?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
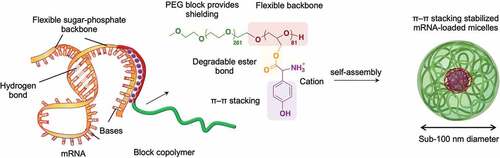
Messenger RNA (mRNA) therapeutics have recently demonstrated high clinical potential with the accelerated approval of SARS-CoV-2 vaccines. To fulfill the promise of unprecedented mRNA-based treatments, the development of safe and efficient carriers is still necessary to achieve effective delivery of mRNA. Herein, we prepared mRNA-loaded nanocarriers for enhanced in vivo delivery using biocompatible block copolymers having functional amino acid moieties for tunable interaction with mRNA. The block copolymers were based on flexible poly(ethylene glycol)-poly(glycerol) (PEG-PG) modified with glycine (Gly), leucine (Leu) or tyrosine (Tyr) via ester bonds to generate block catiomers. Moreover, the amino acids can be gradually detached from the block copolymers after ester bond hydrolyzation, avoiding cytotoxic effects. When mixed with mRNA, the block catiomers formed narrowly distributed polymeric micelles with high stability and enhanced delivery efficiency. Particularly, the micelles based on tyrosine-modified PEG-PG (PEG-PGTyr), which formed a polyion complex (PIC) and π–π stacking with mRNA, displayed excellent stability against polyanions and promoted mRNA integrity in serum. PEG-PGTyr-based micelles also increased the cellular uptake and the endosomal escape, promoting high protein expression both in vitro and in vivo. Furthermore, the PEG-PGTyr-based micelles significantly extended the half-life of the loaded mRNA after intravenous injection. Our results highlight the potential of PEG-PGTyr-based micelles as safe and effective carriers for mRNA, expediting the rational design of polymeric materials for enhanced mRNA delivery.
1. Introduction
Messenger RNA (mRNA) is at the center of many innovative genetic treatments due to its ability to generate a variety of therapeutic proteins in target cells [Citation1]. This interest is driven by mRNA’s predictable expression profile, favorable safety characteristics, ability to express protein in many cell types (even non-dividing cells) [Citation2], simple preparation, and flexible application [Citation3]. Moreover, the recent approvals of SARS-CoV-2 vaccines confirmed the clinical applicability of mRNA [Citation4,Citation5]. However, in applications other than vaccines, the immunostimulatory properties of mRNA, its fragility in physiological environments and its inability to cross the cellular membrane have limited its implementation [Citation3,Citation6]. Thus, the development of safe and effective carriers is crucial to overcome the challenges faced by mRNA in further clinical applications [Citation7].
Nanocarriers based on different materials, such as lipids, polymers and inorganic nanoparticles, are under intense investigation for mRNA delivery [Citation7–11]. Among them, polyion complex (PIC) polymeric micelles using block catiomers with a hydrophilic neutral segment and a cationic block are attractive nanocarriers due to their protective core-shell nanostructure, accessible synthetic procedures and tunable features for enhancing mRNA delivery [Citation3,Citation12,Citation13]. To promote the transport of mRNA to target cells in vivo, it is essential to design micelles capable of overcoming the enzymatic degradation of mRNA [Citation14] and dissociation of the PIC by negatively charged glycans on cell surfaces and plasma [Citation15,Citation16]. Thus, enormous efforts have been dedicated to engineer polymeric structures for promoting both structural and mRNA stabilities [Citation8,Citation17–20]. In this regard, we have recently found that block catiomers containing flexible polycation segments based on polyether backbones stabilized the mRNA-loaded micelles against polyanion and enzymatic attack by enhancing the association with mRNA compared to catiomers having relatively more rigid polycation segments based on polyamide bonds [Citation21]. Such strong polymer-nucleic acid interactions promoted the integrity in biological environments and improved cellular uptake [Citation21,Citation22], demonstrating the relevance of controlling polycation flexibility for improving delivery efficiency. Nevertheless, once the micelles reach the cytosol of the target cells, the strong interactions between the polymers and mRNA should be damped for facilitating mRNA release and boosting protein translation.
Herein, we developed mRNA-loaded micelles based on flexible block catiomers bearing hydrolyzable functional amino groups for improved structural stabilization in in vivo conditions and superior intracellular delivery. The functional groups were inspired by the great contribution of π-π stacking between RNA molecules and the aromatic residues in proteins to the stability of RNA-protein complexes [Citation23,Citation24]. Thus, we modified the side chain of flexible poly(ethylene glycol)-poly(glycerol) (PEG-PG) with tyrosine (Tyr) units for enhancing the structural integrity and protecting mRNA. The Tyr moieties were conjugated via hydrolyzable ester bonds for enabling mRNA release inside the cells, as well as reducing the cytotoxicity of the catiomer. To investigate the contribution of the π-π stacking on the delivery efficiency, we also prepared control block copolymers conjugated with glycine (Gly) and leucine (Leu), which would stabilize the micelle core through its hydrophobic isobutyl residue. Our results showed that the micelles prepared from the Tyr-modified PEG-PG (PEG-PGTyr) displayed excellent stability against polyanion dissociation and effectively protected mRNA in serum due to their PIC and π–π interactions. Moreover, the PEG-PGTyr-based micelles (PEG-PGTyr/m) significantly extended the half-life of the mRNA in the bloodstream after intravenous injection. In addition, PEG-PGTyr/m promoted the transfection in vitro and in vivo upon intramuscular injection, leading to significantly higher protein expression. These findings indicate an innovative approach for improving the stability and functionality of mRNA/polymer assemblies via π–π stacking to generate efficient delivery vehicles.
