Abstract
For computational agent-based simulation, to become a serious tool for investigating biological systems requires the implications of simulation-derived results to be appreciated in terms of the original system. However, epistemic uncertainty regarding the exact nature of biological systems can complicate the calibration of models and simulations that attempt to capture their structure and behaviour, and can obscure the interpretation of simulation-derived experimental results with respect to the real domain. We present an approach to the calibration of an agent-based model of experimental autoimmune encephalomyelitis (EAE), a mouse proxy for multiple sclerosis (MS), which harnesses interaction between a modeller and domain expert in mitigating uncertainty in the data derived from the real domain. A novel uncertainty analysis technique is presented that, in conjunction with a latin hypercube-based global sensitivity analysis, can indicate the implications of epistemic uncertainty in the real domain. These analyses may be considered in the context of domain-specific knowledge to qualify the certainty placed on the results of in silico experimentation.
1. Introduction
Computational agent-based techniques are finding increasing application in the modelling and simulation of complex systems. For biological research they offer a complement to traditional wet-lab research techniques, enabling experimentation that is impractical or even impossible in the real domain [Citation1]. Within agent-based models and simulations (ABMS), individual elements of a system can be explicitly represented and carry their own state [Citation2]. For example, an agent-based model of an infection in a body compartment might explicitly represent individual immune system cells as discrete elements in the model, rather than capturing entire cell populations as single model elements. Agent-based models often explicitly represent the environment (such as spatial orderings) in which agents are placed, which determine the movement and interaction dynamics of the agents [Citation3]. The agent-based simulations that we consider in this article are stochastic in nature; repeated simulation runs with the same parameters will not necessarily yield identical dynamics, as the determination of an agent's behaviour (such as movement) is subject to the generation of pseudo-random numbers from a given distribution.
Our work concerns computational immunology: the modelling and simulation of the immune system. Advances in traditional wet-lab techniques are resulting in vast quantities of data pertaining to the behaviour of very specific system elements under very specific conditions [Citation4]. Modelling and simulation attempts to integrate these data into a coherent whole [Citation5,Citation6], indicating inconsistencies within the data and areas where understanding is lacking. This can often feed back into the wet-lab by directing avenues for experimentation [Citation7]. It allows for the formulation and evaluation of hypotheses concerning system behaviour and provides a means through which these hypotheses can be evaluated in the context of established knowledge [Citation8,Citation9]. Owing to the flexibility of computer code, simulation can facilitate experiments that are impossible to perform in the real domain as a result of either ethical considerations or issues in accessibility [Citation3,Citation10].
The work presented here is conducted in the context of the CoSMoS project,Footnote 1 which seeks to build capacity in complex system simulation construction and analysis. It is developing the CoSMoS process, an approach investigating complex domains through simulation that places emphasis on capturing the domain, and subsequent transitions from explicit models of a domain to executable simulations [Citation11]. Though agent-based modelling and simulation offers great potential in assisting scientific experimentation, its success ultimately depends on the ability to interpret simulation-based results in terms of the original domain. Simulations, however, are artificial and abstract representations of the real domains on which they are based, and hence results do not necessarily translate directly from one domain into the other.
The goal of this article is to explore the problem of relating simulation results back to domain reality, and by way of a case study provide examples of how calibration and statistical techniques can be used to explore uncertainty in simulation results. In doing this, we provide an example of how to qualify the significance of simulation-derived results in terms of the original domain. Like any type of model, ABMS are simplifications; it is intractable to represent every aspect of the real domain in a model, both computationally and because the domain is often not sufficiently well understood. There are many aspects of biological systems for which there exists no consensus in the literature or that remain to be investigated. For example, in 2000 the journal Seminars in Immunology [Citation12] dedicated an entire issue to a debate among leading immunologists regarding the function of the immune system in directing bio-destructive activities towards pathogenic invaders, and not the host [Citation12]. Highlighted was a lack of consensus among leaders in immunology with regard to this fundamental and essential aspect of immune system function. In the context of modelling and simulation, the lack of knowledge regarding a particular aspect of the domain is referred to as epistemic uncertainty [Citation13]. Epistemic uncertainty presents a challenge to the construction, calibration and interpretation of simulations, since the exact nature of a phenomenon of interest may be unclear.
Taken together, abstraction (which dictates that all aspects of the domain that are represented in the simulation must compensate for the actions of those that are not) and epistemic uncertainty complicate the relationship between the real domain and the simulation. Understanding this relationship is critical to relating predictive resultsFootnote 2 arising from ABMS back to the real domain, hence it is important that the implications of epistemic uncertainty and abstraction be appreciated. If the validity of AMBS-generated predictions hinges on aspects of the domain that are not well understood, then caution must be exercised in the interpretation of results.
In the vast majority of cases where simulation results are reported in the computational immunology literature, either the results are assumed to be absolutely representative of the target domain, or no effort to indicate the significance of simulation-derived results in terms of the original domain is made. To the best knowledge of the authors, we present here an original approach to qualify the significance of simulation-derived results in terms of the real domain through the use of uncertainty and sensitivity analyses [Citation14], statistical techniques that elucidate the relationship between a system's inputs and its output. Uncertainty and sensitivity analyses have found recent application in exploring immune system models and simulations, indicating parameters that are influential in dictating simulation behaviours [Citation3,Citation15,Citation16]; however, the use of these techniques in linking simulation results back into the original domain is novel.
Our work is grounded in an immunological case study for the modelling and simulation of experimental autoimmune encephalomyelitis (EAE), a mouse proxy for multiple sclerosis (MS). We make extensive use of a domain expert in mitigating uncertainty in data derived from the real domain. A novel uncertainty analysis technique that examines the simulation's robustness to parameter perturbation is presented, indicating how far a parameter may be perturbed before a scientifically significant change in simulation behaviour is observed. A latin hypercube global sensitivity analysis that determines simulation sensitivity with respect to its various parameters is presented. When considered in the context of domain-specific knowledge, these two analyses can qualify the implications of epistemic uncertainty on simulation-derived predictive results and will contribute to further work in identifying the confidence that may be placed on simulation-derived predictions.
