Abstract
This paper presents the two models, called EM1 and EM2, obtained for the two process configurations considered within the EOLI project. It also gives results of software sensors based on these models to provide on-line estimates of unmeasured process variables, and for which the design has considered in particular uncertainty on the model parameters.
1. Introduction
Within the EOLI project, two different types of models have been proposed to represent the dynamics of two different types of processes. These models have been calibrated and validated on experimental data obtained from the different experimental processes. The first model, referred to as EM1, corresponds to the process run in Mexico. The second model represents the dynamics of the processes run at POLIMI (Milano, Italy), at the LBE (Narbonne, France) and at the UU (Montevideo, Uruguay). Due to slight differences in the process dynamics, two submodels have been identified, i.e. EM2a for the POLIMI process, and EM2b for the LBE process.
Modelling of biochemical systems, and in particular of wastewater treatment processes, is clearly a critical issue, due to the inherent complexity and time variability of such processes as well as the lack of reliable knowledge about the kinetic mechanisms and of precise information and measurement of all the key process variables. However, as it has been illustrated in Citation1, a careful experiment design and identification procedure (briefly summarized in Section 3) can provide mass balance models of increased reliability. A similar procedure has been followed in the present study. The models as they are presented here are indeed the result of an iterative process between model building and process experiments. If the main frame of the models has been built in the initial step on the basis of the partners' experience and available scientific literature of the subject, several modifications have been included at the light of the physical/(bio)chemical evidences provided by the experiments. This has resulted in the design of extra experiments in order to validate the new modelling assumptions.
In this paper, we concentrate on models EM1 and EM2b, which are considered in the second part of the paper for the design of software sensors. Here, only the reaction schemes, the mass balance equations (with the kinetic expresssions), the basic identification procedure and the parameter values are provided. Details about the identification procedures for the different models can be found in the related, more detailed project reports.
The identified models have been used for several purposes in the project: software sensor design, FDI, and control design. In the present manuscript, we concentrate the software sensor design results. As for any biotechnological processes, the on-line monitoring and control of the SBRs is facing two major difficulties: the lack of measuring devices that can provide the values of all the key process components on-line, and the lack of confidence in the dynamical models of the processes (even with our quite careful identification procedure). This motivates design of software sensors that are capable of providing reliable software measurement (estimates) of the key components in spite of the model uncertainty that is typically mainly concentrated in the process kinetics.
2 Dynamical models of two SBRs
2.1 Model EM1
The process consists of a fed-batch reactor with one single aerobic growth reaction and an endogenous respiration reaction:
The specific growth rate is represented by the following Haldane model:
2.2 Model EM2b
The process operation consists of two successive batch steps: an anoxic phase followed by an aerobic phase. The model refers explicitely to these two phases. While in EM2a a two step denitrification/nitrification process is considered, in EM2b only one denitrification step and one step nitrification are considered in the anoxic and aerobic phases, respectively Citation2. Therefore the reaction scheme considered here is the following:
-
Anoxic phase
-
Aerobic phase
In the oxygen balance equation, the term S Omax accounts for the endogeneous respiration via the following relationship:
The kinetic expressions are the following:
3 Parameter identification of models EM1 and EM2b
The identification of the parameters of both models has been performed as follows Citation4,5. First, experiments have been designed so as to cover a sufficiently wide range of process conditions of interest in order to provide experimental data that can possibly provide a dynamical model structure with parameter values as accurate as possible. The collected data have been distributed in two sets: one for parameter calibration, one for model validation. The identification of the model parameters has been processed so as to estimate the kinetic parameters, the yield coefficients and the transfer coefficients independently of each other. This step is very important to guarantee appropriate and reliable parameter estimation. Moreover confidence intervals for each parameter have been computed.
Different configurations have been considered for the parameter calibration (e.g. with or without inlet dissolved oxygen, with or without oxygen sensor dynamics, with or without a modification in the early part of the Haldane kinetics) of model EM1. The following set of parameters ( ) correspond to S O,in = 0, S Os = 6 mg/L and no oxygen sensor dynamics. k 1 has also been given from separate batch experiments.
Table 1. Parameter values for model EM1-A3.
The identified parameter values from the LBE data for model EM2 are given in .
Table 2. Parameter values for model EM2b for LBE data.
4 Software sensor design
4.1 Software sensors for EM1
Software sensors are tools that allow provision of on-line values of key process components that are not accessible for on-line measurement. The software sensors for EM1 are concerned with the on-line estimation of substrate and biomass concentrations from the on-line measurements of dissolved oxygen in presence of uncertainty in the kinetics. The design of software sensors has been concentrating on the on-line estimation of biomass and substrate concentrations mainly in the context of the toxic compounds treatment plant described by model EM1 under the assumption (in line with the statistical analysis of the model parameters) of poor knowledge of the kinetics parameters of the biomass growth (Haldane model). The approach considered for the EOLI project for the EM1 model is a combination of an asymptotic observer with an adaptive observer that provides both state and parameter on-line estimates Citation6.
As a matter of illustration, let us consider the EM1 model where the parameter uncertainty is assumed to be concentrated on the parameter μ0 of the Haldane model. The available on-line measurements are the dissolved oxygen S O and S O,in, the inlet COD concentration S C,in and the inlet flow rate q in.