2. Materials and methods
2.1. Material
n-Butylamine and β-benzyl-L-aspartate-N-carboxyanhydride (BLA-NCA) were obtained from NOF Corporation (Tokyo, Japan). Ethylene oxide was obtained from 3M Japan Co., Ltd. (Tokyo, Japan). Tetrahydrofuran (THF) (super dehydrated, purity >99.5%) and Toluene (super dehydrated, purity >99.5+%) were purchased from Kanto Chemical, Co., Inc. (Tokyo, Japan). Ethanol, diethyl ether, N,N-dimethylformamide (DMF) and piperidine (purity >98.0%) were purchased from Fujifilm Wako Pure Chemical, Co., Inc. (Tokyo, Japan). 1,1,3,3-Tetramethylguanidine (purity >99.0%), 2-methoxyethanol (purity >99.0%) epichlorohydrin (purity >99.0%), triisobutylaluminum, N,N-dimethylformamide (purity >99.5%), 4-dimethylaminopyridine (purity >99.0%), 1-(3-dimethylaminopropyl)-3-ethylcordiimide hydrochloride (purity >98.0%), N-[(9H-fluoren-9-ylmethoxy)carbonyl]glycine (purity >98.0%), N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-leucine (purity >98.0%) and N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-tyrosine (purity >95.0%) were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Sodium hydroxide, hydrochloric acid and HEPES buffer were purchased from Sigma-Aldrich (St. Louis, MO, U.S.A). Fetal bovine serum (FBS), Dulbecco’s Modified Eagle’s Medium and Penicillin-Streptomycin were obtained from Thermo Fisher Scientific (Waltham, MA, U.S.A).
2.2. Cells
Human Embryonic Kidney cells 293 (HEK293) cells and human hepatocellular carcinoma HuH7 cells were obtained from Riken BioResource Center (Tsukuba, Japan).
2.3. Animals
All animal studies described below were approved by the Animal Care and Use Committee of the University of Tokyo (Tokyo, Japan) (KA21–1). Balb/c mice (7-week-old, female) were purchased from Charles River Laboratories (Yokohama, Japan).
2.4. Polymer synthesis
2.4.1. Synthesis of α-methoxy-poly(ethylene glycol)-block-poly(glycerol) copolymer (PEG-PG) block copolymer
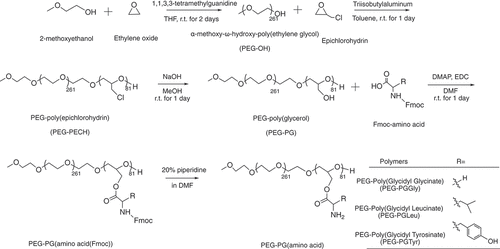
PEG-PLL was prepared as described in a previous paper [Citation25]. The synthesis route of PEG-PG is shown in . First, α-methoxy-ω-hydroxy-poly(ethylene glycol) (PEG-OH) was prepared through ring opening polymerization (ROP). Briefly, the 1,1,3,3-tetramethylguanidine (1.0 mmol, 130 µL) and 2-methoxyethanol (0.10 mmol, 7.9 µL) were dissolved in a solution of 60 mL anhydrous THF. The mixture was stirred for 10 min under argon atmosphere, and then liquid ethylene oxide (27 mmol, 1.4 mL, 0°C) was added to the solution through a cooled syringe. The reaction mixture was stirred at room temperature for 2 days under argon atmosphere and dried under reduced pressure to remove unreacted monomers and THF. Furthermore, epichlorohydrin (10 mmol, 784 µL) in 11 mL of anhydrous toluene was added to the polymer solution. The reaction mixture was stirred for 10 min under Argon atmosphere, followed by the addition of triisobutylaluminum (1.0 µmol, 2 mL) under Argon atmosphere. The reaction was left stirring at room temperature for 1 day. The reaction was then stopped by adding 3 mL of ethanol. The resulting polymer was precipitated in diethyl ether (400 mL) to obtain α-methoxy-poly(ethylene glycol)-block-poly(epichlorohydrin) (PEG-PECH) block copolymer. Then, elemental analysis was performed using a Vario MACRO cube Elemental Analyzer (Elemental, Germany) to determine the molecular weight of the polymer and the length of the PECH block. To prepare PEG-PG, 5 mL of 1 M NaOH (aq) was added to a stirred suspension of PEG-PECH (5.0 µmol,100 mg) in 5 mL methanol. After reacting at room temperature for 1 day, the polymer solution was purified by dialysis against 0.01 M HCl (aq) for 3 times and pure water for 3 times with a 6–8 kD MWCO dialysis membrane. Finally, PEG-PG was collected as a white powder after lyophilization. The polymer was characterized again by elemental analysis using a Vario MACRO cube Elemental Analyzer to confirm the removal of Cl.
Figure 1. Scheme of polymer synthesis. Synthesis scheme of α-methoxy-poly(ethylene glycol)-block-poly(glycerol) (PEG-PG) copolymers from ring opening polymerization and amino-acid conjugated PEG-Poly(Glycidyl Glycinate) (PEG-PGGly), PEG-Poly(Glycidyl Leucinate) (PEG-PGLeu) and PEG-Poly(Glycidyl Tyrosinate) (PEG-PGTyr).
2.4.2. Synthesis of PEG-Poly (Glycidyl Glycinate) (PEG-PGGly), PEG-Poly(Glycidyl Leucinate) (PEG-PGLeu) and PEG-Poly(Glycidyl Tyrosinate) (PEG-PGTyr)
α-Methoxy-poly(ethylene glycol)-block-poly(glycidyl-amino acids) were synthesized by EDC hydrochloride mediated conjugations, as follows: PEG-PG (5.0 µmol,100 mg) was dissolved in 10 mL of DMF, followed by the addition of 4-dimethylaminopyridine (0.44 mmol, 54 mg) and 1-(3-dimethylaminopropyl)-3-ethylcordiimide hydrochloride (4.1 mmol, 637 mg). Then, 4.1 mmol of Fmoc-protected amino acids (1.2 g N-[(9H-fluoren-9-ylmethoxy)carbonyl]glycine; 1.4 g N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-leucine; 1.6 g N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-tyrosine) were added to the solution, and the resulting mixtures were stirred at room temperature for 1 day. The resulting polymer solutions were precipitated in diethyl ether (400 mL) to obtain PEG-PGGly(Fmoc), PEG-PGLeu(Fmoc) and PEG-PGTyr(Fmoc). Finally, the protecting Fmoc group was removed by treatment with a 20% solution of piperidine in DMF at a concentration of 10 mg/mL. The samples were precipitated in diethyl ether (400 mL) to obtain PEG-PGGly, PEG-PGLeu and PEG-PGTyr. The products were characterized by 1H-NMR using a JEOL EX400 spectrometer (Tokyo, Japan) at 400 MHz. The number- and weight-average molecular weight (Mn, Mw), and polydispersity index (PDI =Mw/Mn) of the polymer were tested by gel permeation chromatography using TOSOH HLC-8220 system (Tokyo, Japan) (eluent: 10 mm LiCl containing DMF; temperature: 40°C; flow rate: 0.5 mL/min; detector: refractive index). Polyethylene glycol standard ReadyCal calibration kit (Sigma-Aldrich, St. Louis, MO, U.S.A) with molar masses ranging from ~250 to ~45,000 Da was used for the calibration of the DMF-GPC system.