The article is organized as follows: Section 2 describes EAE, a mouse proxy for MS, which constitutes the domain for this case study. Section 3 describes our calibration technique, along with examples of real domain data that demonstrate epistemic uncertainty and motivate our approach. Section 4 presents the uncertainty and sensitivity analyses employed in this work. Section 5 concludes this article.
2. Experimental autoimmune encephalomyelitis
EAE is an autoimmune disease in mice that serves as a proxy for MS in humans [Citation17,Citation18]. The disease results in the stripping of the insulatory myelin sheath from the neurons of the central nervous system (CNS). An abstract representation of the key cells involved in EAE, and their relationship to one another is presented in EAE is induced in mice by injecting myelin basic protein (MBP – a myelin derivative). MBP is ingested by dendritic cells (DCs) which induce a population of MBP-specific autoimmune CD4 T helper 1 (CD4Th1) cells. The CD4Th1 cell population infiltrates the CNS and secretes molecules that prompt local microglia cells to kill neurons. Neurons killed in this manner are ingested by DCs resident in the CNS. DCs then derive and present MBP from neurons and further induce and activate MBP-specific CD4Th1 cells. In this manner autoimmunity self-perpetuates.
Figure 1. An abstract depiction of the major cells involved in EAE and its associated regulatory recovery. MBP is the target of autoimmunity and is a protein expressed in the CNS. Adapted from [Citation20].
![Figure 1. An abstract depiction of the major cells involved in EAE and its associated regulatory recovery. MBP is the target of autoimmunity and is a protein expressed in the CNS. Adapted from [Citation20].](/cms/asset/59471c42-f05b-4612-a531-cb504840d6e6/nmcm_a_601419_o_f0001g.gif)
The physiological death of CD4Th1 cells, which occurs some time after their activation, leads to their digestion by DCs. Such DCs are then able to induce and activate CD4 regulatory T cells (CD4Treg) and CD8 regulatory T cells (CD8Treg), the former being required for the activation of the latter. Activated CD8Treg cells kill MBP-specific CD4Th1 cells upon direct cell contact. The population-level deletion of CD4Th1 cells permits the expansion of a competing, but normally suppressed, CD4 T helper 2 (CD4Th2) cell population [Citation19], which does not promote autoimmunity.
EAE is a complex disease, characterized by the interaction of two coupled feedback mechanisms: self-perpetuating autoimmunity and the regulation that it promotes. Disease and recovery arise as emergent properties resulting from the interactions of millions of cells of many different types. These interactions between cells occur across several bodily compartments. The complexities of this disease make reasoning about its nature and predicting the result of a particular intervention challenging. EAE is amenable to computational modelling and simulation techniques, where established knowledge and data can be integrated with hypotheses of system operation and executed in the presence of different interventions to indicate how the real system might respond.
3. Calibration
Calibration is an important aspect of in silico experimentation, it seeks to align a simulation's behaviour with that of the target domain, by adjusting parameters and manipulating the simulation's mechanics. However, there exists significant uncertainty and variance in data from the real domain. The stochastic nature of the immune system results in individual experimental animals experiencing vastly different progressions of EAE. Identifying immunological data in a format that a simulation can be calibrated against, and with sufficient samples to constitute a fine-grained representation, can be a challenging task.
highlights the type of data typically derived from EAE experiments with real mice in the laboratory. shows the mean severity of EAE, measured on a five point scale [Citation21], experienced by groups of 6–8 mice of different strains following the same induction of EAE administered in each group. There is considerable variation in EAE progression experienced by each species of mouse. shows the severity of EAE experienced by groups of five mice, with each group undergoing a different intervention. It can be seen that within groups of exactly the same experimental animal undergoing exactly the same intervention, there is considerable variation in the severities of EAE experienced. indicates the number of CD4Th1 cells found in different bodily compartments at various times following immunization in several experimental animals. It can be seen that there is once again significant spread in the data, with several examples where there are no data points lying on the calculated mean values. Acquiring data such as these can require the killing of an experimental animal, and since genetically identical animals undergoing exactly the same EAE induction can experience significantly different responses (), it is difficult to compile a representative progression of EAE in terms of individual cell population number.
Figure 2. Examples of clinical data pertaining to the progression of EAE in mice. EAE severity is scored on a scale of 0–5, ‘1, flaccid tail; 2, hind limb weakness; 3, hind limb paralysis; 4, whole body paralysis; 5, death’ [Citation21]. (a) The mean EAE severity experienced by groups of 6–8 mice of different strains following the same induction of EAE administered in each group. Adapted from [Citation22]. (b) EAE severity experienced by each of 5 mice under different interventions (boxes A–C), over time (in days). Taken from [Citation21]. The interventions involve injecting mice experiencing EAE with varying quantities of Treg cells. (c) An indication of the number of CD4Th1 cells (EI on the y-axis) residing in a selection of organs at various times following the induction of EAE from [Citation23]. Organs examined were lymph nodes (LN), spleen (Sp), blood (Bl), lungs (Lu), bone marrow (BM), liver(Li), mesenteric lymph nodes (mLN) and thymus (Thy).
