetic parameters corresponding to a cost function Jml
= 65760, the minimization (7) leads to a cost function Jml
= 308 in 50 000 iterations. This number of iterations might appear quite large at first sight, but the computational cost is very modest as each iteration only requires two cost function evaluations (each of these evaluations involves five model simulations corresponding to the five experimental batches used in this identification phase). On the other hand, standard centred finite difference approximations would require 56 cost function evaluations per iteration!
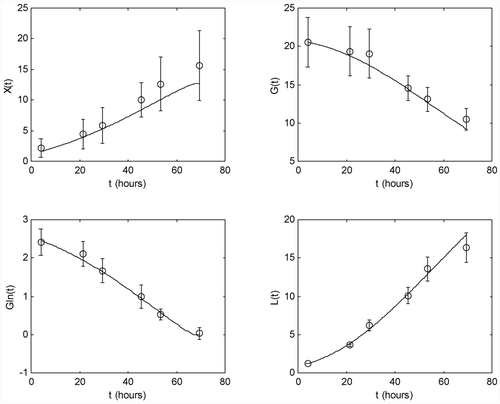
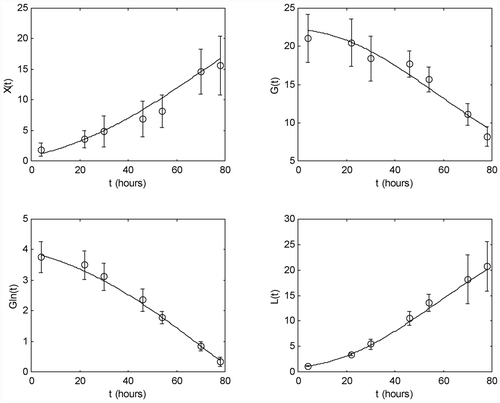
The parameter estimates are listed in . compares the measurement data of one of the five experiments used in the parameter identification procedure with the model prediction (direct validation), whereas shows the same kind of comparison with the measurement data of one of the remaining two experiments (cross-validation). In these graphs, the circled points are the measured data and the bars represent the 99% confidence intervals. The solid lines are the concentration trajectories predicted by the identified model.
Table 1. Parameter estimates.
These figures demonstrate the excellent model agreement.
4 Conclusion
The simultaneous perturbation approach developed by Spall Citation1,Citation2 is a very powerful technique, which allows an approximation of the gradient of the objective function to be computed by effecting simultaneous random perturbations in all the parameters. Therefore, this approach is particularly well-suited to problems involving a relatively large number of design parameters. In this study, a first-order SP algorithm is described and applied to the maximum-likelihood estimation of the kinetic parameters and initial conditions of a bioprocess model from experimental measurements of a few macroscopic components. These applications, as well as previous studies by the authors Citation7, demonstrate the usefulness of the proposed SP-algorithm.
Acknowledgements
The authors are very grateful to Professor J. Wérenne (Université Libre de Bruxelles, Departement of Animal Cell Biotechnology) and Mr M. Cherlet for providing them with the measurement data relating to the CHO-animal cell cultures.
References
- Spall , J. C. 1992 . Multivariate stochastic approximation using a simultaneous perturbation gradient approximation . IEEE Transactions Automatic Control , 37 : 332 – 341 .
- Spall , J. C. 2000 . Adaptive stochastic approximation by the simultaneous perturbation method . IEEE Transactions Automatic Control , 45 : 1839 – 1853 .
- Sadegh , P. 1997 . Constrained optimization via stochastic approximation with a simultaneous perturbation gradient approximation . Automatica , 33 : 889 – 892 .
- Bastin , G. and Dochain , D. 1990 . On-line Estimation and Adaptive Control of Bioreactors , Amsterdam : Elsevier .
- Bogaerts , Ph. and Hanus , R. 2000 . Focus on Biotechnology, Vol. IV, Engineering and Manufacturing for Biotechnology , Edited by: Thonart , P h. and Hofman , M. Dordrecht, , The Netherlands : Kluwer Academic Publishers .
- Bogaerts , Ph. and Vande Wouwer , A. 2001 . “ Systematic generation of identifiable macroscopic reaction schemes ” . In Proceedings IFAC CAB8 Quebec, , Canada
- Vande Wouwer , A. , Renotte , C. and Remy , M. 2003 . Application of stochastic approximation techniques in neural modelling and control . International Journal of Systems Science , 34 : 851 – 863 .