2.4.3. Synthesis of Homo-poly(α,β-aspartic acid) (Homo-P(asp))
n-Butylamine and β-benzyl-L-aspartate-N-carboxyanhydride (BLA-NCA) were dissolved in super-dehydrated DMF under Argon flow. The mixture was then stirred at room temperature for 3 days. The resulting polymer solution was precipitated in diethyl ether to obtain homo-poly(β-benzyl-L-aspartate) (Homo-PBLA). Deprotection of the benzyl groups was performed by dissolving the prepared Homo-PBLA in a 0.5 N NaOH solution and stirring at room temperature for 1 h. The reaction solution was then dialyzed against pure water and lyophilized. The final product was characterized by 1H-NMR using a JEOL EX400 spectrometer (Tokyo, Japan) at 400 MHz (Supplementary Figure S1).
2.5. In vitro transcribed mRNA
Gaussian luciferase (Gluc) and Firefly luciferase (Fluc) mRNA were prepared from in vitro transcription. First, plasmid DNA temples were prepared by inserting the corresponding protein-coding sequences and 120 bp poly A/T sequence into the pSP73 vector (Promega, Madison, WI, U.S.A). Plasmid DNA was then linearized and transfected to produce Gluc and Fluc mRNA using mMESSAGE mMACHINE T7 Ultra Kit (Thermo Fisher Scientific, Waltham, MA, U.S.A). The resulting mRNA was purified with RNeasy Mini Kit (Qiagen, Hilden, Germany). mRNA concentration was finally determined by NanoDrop 3300 spectrophotometer (Thermo Fisher Scientific).
2.6. mRNA labelling
Gluc mRNA was labeled using Label IT Nucleic Acid Labeling Kit according to the manufacturer’s protocol. Briefly, 25 µg mRNA was mixed with 10× labeling buffer A and Label IT® regent at 1:2 (v:w) ratio of label reagent to mRNA. The resulting solution was incubated at 37°C for 90 min. Labeled mRNA was purified by using ethanol precipitation.
2.7. Hydrolysis of pendant moieties in PEG-PGGly, PEG-PGLeu and PEG-PGTyr
The detachment of the amino acids from PEG-PGGly, PEG-PGLeu and PEG-PGTyr was investigated by incubating 10 mg of the polymers in 10 mL pH 7.4 phosphate buffered saline (10 mM phosphate with 150 mm NaCl) and 10 mL pH 5.0 sodium acetate buffered saline (10 mM sodium acetate with 150 mM NaCl) at 37°C. At defined time intervals, the polymer solutions were placed in a Vivaspin 500 protein concentrator spin column (molecular weight cutoff (MWCO): 3,000 Da) and centrifuged at 5,000 rpm for 1 h to separate the detached amino acids from the polymers. Finally, the solution was lyophilized and the amount of amino acids was quantified by measuring the concentration of primary amines by fluorometric assay with fluorescamine.
2.8. Cytotoxicity
HEK293 cells were seeded on 96-well plates at 5 × 103 cells/well. After 24 h, the cells were washed twice with PBS. Then, PEG-PGTyr, PEG-PGLeu, PEG-PGGly and PEG-PLL were added to cell culture media at concentrations of 0.02, 0.04, 0.08, 0.2, 0.4, 0.8, 1 mg/mL. After 24-h incubation, the cell viability was determined by cell-counting kit-8 (CCK-8; Dojindo Molecular Technologies Inc., Tokyo, Japan) assay following the manufacturer’s protocol.
2.9. Micelle preparation and characterization
mRNA and PEG-PGGly, PEG-PGLeu and PEG-PGTyr were dissolved in 10 mM HEPES buffer (pH 7.3) at 40 ng/µL and 1 mg/mL, respectively. The mRNA-loaded micelles were prepared via mixing freshly prepared polymer solution with mRNA solution at 1:1 volume. The polymer and mRNA amounts were fixed at the molar ratio of primary amines in polymer and phosphates in mRNA (N/P ratio) from 1 to 5. After mild vortexing, the resulting micelle solution was incubated at 4°C for 1 h to allow the micelles to stabilize. Then, electrophoretic analysis of micelles at varying N/P ratios ranging from 1 to 5 on 1% agarose gel (15 µL sample solution containing 500 ng of mRNA was applied to each well and mRNA was visualized using Midori Green Direct dye). The Z-average diameter and polydispersity index (PDI) of the micelles were characterized by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern Instruments Ltd., UK).
2.10. Micelle stability against polyanions
Micelles containing Cy5-labeled mRNA (20 ng/µL) were incubated with heparin at different S/P ([sulfate in heparin]/[phosphate in mRNA]) ratios. After 6-h incubation, fluorescence correlation spectroscopy (FCS) measurements were performed using an LSM-780 confocal laser scattering microscope (CLSM; Carl Zeiss AG, Oberkochen, Germany) with He-Ne laser (633 nm) scanning. The Cy5 dye (Lumiprobe Co., U.S.A) was used as a standard to calculate the diffusion coefficient of the Cy5-labeled mRNA and the micelles loading Cy5-labeled mRNA. The following theoretical model, which describes a general case of a sample containing M populations of fluorescent particles was used for fitting of autocorrelation curve. Each population of fluorescent particles was characterized by its diffusion time ,
is the molecular brightness,
is the fraction of the particle number and
is the structure parameter [Citation26].