![Figure 2. Examples of clinical data pertaining to the progression of EAE in mice. EAE severity is scored on a scale of 0–5, ‘1, flaccid tail; 2, hind limb weakness; 3, hind limb paralysis; 4, whole body paralysis; 5, death’ [Citation21]. (a) The mean EAE severity experienced by groups of 6–8 mice of different strains following the same induction of EAE administered in each group. Adapted from [Citation22]. (b) EAE severity experienced by each of 5 mice under different interventions (boxes A–C), over time (in days). Taken from [Citation21]. The interventions involve injecting mice experiencing EAE with varying quantities of Treg cells. (c) An indication of the number of CD4Th1 cells (EI on the y-axis) residing in a selection of organs at various times following the induction of EAE from [Citation23]. Organs examined were lymph nodes (LN), spleen (Sp), blood (Bl), lungs (Lu), bone marrow (BM), liver(Li), mesenteric lymph nodes (mLN) and thymus (Thy).](/cms/asset/82598254-2575-43cb-a372-af9e50be9839/nmcm_a_601419_o_f0002g.gif)
![Figure 2. Examples of clinical data pertaining to the progression of EAE in mice. EAE severity is scored on a scale of 0–5, ‘1, flaccid tail; 2, hind limb weakness; 3, hind limb paralysis; 4, whole body paralysis; 5, death’ [Citation21]. (a) The mean EAE severity experienced by groups of 6–8 mice of different strains following the same induction of EAE administered in each group. Adapted from [Citation22]. (b) EAE severity experienced by each of 5 mice under different interventions (boxes A–C), over time (in days). Taken from [Citation21]. The interventions involve injecting mice experiencing EAE with varying quantities of Treg cells. (c) An indication of the number of CD4Th1 cells (EI on the y-axis) residing in a selection of organs at various times following the induction of EAE from [Citation23]. Organs examined were lymph nodes (LN), spleen (Sp), blood (Bl), lungs (Lu), bone marrow (BM), liver(Li), mesenteric lymph nodes (mLN) and thymus (Thy).](/cms/asset/2d690afb-3bd4-4869-9650-f1de7884f4b6/nmcm_a_601419_o_f0009g.gif)
Data of this nature can be challenging to calibrate a simulation against. A stochastic computer simulation can be run many hundreds or thousands of times in order to obtain highly representative averaged values for a particular metric of interest. The same fidelity of data is not available in the immunological literature, where the number of samples obtained in acquiring an average rarely exceeds 10. Whereas a computational simulation can readily provide exact numbers of a particular cell type over time, the data of only indicate their number, there is not an easily established exact mapping between the metric used and the actual number of cells observed by the instrument.
3.1. Calibration process
Calibration of the EAE simulator was undertaken as a collaborative effort between the modeller (Mark Read) and the domain expert (Vipin Kumar). The domain expert provides a consistent and comprehensive single source of data against which to calibrate, which helps mitigate the uncertainty and variation in the data surrounding EAE. In this manner, the simulation is calibrated against the domain expert's understanding of EAE. The input of the domain expert in this calibration procedure serves to keep the simulation well grounded in the domain.
The process through which the simulation was calibrated is as follows: The simulation is executed with a ‘best guess’ set of parameters and representative median results are collected. The domain expert identifies aspects of simulation dynamics that deviate from his perspective of the real system. Both domain expert and modeller discuss to identify the source of the deviation in the simulation. Both parties' input are invaluable: the domain expert brings a wealth of domain-specific understanding, whilst the modeller, having built the simulation, has a detailed understanding and intuition of how this information has been abstracted and how the simulation operates. Potential avenues of model and simulation amendment and development are identified, and each one is independently integrated into the simulation in turn. In each case the simulation is then executed once more to obtain representative median results. Subsequent interactions between domain expert and modeller re-examine the results and decide upon which amendments are to be permanently adopted into the simulation. As such, the calibration process is iterative.
Throughout this calibration process, the simulation's behaviour is examined under two circumstances: that of normal EAE progression and recovery following induction of EAE; and the progression of EAE with the regulatory network disabled.Footnote 3 It is important to calibrate the simulation against these two different circumstances. An incorrect model can be fitted to the data derived from a single biological circumstance, such that its dynamics reflect that observed in vivo. We believe it is less likely that an incorrect model can replicate in vivo dynamics of several different biological circumstances in turn without having to drastically manipulate its parameters or change the underlying model. The parameter manipulations would reflect a need to compensate for the model's incomplete capture of the real domain. If the complexities and mechanics of the real domain are adequately represented in the simulation, then both biological circumstances (presence and absence of regulatory activity) should be replicated without having to tweak parameters. The end goal of simulating EAE is to use the simulation to perform experimentation that are intractable in the real domain. More confidence can be held in the results of predictive in silico experimentation if the simulation correctly represents multiple disparate real-world experimental circumstances.
3.2. Baseline behaviour
The calibration process results in the simulation being brought into a state that both domain expert and modeller accept as representing a normal, baseline behaviour. This baseline behaviour, shown for both the presence and absence of regulatory activity in , is generated by both a set of parameter values and the simulation mechanics for which they are appropriate. As explained in Section 1, the simulation is an abstract representation of the real domain; all elements present in the simulation compensate for the action of elements of the real domain that are not represented. Were the simulation's mechanics to be altered in some way, then the simulation's parameters may not represent exactly the same aspects as they did previously. Their role in compensating for model abstractions would be changed, and as a result, it may be necessary to recalibrate the simulation.
Figure 3. The baseline behaviour of the EAE simulation. (a) The baseline behaviour of the EAE simulation, following calibration, in the presence of regulation. Graph shows the number of effector T cells in the system over time and the number of neurons killed per hour (×10 for clarity). This is the median of 1000 simulation runs. (b) The baseline behaviour of the EAE simulation, following calibration, with regulatory activity disabled by preventing CD8Tregs from killing CD4Th1 cells. This represents the median behaviour of 1000 individual runs such as that depicted here. (c) An example of a single simulation run, in the absence of regulatory activity. The number of CD4Th1 cells declines heavily to a minimum at around day 80, but rises again thereafter. This reflects the relapsing nature of EAE seen in absence of regulation in some mice. The median progressions shown in and (b) are compiled from unique individual executions such as that of (c).