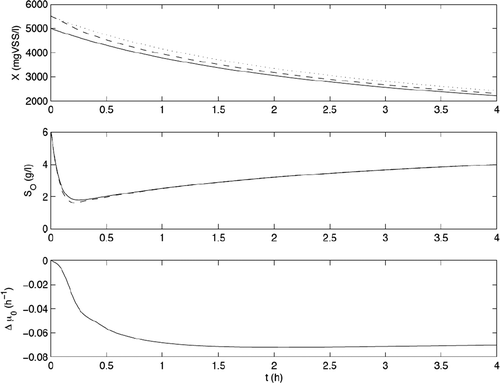
Since the operation time is intrinsically limited, one major challenge in the software sensor design is to be able to act on the convergence rate of the observer so that estimates of the unmeasured biomass and substrate can convergence fast enough to their true experimental values. Asymptotic observers are robust observers with respect to the lack of knowledge about the process kinetics but their convergence rate is fully dependent on the operating conditions (via the inlet flow rate). This explains why an alternative is proposed in the EOLI project that combines an asymptotic observer with an adaptive observer. Let us concentrate here on the case where the priority is to obtain a fast estimate of the biomass concentration X.
For the asymptotic observer, we first define the following auxiliary variables:
The adaptive observer is based on the assumption that a mean value of μ0 (denoted
) is known. The adaptive observer will therefore provide an
estimate of the deviation from this mean (possibly wrong) value. The design of the observer is based on the mass balance equations of the biomass concentration X and of the dissolved oxygen concentration S
O:
Note that the estimates of the asymptotic observer are used in the above adaptive observer; they play a key role in the stability analysis of the observer. C 1, C 2 and C 3 are the design parameters, in other words the knobs on which one can act to modify the convergence rate of the estimation of X. A careful mathematical stability analysis shows that they have to be selected as follows:
In the above equations λ1, λ2, λ3 are the eigenvalues associated to the adaptive observer. They are indeed the true knobs for selecting the convergence rate. They have to be negative (to guarantee the stability of the adaptive observer). When referring to simple dynamics views, large values of the eigenvalues mean fast convergence. However other dynamical effects (like the zero dynamics) may interfere and deteriorate the convergence in presence of large eigenvalues. Moreover, the presence of noise on the measurement may also limit the values of the eigenvalues. The appropriate selection of the design parameters is therefore a job that has to be performed carefully, in particular via exhaustive numerical simulations.
Alternative versions of the above EOLI observer can be deduced, e.g. by considering other parameter uncertainty assumptions or for the on-line estimation of the substrate concentration with convergence faster than with the asymptotic observer.
The EOLI observer has been tested in numerical simulation under various conditions. illustrates the performance of the EOLI observer in typical operating conditions of the fedbatch process of Mexico. The intial values of the biomass concentration and of the substrate concentration have been set with 10% error with respect to their values. The eigenvalues have been set to −5. Note that the adaptive observer (dashed line: - -) is able to converge faster to the true value of X (straight line) than the asymptotic observer (dotted line: …).
4.2 Software sensors for EM2b
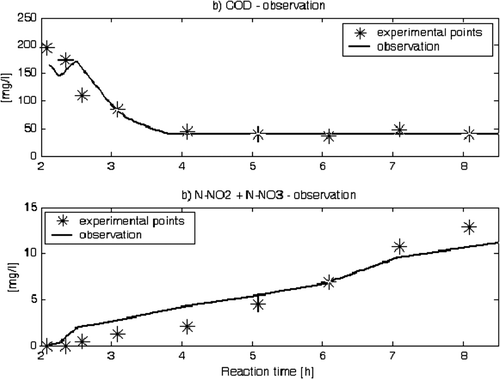
In the present instance, the software sensor design has concentrated in particular on the estimation of the organic matter concentration S C and of the nitrate–nitrite concentrations S NO from the on-line measurements of oxygen S O and of ammonia nitrogen S NH in the aerobic phase Citation7.
The estimation approach that has been considered here is an asymptotic observer Citation4. It is based on the following auxiliary variables z 1 and z 2:
Note that this estimator requires the knowledge of both Z
1(0) and Z
2(0) in EquationEquations (39) and Equation(40)
. This can be solved as follows. At the beginning of the aerobic phase, there is no nitrite nor nitrate in the reactor. Thus S
NO(0) = 0. Since S
NH is measured,
is known. Since S
O is also measured, one has only to assume that the initial value of the organic carbon to be removed in one cycle is known (
). It must then be assumed that an approximate value of S
C(0) is known in order to use this estimator. In such a case, the results shown in
illustrates the effectiveness of the method.
Acknowledgements
This paper includes results of the EOLI project that is supported by the INCO program of the European Community (Contract number ICA4-CT-2002-10012). It also presents research results of the Belgian Programme on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science, Technology and Culture. The scientific responsibility rests with its authors.
References
- Bernard , O. , Hadj-Sadok , Z. , Dochain , D. , Genovesi , A. and Steyer , J. Ph. 2001 . Dynamical model development and parameter identification for an anaerobic wastewater treatment process . Biotechnol. Bioeng , 75 : 424 – 438 .
- Betancur , M. , Dochain , D. and Fibrianto , H. 2004 . WP 2: Model selection and parameter identification. EOLI Project, Deliverable D2.3: Validated model
- Henze , M. , Grady , C. P.L. , Gujer , W. , Marais , G.v.R. and Matsuo , T. 1987 . Activated sludge model No. 1, IAWPRC Scientific and Technical reports, No. 1 , London : IAWPRC .
- Bastin , G. and Dochain , D. 1990 . On-Line Estimation and Adaptive Control of Bioreactors , Amsterdam : Elsevier .
- Dochain , D. and Vanrolleghem , P. A. 2001 . Dynamical Modelling and Estimation in Wastewater Treatment Processes , London : IWA Publishing .
- Dochain , D. 2003 . State observers for processes with uncertain kinetics . Int. J. Control , 76 : 1483 – 1492 .
- Mazouni , D. , Ignatova , M. and Harmand , J. 2005 . A multi model approach for the monitoring of carbon and nitrogen concentrations during the aerobic phase of a biological SBR . Proceedings of the IFAC 2005 World Congress . 2005 , Prague.