2.11. Hydrolysis of esters and esterase-responsive mRNA release
Micelles (mRNA concentration: 50 ng/μL) containing Cy5-labeled GLuc mRNA were incubated in HEPES buffer (pH 7.4) with and without esterase. The final esterase concentration was 2.85 µg/mL. After 24-h incubation, the micelles were incubated with heparin at different S/P ([sulfate in heparin]/[phosphate in mRNA]) ratios, the final mRNA concentration was 20 ng/µL. After 1-h incubation, fluorescence correlation spectroscopy (FCS) measurements were performed as described in 2.10.
2.12. Micelle stability in serum
Micelles containing GLuc mRNA were incubated with 50% fetal bovine serum (FBS; Thermo Scientific Fisher Inc., U.S.A) at 37°C for 15 min. The final mRNA concentration was 50 ng/μL. The mRNA was then extracted by the RNeasy Mini Preparation Kit (Qiagen, Hilden, Germany). The mRNA was then reverse-transcribed with the ReverTra Ace qPCR RT Master Mix kit (Toyobo Life Science, Osaka, Japan), followed by quantitative real-time PCR (qRT-PCR) using an ABI Prism 7500 Detector (Applied Biosystems, Foster City, CA, U.S.A) and a primer pair for GLuc (Forward: TGAGATTCCTGGGTTCAAGG, Reverse: GTCAGAACACTGCACGTTGG).
2.13. π–π stacking assessment by Tyr fluorescence quenching
A solution of PEG-PGTyr (200 μL; 1 mg/mL) was placed in 96-well plates. Next, mRNA (40 ng/µL) and Homo-PAsp solution (1 mg/mL) was added to the polymer at N/P = 3 and N/[COO] = 3, respectively. The fluorescence intensity of Tyr in the solution was then evaluated at the excitation wavelength (λex) of 280 nm and the emission wavelength (λem) of 350 nm by multimode microplate reader (Tecan Group Ltd., Switzerland).
2.14. Cellular uptake and endosome escape study
HuH-7 cells were seeded on an eight-well chambered borosilicate cover glass (Lab Tek) a day before transfection at a density of 10,000 cells/well and incubated for 24 h in DMEM containing 10% FBS and 1% penicillin/streptomycin under 5% CO2 at 37°C. Next, naked Cy5-labeled mRNA and Cy5-labeled mRNA-loaded micelles were applied to each well (700 ng mRNA per well, relative fluorescence intensity: 400 RFU). The cellular uptake of the micelles was evaluated after 8 h using an LSM-780 with a 40 × objective (C-Apochromat, Carl Zeiss, Germany). The lysosomes were stained by LysoTracker Green, and the nuclei were stained by Hoechst 33,342. To evaluate the intracellular distribution of mRNA loaded in micelles, the rate of colocalization of the Cy5 signal with that of LysoTracker Green was quantified.
2.15. In vitro transfection
HuH-7 cells were seeded in a 96-well plate a day before transfection at a density of 30,000 cells/well. After 24-h incubation in DMEM containing 10% FBS and 1% penicillin/streptomycin under 5% CO2 at 37°C, the cell culture medium was replenished with media containing GLuc mRNA, PEG-PGGly/m and PEG-PGTyr/m containing 1000 ng of Gluc mRNA. After 24 h, the luminescence intensities of 50 μL supernatant were evaluated by GloMax 96 Microplate Luminometer (Promega, Madison, WI, U.S.A) using Renilla Luciferase Assay System (Promega, Madison, WI, U.S.A).
2.16. Blood circulation
Micelle solutions (200 μL) containing 4 μg of mRNA were administered to BALB/c mice (female, 7 weeks old, Charles River Laboratories Japan Inc., Kanagawa, Japan) by intravenous injection. Blood (2 μL) was collected from the tail vein of each mouse after 2.5, 5 and 10 min of administration, and the isolation of mRNA from the blood was performed by immediately mixing the blood samples with 350 μL 1% 2-mercaptoethanol containing RLT buffer from the RNeasy Mini Kit (Qiagen, Hilden, Germany). mRNA was then purified using RNeasy Mini Kit (Qiagen, Hilden, Germany), followed by reverse transcription of mRNA using ReverTra Ace qPCR RT Master Mix kit (Toyobo Life Science, Osaka, Japan). Then, qRT-PCR analysis was performed as described in point 2.12.
2.17. In vivo transfection
Micelle solutions (70 μL) containing 5 μg of firefly luciferase mRNA (Fluc) were administered by intramuscular injection to Balb/c mice (female, 7 weeks-old; Charles River Laboratories Japan, Inc). After 9 h of administration, the mice were intraperitoneally injected with 200 μL 50 mg/mL luciferin solution. The mice were anesthetized with isoflurane via inhalation in a box chamber, and the luciferase expression was evaluated after 15 min using an in vivo bioluminescence imaging system (IVIS Spectrum SP-BFM-T1, PerkinElmer, Waltham, MA, U.S.A).
3. Results
3.1. Block copolymer synthesis
α-Methoxy-ω-hydroxy-poly(ethylene glycol) (PEG-OH) was synthesized via ROP initiated by the terminal hydroxyl of 2-methoxyethanol, with an organometallic catalyst (triisobutylaluminum). The synthesized PEG-OH showed a unimodal molecular weight distribution (Supplementary Figure S3a) with 261 units as determined by 1H-NMR (Supplementary Figure S2). Next, the PEG-OH was used as the initiator for the ROP of epichlorohydrin. Triisobutylaluminum was again used as a catalyst. The resulting PEG-PECH polymer showed a unimodal molecular weight distribution (Suplementary Figure S3b). Elemental analysis of the chloride atoms in the PECH segments determined the number of PECH units in the block copolymer to be 81 (Supplementary Table S1). After treating the polymer with NaOH, the chloride groups on the side chain were completely converted to hydroxyl groups, resulting in PEG-PG as confirmed by the disappearance of the Cl atoms in the elemental analysis. Unimodal molecular weight distribution is also observed for PEG-PG (Suplementary Figure S3c). The hydroxyl groups of the PEG-PG were conjugated with three different Fmoc-protected amino acids (Gly-Fmoc, Leu-Fmoc and Tyr-Fmoc) via condensation reaction to form hydrolyzable ester bonds. Thus, the amino acid-modified block copolymers were obtained by deprotecting the Fmoc groups of the amino acids, resulting in PEG- PEG-PGGly, PEG-PGLeu and PEG-PGTyr. The conjugation rates of glycine, leucine and tyrosine to PEG-PG were calculated to be 92.5%, 98.7% and 93.7% by 1H-NMR, respectively ( and Supplementary Figure S4). The block copolymers also maintained narrow molecular weight distributions (Mw/Mn), ranging from 1.03 to 1.08 ().