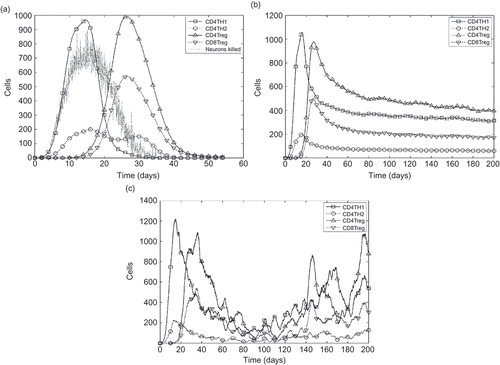
The baseline simulation serves as an accepted ‘normal’ state against which to contrast the results of experimentation with the simulation. The aspects of the system that in silico experiments investigate are usually represented as parameters in the system.
4. Exploring the EAE simulation with uncertainty and sensitivity analyses
4.1. Aleatory and epistemic uncertainty
There are two sources of uncertainty in the simulation that must be analysed before the significance of predictive results arising from the simulation can be appreciated: aleatory and epistemic uncertainty. Aleatory uncertainty results from inherent stochasticity in a system [Citation13] and is present in both the biological system (as can be observed in (b) and (c)) and the simulation. In order to acquire representative predictive results from a stochastic simulation, it is necessary to acquire many samples in the form of simulation executions. Appreciation of the spread of data is important for understanding when some experiment in the simulation has generated scientifically significant results.
Epistemic uncertainty results from a lack of knowledge about the value that a particular parameter should be assigned [Citation13]. There are many immunological details that are not well understood, such as the decay rates of certain molecules, or the exact average time that a particular cell type might remain in a particular state. Further to this, the simulation's abstract nature dictates that there is not a direct translation from these details into simulation parameters, since, as previously stated, everything present in the simulation must compensate for all those aspects of the real domain that are absent.
In order to fully appreciate the results of experimentation with the simulation, it is necessary to investigate the simulation's robustness to parameter alterations. The results of in silico experimentation can be considered to be of greater significance if the predictions that they delineate hold under perturbation of the simulation's parameters. If a prediction relies on a parameter holding a very specific value within its biologically plausible range, then the prediction cannot be held with great confidence. If a prediction breaks only when parameter values are perturbed to outside their biologically accepted range, then more confidence can be held that the prediction is representative of the real system.
Biologically plausible domains for parameter values are not clearcut and are accepted with some informal degree of confidence. This can arise from the different experimental set-ups through which results are obtained. For example, results arising from in vitro experiments conducted in test tubes may not be completely representative of the real system since the elements under investigation have been removed from their natural environment, yet it may be impossible to perform such experiments in vivo. Additional information useful in qualifying the certainty of simulation predictions is the relative contribution that each parameter makes to the simulation's system-wide dynamics. Epistemic uncertainty surrounding parameters that are of little consequence to simulation behaviour is less of an issue than uncertainty surrounding parameters of greater influence.
Uncertainty and sensitivity analysis techniques are closely related statistical techniques that attribute variation in a system's outputs to variation in its inputs [Citation14]. Uncertainty analysis investigates the effect of uncertainty in parameter values on output behaviour, whereas sensitivity analysis investigates the relative sensitivity of a simulation's output to its individual inputs. We have employed two uncertainty and sensitivity analysis techniques, one that qualifies the robustness of the simulation to parameter alteration, and one that determines the relative influence of parameters on simulation behaviour, discussed in Sections 4.3 and 4.4, respectively.
4.2. Uncertainty and sensitivity analysis responses
Uncertainty and sensitivity analysis techniques perform their analysis on a single variable that is assumed as the output of a system [Citation14]. The results of analyses can only be appreciated in terms of this variable, typically termed the response. For the purposes of analysing the EAE simulation, nine responses are assumed, and the uncertainty and sensitivity analysis techniques are run on each. shows these nine responses, namely, the maximum number of effector T cells reached over the simulation's execution for each of the four species of T cell, the times at which those four peaks occurred and finally the number of effector CD4Th1 cells that remain at 1000 hours.
Figure 4. The nine responses adopted to analyse the EAE simulation, shown on a graph of the number of effector T cells over time. For each of the four T cell species, both the maximum number reached and the time at which that number was reached are assumed as a separate response (these are indicated as crosshairs on the diagram). The ninth response is the number of autoimmune CD4Th1 effector cells remaining at 1000 hours.
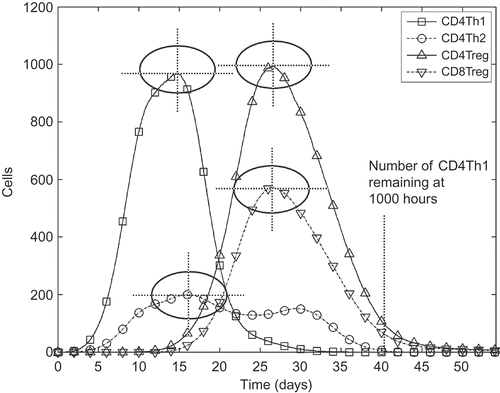
Analysing the simulation through these nine responses yields detailed information on how its parameters affect each of the major T cell populations independently, rather than attempting to condense a complex disease into a single variable. The choice of the nine responses was approved by the domain expert as being of biological relevance and interest in the context of EAE. For example, the last response, the number of CD4Th1 cells remaining at 1000 hours, is an effective means of determining whether efficient regulation of the CD4Th1 population has taken place. This cannot other eight responses.
4.3. Robustness analysis
In order to assess the robustness of the simulation to parameter perturbation, we have devised a one at a time uncertainty analysis technique, whereby each parameter is adjusted independently of the others, which remain at their baseline values. This baseline may be the result of calibration, as is the case for the examples presented here, or may be a point in parameter space that reflects a point of interest in making a prediction of the system; in this manner our technique would entail deliberately breaking that prediction. When considered in the context of biologically accepted ranges for parameter values, the results of this analysis help to qualify the validity of simulation results.