Table 1. Characterization of PEG-PGGly, PEG-PGLeu and PEG-PGTyr.
3.2. Detachment of amino acids and cytotoxicity of polymers
The hydrolytic degradation of the PEG-PGGly, PEG-PGLeu and PEG-PGTyr was evaluated by dissolving the polymers at pH 7.4 10 mM PBS and pH 5.0 10 mM sodium acetate at 37°C. The degradation was monitored by measuring the amount of free amino acid in the solution as a function of exposure time. Comparable detachment of amino acids from the polymers was observed at conditions mimicking extracellular pH (pH 7.4) and endosomal pH (pH 5.0) (). Thus, after 6-h incubation, more than 25% of the conjugated amino acids were detached, while at 24 h around 50% of the amines were detached. The control PEG-PLL remained stable throughout the experiment . The detachment of the amino acids would be beneficial for releasing the mRNA after cellular uptake, as well as reducing the cytotoxicity by decreasing the positive charges in the polycation segment.
Figure 2. Evaluation of amino acid detachment and cytotoxicity of polymers. Remaining amines in the block copolymers after incubation in pH 7.4 10 mM PBS (a) and pH 5.0 10 mM sodium acetate (b) at 37°C determined by fluorometric assay with fluorescamine. Data are presented as the average ± S.D. (n = 3). (c) Cytotoxicity of polymers against HEK293 cells. The cells were incubated with the polymers for 24 h. Data are presented as the average ± standard deviation (n = 3).
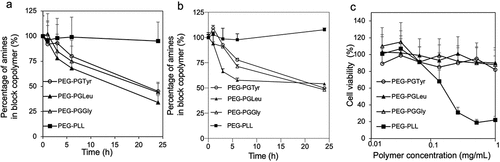
In this regard, the cytotoxicity of the block copolymers was evaluated in HEK293 cells after 24-h incubation. The results showed that the cell viability was not affected by PEG- PEG-PGGly, PEG-PGLeu and PEG-PGTyr (). In contrast, the addition of conventional PEG-PLL to the cells resulted in a much lower cell viability. These results support the enhanced safety of PEG-PGGly, PEG-PGLeu and PEG-PGTyr block copolymers.
3.3. Micelle formation and characterization
PIC micelles were assembled after mixing PEG-PGGly, PEG-PGLeu and PEG-PGTyr, with mRNA in 10 mM HEPES buffer (pH 7.3) at different N/P ratios. The condensation of the mRNA by polymers at various N/P ratios was analyzed by gel retardation assay and DLS measurement. During electrophoresis, the micelles, which are less negatively charged and heavier than free mRNA, are retained in the wells. In contrast, uncomplexed mRNA can migrate into the gel. Supplementary Figure S5 shows that the N/P ratio of 3 was sufficient to condense the mRNA in PEG-PGGly/m. Also, the PDI of the PEG-PGGly/m decreased as the N/P increased from 1 to 3 (Supplementary Figure S6b). The complexation was more efficient with PEG-PGLeu/m and PEG-PGTyr/m. mRNA was complexed and maintained in the wells even at N/P ≥2 (Supplementary Figure S5). For better comparison, the derived count rate of micelles at N/P 2-5 are normalized to N/P 1. At N/P = 3, micelles with a z-averaged diameter of around 60 nm, and PDI below 0.2 were obtained for all the polymers (Supplementary Figure S6, ). At N/P equal to and higher than 3, the normalized the derived count rate of all the micelles reached a plateau, supporting the formation of the micelles (Supplementary Figure S6). These results are in agreement with our previous reports on mRNA-loaded PIC micelles [Citation21,Citation22].
Table 2. Size of micelles determined by DLS.
3.4. Micelle stability and esterase-responsive mRNA release
PIC-based carriers are susceptible to be disassociated by polyion exchange when exposed to anionic macromolecules (i.e. glycosaminoglycans on cell surfaces) [Citation16,Citation27], which can lead to the premature release and degradation of mRNA in the extracellular space. Thus, we incubated our mRNA-loaded micelles with negatively charged heparin at different [sulfate in heparin]/[phosphate in mRNA] (S/P) ratios for 6 h to compare their stability against polyanions. In this experiment, we used Cy5-labeled mRNA to trace the diffusion coefficient by FCS [Citation28]. Thus, when Cy5-labeled mRNA is loaded into the micelles, the diffusion coefficient decreases (). However, when the mRNA is released, the diffusion coefficient increases. For PEG-PGTyr/m, the diffusion coefficient remained stable at S/P = 2, indicating that the micelles maintained their integrity even when the amount of negative charges in the polyanion doubles that in mRNA. On the other hand, the diffusion coefficient of PEG-PGGly/m started to increase at S/P = 1. Moreover, at S/P = 2, around 50% of the mRNA in PEG-PGGly/m has been released. These results indicate that PEG-PGGly/m is less stable than PEG-PGTyr/m. The introduction of the hydrophobic isobutyl groups in PEG-PGLeu/m improved the stability, but to a lower extent when compared to PEG-PGTyr/m (). These results suggest the importance of the aromatic residue in Tyr for stabilizing the micelles against polyanions.
Figure 3. Stability and release of mRNA-loaded micelles. (a) Diffusion coefficient of fresh micelles after incubation with heparin at different S/P (sulfate in heparin/phosphate in mRNA) ratios for 6 h. (b) After 24-h incubation in HEPES buffer (pH 7.4), the micelles were incubated with heparin at different S/P (sulfate in heparin/phosphate in mRNA) ratios for 1 h, diffusion coefficient was recorded. (c) After 24-h incubation with esterase, micelles were then incubated with heparin at different S/P (sulfate in heparin/phosphate in mRNA) ratios for 1 h, diffusion coefficient was recorded. The upper and lower dashed lines represent the diffusion coefficients of the free mRNA and mRNA loaded micelles, respectively. Data are presented as the mean ± S.D. (n = 10). (d) Remaining mRNA percentage after incubating mRNA-loaded micelles with 50% FBS at 37°C for 15 min, followed by reverse transcription and qRT-PCR quantification. Data are presented as the mean ± S.D. (n = 3). Statistical significance was calculated by one-way ANOVA test. The difference was considered statistically significant with *p < 0.05, ***p < 0.001.