We employ the Vargha–Delaney A test [Citation24], which is a non-parametric effect magnitude test, to determine when a parameter adjustment has resulted in a scientifically significant change in simulation behaviour from the baseline. The A test compares two population distributions and returns a value in the range [0.0,1.0] that represents the probability that a randomly chosen sample taken from population A is larger than a randomly chosen sample from population B. A value of 0.5 indicates no difference, whereas values above 0.71 and below 0.29 indicate a ‘large’ difference in the distributions [Citation24]. details which A test scores relate to various magnitudes of difference between two populations.
Table 1. The magnitude of effect size indicated by A test score [24]
Our choice in using a non-parametric test is to avoid the assumption that underlying distributions are normally distributed. We use an effect magnitude test to determine scientific significance in place of statistical significance since a sufficiently large number of samples can always reveal a statistical significance, unless the variable of interest has no effect.
4.3.1. Experimental methodology
The baseline assumed in these examples is the result of the calibration process described in Section 3.1, though any point of interest in parameter space may be used. Each parameter is considered in turn and is perturbed around its baseline value. For each perturbation, the simulation is executed n number of times. The nine responses are calculated for each of the n simulation runs, and together form nine distributions. These can each be compared using the A test to similar distributions obtained from executing the simulation using baseline values n times.
(a) shows a graph depicting the robustness analysis on a particular parameter of the simulation. The effect magnitude of the parameter being assigned various values around the default value can be observed from the nine lines representing each of the nine responses. The y-axis shows the A test scores for each response under each parameter value, when compared to the default. The default value for this parameter is 100 and is where all the response lines converge. The A test score here is 0.5, as the default parameter value distribution data are being compared to themselves. Horizontal lines are drawn at A test score values of 0.71 and 0.29. These are the boundaries for differences in distributions that are considered ‘large’ [Citation24]. If a response line exceeds or falls below either of these respective boundaries, then a scientifically significant change in that response is observed for the particular parameter value that generated it.
Figure 5. Example robustness analysis of a parameter in the EAE simulation. Perturbation of this parameter has a large effect on all the responses. The parameter investigated here dictates the rate of TNF-α, a substance harmful to neurons in sufficient concentration, secretion by activated microglia cells in the CNS. (a) A test scores for the response distributions arising from parameter perturbations. The default value for this parameter is 100 and is where all lines on the graph converge. (b) The distribution of response data (maximum number of CD4Th1 cells reached) for each parameter perturbation. A dash, a circle and a cross above each box respectively indicate the default value, a change in distribution not considered large and a change that is considered large.
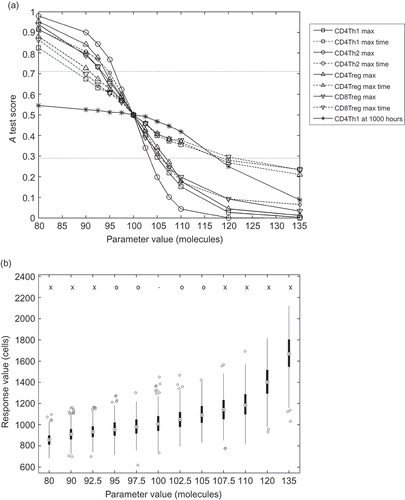
(b) shows the effect of the parameter adjusted in (a) on a particular response, as a box and whisker plot. The distribution of response values that arise from the n simulation executions for each parameter value is indicated by the boxes.
Data illustrated in this manner give a clear indication of exactly how the parameter of interest influences each response.
4.3.2. Mitigating aleatory uncertainty
The intention of this uncertainty analysis technique is to determine the robustness of the simulation to parameter perturbations. When considered in the context of epistemic uncertainty, a robust simulation may produce predictive results in which a relatively high degree of confidence may be placed. If the simulation's behaviour is shown to be fragile when parameters are perturbed to specific values that lie within the range of values reported in the domain's literature, then predictions arising from the simulation cannot be held with a high degree of confidence; the prediction mayFootnote 4 rely on the simulation holding a parameter value more specific than what the literature can dictate as appropriate. The simulation results might be an artefact of underspecified parameter values.
When performing this uncertainty analysis technique, it is important to obtain sufficient samples such that the response distributions obtained through parameter perturbation may be considered representative of the simulation's nature, rather than the result of aleatory uncertainty arising from the stochastic nature of the simulation. Uncertainty and sensitivity analyses techniques may be applied to any system, including systems that are costly to run, such as the computational expense of the present EAE simulation. Finding the relationship between sample size and the effect of aleatory uncertainty is important for balancing requirements (e.g. desired fidelity of data) and resources (e.g. computational resource required).
The following procedure is used to investigate the relationship between the number of samples obtained and the effect of aleatory uncertainty. The baseline parameter set-up is run 20 times, forming 20 dummy parameter permutations. These dummy permutations have no effect on the simulation's behaviour, since no parameter values are changing. This procedure can quantify the effect of aleatory uncertainty on A test scores, by examining the scores between the first and remaining 19 distributions generated. By repeating the experiment for different sample sizes, the number of samples required to reduce the noise in A test scores that arise from aleatory uncertainty to an acceptable level can be determined. (a)–(c) shows the A test scores across 20 dummy parameters using sample sizes of 5, 50 and 500, respectively. shows how the maximum and median A test scores derived from these 20 dummy parameters are affected by the sample size used, for the maximum number of CD4Th1 cells reached during simulation execution response. plots how the sample size affects the maximum A test scores achieved for each response. For both and , scores below 0.5 have been assigned their corresponding value above 0.5.
Figure 6. The maximum A test score achieved for each response over 20 dummy parameter experiments. The sample size represents the number of simulation runs used in compiling the response distributions for each dummy parameter experiment. Note that all scores below 0.5 are assigned corresponding values above 0.5 before maximum are performed; we are interested only in the magnitude of effect, not its direction above or below 0.5. The three effect magnitude boundaries for the A test are indicated.
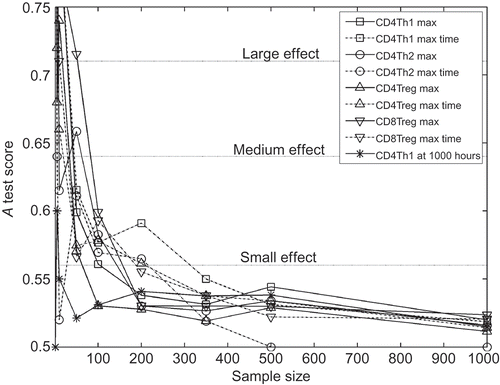
Figure 7. The effect of aleatory uncertainty on the results of the robustness analysis, for various sample sizes used when obtaining representative results from the simulation. The x-axes are labelled ‘dummy parameter’ as no parameters are actually being changed. The tests are designed to ascertain how the number of samples (simulation executions) from which median data are compiled affects the consistency of results. A test scores for 20 dummy parameter permutations, using sample sizes of (a) 5, (b) 50 and (c) 500.
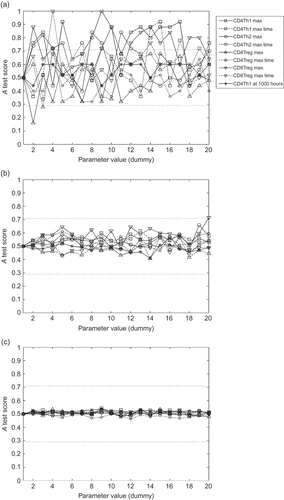
Table 2. An example of how sample size can reduce aleatory uncertainty, as measured by A test scores
indicates that a sample size of at least 350 is required to reduce the magnitude of aleatory uncertainty to an effect size less than ‘small’ (see ), for all responses. Hence, when using sample sizes of 350 simulation executions or more, results that deliver effect magnitudes less than or equal to ‘small’ should be discarded; one cannot determine whether the results are genuinely representative of the simulation's behaviour, or the result of aleatory uncertainty.
4.4. Determining the influence of parameters
The robustness analysis technique reported above is effective at providing an indication for how far a parameter may be adjusted before some significant deviation in simulation behaviour occurs. A shortcoming of the technique is that it does not elucidate non-linear effects between parameter values that are revealed only by adjusting two or more simultaneously; a particular parameter's influence on simulation behaviour may vary in magnitude depending on the value held by another parameter.
A global sensitivity analysis technique is used to provide a more representative indication of the relative influence of parameters on simulation behaviour, since it does highlight non-linear effects. Global sensitivity analysis refers to an experimental set-up where all parameters are perturbed simultaneously, as opposed to one at a time methods. Identifying which parameters exhibit the greatest influence over simulation behaviour can help qualify the implications of epistemic uncertainty for prediction validity. Significant uncertainty in a parameter's value is of greater consequence if that parameter has a large effect on simulation dynamics than if it is found to have a small effect.
4.4.1. Experimental methodology
A latin hypercube design [Citation25] is used to select k number of samples from the simulation's parameter space. (a) shows an example latin hypercube design over two parameters, taking 10 samples. The domain for each parameter over which samples are to be taken is defined and divided into k sections. In the example the 10 sections are uniformly distributed, but that need not be the case if information from the real domain dictates otherwise [Citation14]. For systems that are costly to run, latin hypercube design ensures an efficient coverage of parameter space using a minimal number of samples [Citation14]. A single sample exists in each segment of each parameter's domain.
Figure 8. Examples of latin hypercube design approach to determining relative parameter influence on simulation behaviour. The horizontal and vertical lines on (b) and (c) indicate the default parameter value and its associated response value. (a) An example latin hypercube design for two parameters. Ten samples are taken from the parameter space, within the specified domain of each parameter. Each of the 10 sub-domains (indicated by dotted lines) contains a sample, ensuring that the full domain of each parameter is explored. (b) This simulation parameter has a large influence on the time at which the maximum number of CD4Treg effector cells is reached during simulation execution. The rank correlation coefficient is 0.65. The parameter analysed here dictates the mean time that a T cell spends in a proliferative state before differentiating into an effector cell. (c) This simulation parameter has a small influence on the time at which the maximum number of CD4Treg effector cells is reached during simulation execution. The rank correlation coefficient is −0.001. The parameter analysed here is the rate of type 1 cytokine (an immune system messenger molecule) secretion by activated CD8Treg cells.
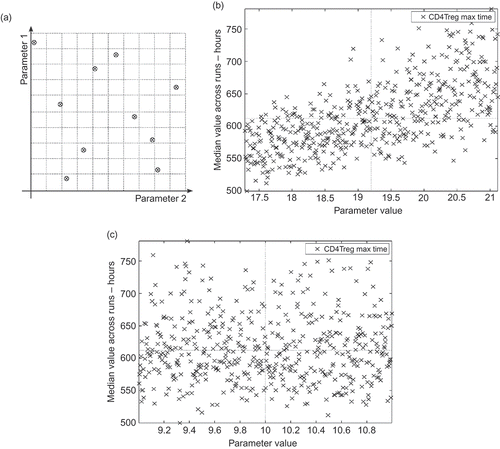
The simulation is run n times at each of the k sample points, and each of the nine responses is calculated for each run. Median values for each response at each sample point are calculated. The samples are ordered according to their value for a particular parameter of interest, and for each response a plot of median response value against each sample's value for the parameter of interest is generated. If the latin hypercube is of good design, having chosen samples such that correlations between parameters are minimized, then this ordering of samples should leave a minimum correlation between any other parameters. Any correlation between median response values and parameter values for each plot can be attributed to the effect of that parameter. Parameters that have a significant influence on simulation behaviour will yield bigger correlations than those that have less influence.