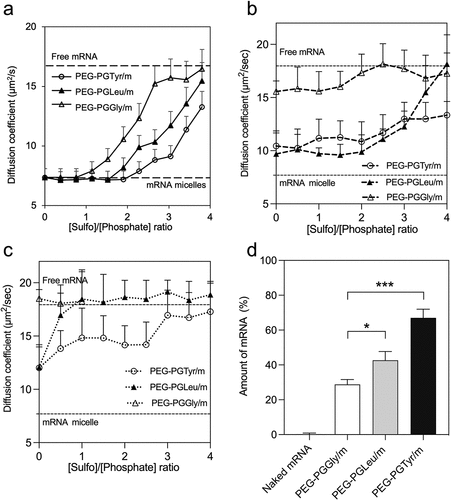
We have characterized the polyanion stability of the micelles after incubating them for 24 h in HEPES buffer (pH 7.4) with and without esterase. After 24-h incubation in HEPES, the diffusion coefficients of micelles were increased even at sulfate-to-phosphate ratio (S/P) = 0 (), suggesting that the molecular weight of the micelles has been decreased. All the micelles were dissociated at lower concentration of polyanions, which was expressed as the S/P in heparin and mRNA, respectively (). When the micelles were exposed to esterase, the micelle dissociation occurred at lower S/P (). The results showed that PEG-PGGly/m incubated with esterase was already dissociated even at S/P = 0. PEG-PGLeu/m and PEG-PGTyr/m incubated with esterase were more stable than PEG-PGGly/m incubated with esterase, though they dissociated at lower S/P than the micelles without esterase. These data confirm the importance of the amino acids for stabilizing the micelles, and indicate that the side groups of the polymers can be hydrolysed to accelerate the release of mRNA. Also, the micelles exhibited esterase-promoted mRNA release, which could have considerable advantages for intracellular activation in some cells, for example, cancer cells [Citation29–31].
3.5. mRNA protection ability against serum
mRNA therapeutics should also be protected against enzymatic attack following in vivo administration. To estimate the ability of the micelles to protect the mRNA cargo, we further incubated the micelle in 50% FBS, which simulates the harsh in vivo conditions [Citation32]. The integrity of mRNA was then determined by quantitative real-time PCR (qRT-PCR). After 15 min incubation, around 70% of the mRNA was still detectable in PEG-PGTyr/m. In contrast, approximately 40% and 30% of the mRNA were detectable in PEG-PGLeu/m and PEG-PGGly/m, respectively (). The stability enhancement was more pronounced in PEG-PGTyr/m compared to PEG-PGLeu/m, which is stabilized by the hydrophobicity of the isobutyl groups. These results suggest that PEG-PGTyr achieved a remarkable stabilization effect, which is probably related to the ability of the polymer to form π–π stacking with mRNA.
3.6. π–π stacking assessment by Tyr fluorescence quenching
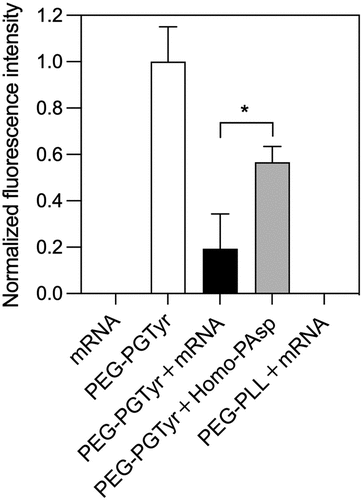
In order to determine the π–π stacking between PEG-PTyr and mRNA in the micelles, we investigated the fluorescence quenching of Tyr moieties in the polymer. Tyr groups have intrinsic fluorescence (λex ~280 nm, λem ~350 nm), and π–π stacking can quench the fluorescence by electron transfer [Citation33,Citation34]. Thus, we investigated the change of fluorescence signal from the Tyr groups of PEG-PGTyr polymers after forming micelles with mRNA. For comparison, we also assembled micelles of PEG-PGTyr polymers with anionic Homo-PAsp, which does not have aromatic residues. We also checked the fluorescence signal of mRNA alone and PEG-PLL/m loading mRNA as controls for the conducted fluorescence emission measurements. The results showed that no fluorescence was detected in these controls (). There is a strong decrease in Tyr fluorescence intensity in PEG-PGTyr/m loading mRNA. The fluorescence quenching of PEG-PGTyr/m was significantly higher than that of the micelles loading Homo-PAsp (). These results support the π–π stacking between the aromatic Tyr groups and mRNA.
Figure 4. π–π stacking assessment by Tyr fluorescence quenching. Normalized fluorescence intensity of PEG-PGTyr polymer and corresponding fluorescence intensities of the mRNA, polymer and micelles. Data are presented as the mean ± S.D. (n = 3). Statistical significance was conducted using a two-tailed Student’s t-test. The difference was considered statistically significant with *p < 0.05.
3.7. In vitro activity
We then explored the in vitro performance of the mRNA-loaded PIC micelles. The ability of the micelles for intracellularly delivering mRNA was studied in Huh7 cells. The uptake was visualized by CLSM using micelles loading Cy5-labeled mRNA. Thus, by quantifying the Cy5 fluorescence intensity, we found that micelles improved mRNA uptake in vitro remarkably (). Moreover, the cellular uptake of PEG-PGTyr/m was about twofold higher than that of PEG-PGGly/m (). The cellular uptake enhancement of PEG-PGTyr/m could be associated with their high stability against polyanion exchange, which may allow them to effectively overcome the glycan barrier on the cell surface.