(b) and (c) illustrates this technique with two examples from the EAE simulator. The samples are ordered according to the parameter of interest, and the median values for the time at which the maximum number of CD4Treg cells is reached are plotted. There are 300 samples, and the simulation was run 300 times at each sample point, to minimize the effect of aleatory uncertainty.Footnote 5 The variation in the data arises from the pseudo-random values assigned to all other parameters but the one of interest, once the samples have been ordered. In (b), the parameter being examined is of such influence that despite all this movement in other simulation parameters, a clear trend does emerge. This is not the case for the parameter analysed in (c). As such, large epistemic uncertainty in the domain literature relating to the parameter of (b) must be given more consideration when interpreting simulation-derived results into the original domain than epistemic uncertainty concerning the parameter of (c).
5. Discussion
While computational modelling and simulation techniques such as ABMS have the ability to assist and complement more traditional wet-lab experimentation, there are several challenges that must be addressed before this potential can be fully realized. The complexities of biological systems dictate that models and simulations must adopt simplifying assumptions and abstractions in their representations. It is usually intractable to fully represent a complex system in a model or simulation, and there typically exist many aspects of the real domain that are not well understood. Epistemic uncertainty in the real domain can prevent the exact determination of simulation parameter values. It is critical in relating the results of in silico experimentation back to the real domain that the implications of these uncertainties, abstractions and assumptions be appreciated. To address this within the context of an EAE case study, we describe a method for calibrating the simulation with input from a domain expert and explore suitable statistical techniques to tackle uncertainty in simulation results.
The value of input from a domain expert in calibration cannot be understated, as it helps mitigate the uncertainties and inconsistencies found in data pertaining to the domain. We also note the importance of considering simulation dynamics under multiple circumstances when calibrating. The different circumstances arise from interventions applied to the real domain that expose the complexity of the system, which the simulation intends to capture. Comparing simulation dynamics under these different interventions with the dynamics of the real system helps avoid the pitfall of fitting an incorrect model to a single set of biological data.
Our novel sensitivity analysis technique makes use of the Vargha–Delaney A test in assessing a simulation's robustness to parameter perturbation. In addition, a global sensitivity analysis technique that makes use of latin hypercube design quantifies the relative influence of each parameter on simulation dynamics. When considered in the context of information from the real domain (not shown in this article), these analyses can help qualify the certainty of predictions arising from in silico experimentation. Predictions that rest on a highly influential parameter holding a value within a range more specific than the biological literature can attest to undermine the certainty of that prediction.
Though Vargha and Delaney present guidelines for the A test scores that correspond to large differences between two sets of results, these scores are not well justified in the real domain of EAE investigated here. Ongoing work is examining how to assign an EAE severity score, a measure used in the wet-lab, to the execution of the simulation. Such an analysis will allow results from in silico experimentation to be better related to activities being conducted in the wet-lab. This is challenging because the EAE severity score is a subjective analysis of the level of autoimmunity being experienced, and results from observing the entire experimental animal, many aspects of which are not represented in the simulation.
6. Materials and methods
6.1. EAE simulation and statistical analysis
The EAE simulation used in this articleFootnote 6 is coded in Java and makes use of the MASON [Citation26,Citation27] library to provide simulation infrastructure. Random number seeds for simulation execution are supplied such that each simulation execution makes use of a unique seed. All statistical analyses are performed using Matlab. Non-parametric statistics are used throughout.
6.2. Compilation of median-averaged graphs
The EAE simulation provides data on specific metrics of interest for every hour of simulated time. Unless otherwise stated averaged graphs are compiled from 500 simulation executions. Each sample in simulated time for each metric is treated as a dependent statistical variable. The 500 simulation runs form a distribution of 500 sample points for each of these variables, and it is from here that median values are extracted.
6.3. Compilation of response distributions
The nine responses identified in and Section 4.2 are calculated for each individual simulation execution. Analyses of experimentation making use of multiple (n) simulation executions treats each of the nine responses as a dependent variable, drawing a population of n samples for each response from the n simulation executions. Note that the median response value as drawn from a population of n simulation runs will not necessarily equal the single response value derived from compiling the median execution of n simulation executions (as described above, in creating averaged graphs).
6.4. Calculating the A test
The following Matlab code is used to generate A test scores in this article. Note that it is only valid if the two distributions being compared contain the exact same number of samples.
function A = Atest(X,Y) | |||||
[p,h,st] = ranksum(X,Y,'alpha',0.05); | |||||
N = size(X,1); M = size(Y,1); | |||||
A = (st.ranksum/N – (N + 1)/2)/M; | |||||
Acknowledgements
Paul Andrews is funded by EPSRC grant EP/E053505/1. Work in Dr. Kumar's laboratory has been supported by grants from the National Institutes of Health, USA.
Notes
1. The EPSRC funded Complex System Modelling and Simulation infrastructure (CoSMoS) project. http://www.cosmos-research.org/.
2. With regard to simulation, predictive results are those which highlight possible scenarios that could arise in the real domain being modelled.
3. This is achievable in the real domain through a number of different interventions [21], whilst in the simulation it is trivial to revoke the ability of CD8Tregs to kill CD4Th1 cells, hence disabling regulatory activity.
4. One may not be certain that the prediction definitely does rely on this, since abstraction dictates that the relationship between simulation parameters and domain parameters is not exact. The analysis can, however, indicate that caution must be exercised when interpreting simulation results.
5. Samples from parameter space obtained using latin hypercube sampling can be computationally expensive to execute in the simulation. For example, parameters might be chosen that generate a lot of cells that do not die quickly. As such, only 300 simulation executions are performed here.
6. Available upon request from the corresponding author.
References
- Kirschner , D.E. and Linderman , J.J. 2009 . Mathematical and computational approaches can complement experimental studies of host-pathogen interactions . Cell. Microbiol. , 11 : 531 – 539 .