Figure 5. In vitro delivery of mRNA-loaded micelles. (a) Representative CLSM images of HuH7 cells incubated with micelles for 8 h (scale bar = 20 μm; Blue: Hoechst; Red: Cy5-labeled mRNA; Green: Lysotracker; yellow: colocalization of mRNA and Lysotracker). (b) Cellular uptake quantified by the mean fluorescence intensity of Red channel from 20 cells in each sample. Data are presented as the mean ± S.D. (n = 20). (c) Colocalization coefficients of Red-Green channels quantified from 10 cells in each sample. Data are presented as the mean ± S.D. (n = 10). (d) Transfection in HuH7 cells after 24 h treatment with micelles encapsulating GLuc mRNA. Data are presented as the mean ± S.D. (n = 4). Statistical significance was conducted using one-way ANOVA and Tukey’s post hoc tests (b) and two-tailed Student’s t-test (c,d). The difference was considered statistically significant with **p < 0.01, ***p < 0.001, denoted with asterisks.
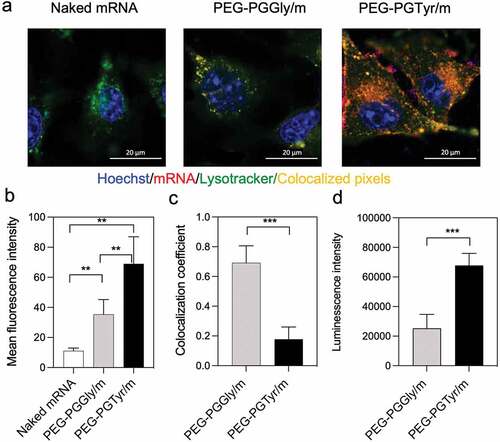
The ability of the micelles to escape from the endosomes was then evaluated by studying the endosomal colocalization. The endo/lysosomes were labeled with LysoTracker Green, and the mRNA in the endosomes was visualized as yellow pixels in the CLSM images (). The colocalization rate of mRNA and endo/lysosomes was quantified by the colocalization coefficient. A low colocalization coefficient indicates a high endosome escape ability. After 8-h incubation of the micelles with the HuH7 cells, PEG-PGTyr/m demonstrated a strong endosomal escape with decreased colocalization coefficient (), which was significantly lower than that of PEG-PGGly/m. The mechanism of the enhanced endosomal escape of PEG-PGTyr/m may be related to the hydrophobic Tyr residue, which may facilitate the destabilization of the endosomal membrane destabilization, as recently observed for carriers bearing aromatic amino acids [Citation35–37].
The transfection ability of the micelles was studied in Huh7 cells after incubation with GLuc mRNA-loaded micelles. Herein, we used a naturally secreted Gluc as a reporter to assess gene expression in culture cells. The protein translation ability was determined by assessing the bioluminescence signal. PEG-PGTyr/m resulted in higher transfection efficiency compared to the PEG-PGGly/m (). We believe that this result can be associated with the enhanced stability against serum by π–π interaction and the improved endosomal escape of the PEG-PGTyr/m.
3.8. mRNA delivery performance in vivo
The ability of the micelles to deliver mRNA was further tested in mice. We first studied the capacity of the micelles to extend the circulation in the bloodstream, which is the ultimate experiment to determine the stability improvement of the micelles. Thus, a qRT-PCR-based assay was applied to assess the mRNA integrity in blood after intravenously injecting the micelles. The results showed that PEG-PGTyr/m prolonged the circulation of mRNA in the blood, with almost 40% of the injected dose remaining at 2.5 min after injection (). This value is twofold higher than that of the PEG-PGGly/m, indicating the superior performance of the PEG-PGTyr/m during blood circulation. Moreover, our previous report showed that mRNA-loaded polymeric micelles based on flexible catiomers achieved around 25% of the injected dose at 2.5 min, which is in agreement with the PEG-PGGly/m [Citation21]. Also, PEG-PLL/m resulted in a significantly lower mRNA amount than the PEG-PGGly/m and PEG-PGTyr/m, with 8.5% of the injected dose remaining at 2.5 min after injection (). Free mRNA cannot be detected in blood 2.5 min after injection due to its fast degradation [Citation32]. At longer time points, the mRNA of PEG-PGTyr/m was still detectable, attaining almost 10% of the injected dose at 10 min after injection. These findings confirmed the high stability of PEG-PGTyr/m in biological settings, and could be attributed to the interactions of the tyrosyl groups and mRNA’s bases in the core of the micelles.
Figure 6. In vivo performance of mRNA-encapsulating micelles. (a) Remaining mRNA amount in the bloodstream of mice after intravenous injection of mRNA-loaded micelles containing 4 μg of mRNA. Data were shown as means ± S.D. (n = 5). Statistical significance was conducted using one-way ANOVA and Tukey’s post hoc tests. (b) Representative in vivo bioluminescence images 9 h after intramuscular injection of Fluc mRNA, PEG-PGGly/m and PEG-PGTyr/m (5 μg mRNA per mouse, n = 3). (c) Quantification of the luminescence signals from the images in b. Data were shown as means ± S.D. (n = 3). Statistical significance was conducted using a two-tailed Student’s t-test. The difference was considered statistically significant with *p < 0.05, **p < 0.01, ***p < 0.001, denoted with asterisks.
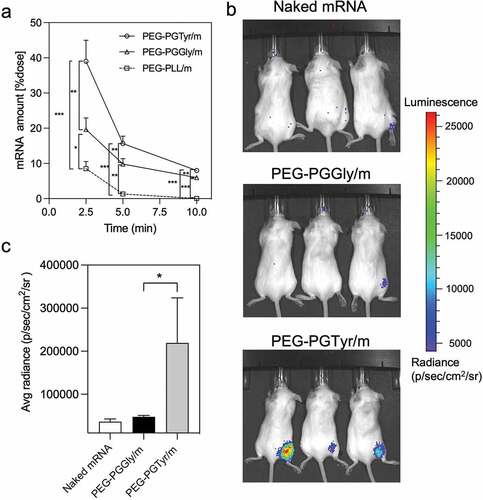
The in vivo mRNA transfection was evaluated in mice after intramuscular administration of Fluc mRNA-loaded PIC micelles. Here, we used Fluc as a reporter to study gene expression in mice due to its non-secreted nature, which makes it beneficial to study the position of protein expression. Nine-hours after administration, the bioluminescent signals were seen at the injected site (). PEG-PGTyr/m treated mice showed strong bioluminescent signals, while almost no bioluminescence was detected from PEG-PGGly/m and naked mRNA (). These results clearly support the ability of PEG-PGTyr/m to improve the efficacy of mRNA delivery in vivo after intramuscular injection, suggesting the potential of this system for vaccination use.