- Bauer , A.L. , Beauchemin , C.A. and Perelson , A.S. 2009 . Agent based modeling of host-pathogen systems: The successes and challenges . Inf. Sci. , 179 : 1379 – 1389 .
- Ray , J.C.J. , Flynn , J.L. and Kirschner , D.E. 2009 . Synergy between individual TNF-dependent functions determines granuloma performance for controlling Mycobacterium tuberculosis infection . J. Theor. Biol. , 182 : 3706 – 3717 .
- Cohen , I.R. 2007 . Modeling immune behavior for experimentalists . Immunol. Rev. , 216 : 232 – 236 .
- Efroni , S. , Harel , D. and Cohen , I.R. 2003 . Toward rigorous comprehension of biological complexity: Modeling, execution, and visualization of thymic T-cell maturation . Genome Res. , 13 : 2485 – 2497 .
- Cohen , I.R. 2007 . Real and artificial immune systems: Computing the state of the body . Nat. Rev. Immunol. , 7 : 569 – 574 .
- Young , D. , Stark , J. and Kirschner , D. 2008 . Systems biology of persistent infection: Tuberculosis as a case study . Nat. Rev. Microbiol. , 6 : 520 – 528 .
- Swerdlin , N. , Cohen , I.R. and Harel , D. 2008 . The lymph node B cell immune response: Dynamic analysis in silico . Proc. IEEE , 96 : 1421 – 1443 .
- Chakraborty , A.K. and Das , J. 2010 . Pairing computation with experimentation: A powerful coupling for understanding T cell signalling . Nat. Rev. Immunol. , 10 : 59 – 71 .
- Kitano , H. 2002 . Computational systems biology . Nature , 420 : 206 – 210 .
- Andrews , P.S. , Polack , F.A.C. , Sampson , A.T. , Stepney , S. and Timmis , J. 2010 . “ The CoSMoS Process Version 0.1: A Process for the Modelling and Simulation of Complex Systems ” . In YCS-2010-453, Department of Computer Science , York : University of York .
- Langman , R.E. and Cohn , M. 2000 . Editorial introduction . Semin. Immunol. , 12 : 159 – 162 .
- Helton , J.C. 2008 . “ Uncertainty and sensitivity analyss for models of complex systems ” . In Computational Methods in Transport: Verification and Validation , Edited by: Barth , T.J. , Griebel , M. , Keyes , D.E. , Nieminen , R.M. , Roose , D. and Schlick , T. 207 – 228 . Berlin , Springer : Heidelberg .
- Saltelli , A. , Chan , K. and Scott , E.M. , eds. 2000 . Sensitivity Analysis, Wiley Series in Probability and Statistics , New York : Wiley .
- Marino , S. , Hogue , I.B. , Ray , C.J. and Kirschner , D.E. 2008 . A methodology for performing global uncertainty analysis and sensitivity analysis in systems biology . J. Theor. Biol. , 254 : 178 – 196 .
- Beauchemin , C. , Samuel , J. and Tuszynski , J. 2005 . A simple cellular automaton model for inuenza A viral infections . J. Theor. Biol. , 232 : 223 – 234 .
- Kumar , V. 2004 . Homeostatic control of immunity by TCR peptide-specific Tregs . J. Clin. Invest. , 114 : 1222 – 1226 .
- Kumar , V. and Sercarz , E. 2001 . An integrative model of regulation centered on recognition of TCR peptide/MHC complexes . Immunol. Rev. , 182 : 113 – 121 .
- Smith , T.R.F. , Maricic , I. , Ria , F. , Schneider , S. and Kumar , V. 2010 . CD8α + dendritic cells prime TCR-peptide-reactive regulatory CD4+FoxP3− T cells . Eur. J. Immunol. , 40 ( 7 ) : 1906 – 1915 .
- Read , M. , Andrews , P.S. , Timmis , J. and Kumar , V. August 2009 . “ A Domain Model of Experimental Autoimmune Encephalomyelitis ” . In Workshop on Complex Systems Modelling , Edited by: Simulation , S. , Stepney , P. , Welch , P.S. , Andrews and Timmis , J. August , 9 – 44 . Beckington , , UK : Luniver Press .
- Kumar , V. , Stellrecht , K. and Sercarz , E. 1996 . Inactivation of T cell receptor peptide-specific CD4 regulatory T cells induces chronic experimental autoimmune encephalomyelitis . J. Exp. Med. , 184 : 1609 – 1617 .
- Madakamutil , L.T. , Maricic , I. , Sercarz , E.E. and Kumar , V. 2008 . Immunodominance in the TCR repertoire of a TCR peptide-specific CD4+ Treg population that controls experimental autoimmune encephalomyelitis . J. Immunol. , 180 : 4577 – 4585 .
- Menezes , J.S. , Elzenvan den , P. , Thornes , J. , Huffman , D. , Droin , N.M. , Maverakis , E. and Sercarz , E.E. 2007 . A public T cell clonotype within a heterogeneous autoreactive repertoire is dominant in driving EAE . J. Clin. Invest. , 117 : 2176 – 2185 .
- Vargha , A. and Delaney , H.D. 2000 . A critique and improvement of the CL common language effect size statistics of McGraw and Wong . J. Edu. Behav. Stat. , 25 : 101 – 132 .
- McKay , M.D. , Beckman , R.J. and Conover , W.J. 1979 . A comparison of three methods for selecting values of input variables in the analysis of output from a computer code . Technometrics , 21 : 239 – 245 .
- Balan , G.C. , Cioffi-Revilla , C. , Luke , S. , Panait , L. and Paus , S. MASON: A Java Multi-Agent Simulation Library . Proceedings of the Agent 2003 Conference . Argonne , IL . Argonne National Laboratory .
- Luke , S. , Cioffi-Revilla , C. , Panait , L. and Sullivan , K. MASON: A New Multi-Agent Simulation Toolkit . Proceedings of the 2004 Swarmfest Workshop . Fairfax , VA . George Mason University .