4. Conclusion
Our study showed the enhanced mRNA delivery efficiency of polymeric micelles based on flexible block catiomers modified with functional amino acid moieties. We demonstrated that this system has minimal cytotoxicity due to the inclusion of hydrolyzable ester bonds between the amino groups and the polycation backbone. Moreover, the polymers having pendant tyrosyl moieties stabilized the micelle structure and effectively protected mRNA in the core by promoting π–π stacking with mRNA, which resulted in improved in vitro and in vivo delivery efficiency. These findings provide an innovative strategy for improving the stability and functionality of mRNA/polymer assemblies with high efficacy, and promote the rational design of polymeric materials for enhanced mRNA delivery.
Supplemental Material
Download PDF (648.3 KB)Acknowledgement
This work was supported by the Project for Cancer Research and Therapeutic Evolution (P-CREATE) (Project No. 16 cm0106202h0001; H.C.) from the Japan Agency for Medical Research and Development (AMED), the GAP Fund from The University of Tokyo (H.C.) and the Grants-in-Aid for Scientific Research B (20H04524; H.C.). The study was also partially supported by Grants-in-Aid for Early-Career Scientists (21k18070; T.M.) from the Japan Society for the Promotion of Science (JSPS), GSK Japan Research Grant 2020 from GlaxoSmithKline plc (T.M.) This research was also partially supported by Grant-in-Aid for JSPS Research Fellow (No. 20J13458 to W.Y.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and a fellowship of the Chugai Foundation for Innovative Drug Discovery Science: C-FINDs (W.Y.).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/14686996.2023.2170164.
Additional information
Funding
References
- Chabanovska O, Galow AM, David R, et al. mRNA - a game changer in regenerative medicine, cell-based therapy and reprogramming strategies. Adv Drug Deliv Rev. 2021 Dec;179:114002.
- Lin CY, Perche F, Ikegami M, et al. Messenger RNA-based therapeutics for brain diseases: an animal study for augmenting clearance of beta-amyloid by intracerebral administration of neprilysin mRNA loaded in polyplex nanomicelles. J Control Release. 2016 Aug 10;235:268–13.
- Sahin U, Kariko K, Tureci O. mRNA-based therapeutics–developing a new class of drugs. Nat Rev Drug Discov. 2014 Oct;13(10):759–780.
- Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020 Dec 31;383(27):2603–2615.
- Shroff RT, Chalasani P, Wei R, et al. Immune responses to two and three doses of the BNT162b2 mRNA vaccine in adults with solid tumors. Nat Med. 2021 Nov;27(11):2002–2011.
- Tavernier G, Andries O, Demeester J, et al. mRNA as gene therapeutic: how to control protein expression. J Control Release. 2011 Mar 30;150(3):238–247.
- Kowalski PS, Rudra A, Miao L, et al. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol Ther. 2019 Apr 10;27(4):710–728.
- Uchida S, Perche F, Pichon C, et al. Nanomedicine-Based approaches for mRNA delivery. Mol Pharm. 2020 Oct 5;17(10):3654–3684.
- Yang Y, Zhang M, Song H, et al. Silica-based nanoparticles for biomedical applications: from nanocarriers to biomodulators. Acc Chem Res. 2020 Aug 18;53(8):1545–1556.
- Levacic AK, Berger S, Muller J, et al. Dynamic mRNA polyplexes benefit from bioreducible cleavage sites for in vitro and in vivo transfer. J Controlled Release. 2021 Nov 10;339:27–40.
- Cheng Q, Wei T, Farbiak L, et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-cas gene editing. Nat Nanotechnol. 2020 Apr;15(4):313–320.
- Cabral H, Miyata K, Osada K, et al. Block copolymer micelles in nanomedicine applications. Chem Rev. 2018 Jul 25;118(14):6844–6892.
- Jarak I, Pereira-Silva M, Santos AC, et al. Multifunctional polymeric micelle-based nucleic acid delivery: current advances and future perspectives. Appl Mater Today. 2021 Dec 25;25:101217.
- Kartha G, Bello J, Harker D. Tertiary structure of ribonuclease. Nature. 1967 Mar 4;213(5079):862–865.
- Burke RS, Pun SH. Extracellular barriers to in vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver. Bioconjug Chem. 2008 Mar;19(3):693–704.
- Zuckerman JE, Choi CH, Han H, et al. Polycation-siRNA nanoparticles can disassemble at the kidney glomerular basement membrane. Proc Natl Acad Sci USA. 2012 Feb 21;109(8):3137–3142.
- Dirisala A, Uchida S, Tockary TA, et al. Precise tuning of disulphide crosslinking in mRNA polyplex micelles for optimising extracellular and intracellular nuclease tolerability. J Drug Target. 2019 Jun-Jul;27(5–6):670–680.
- Yoshinaga N, Uchida S, Dirisala A, et al. mRNA loading into ATP-responsive polyplex micelles with optimal density of phenylboronate ester crosslinking to balance robustness in the biological milieu and intracellular translational efficiency. J Controlled Release. 2021 Feb 10;330:317–328.
- Yoshinaga N, Uchida S, Dirisala A, et al. Bridging mRNA and polycation using mRNA Oligonucleotide derivatives improves the robustness of polyplex micelles for efficient mRNA delivery. Adv Healthc Mater. 2022 May;11(9):e2102016.
- Yang W, Chen P, Boonstra E, et al. Polymeric micelles with pH-responsive cross-linked core enhance in vivo mRNA delivery. Pharmaceutics. 2022 Jun 6;14(6):1205.
- Miyazaki T, Uchida S, Nagatoishi S, et al. Polymeric nanocarriers with controlled chain flexibility boost mRNA delivery in vivo through enhanced structural fastening. Adv Healthc Mater. 2020 Aug;9(16):e2000538.
- Miyazaki T, Uchida S, Hatano H, et al. Guanidine-phosphate interactions stabilize polyion complex micelles based on flexible catiomers to improve mRNA delivery. Eur Polym J. 2020 Nov 5;140:110028.
- Wilson KA, Holland DJ, Wetmore SD. Topology of RNA-protein nucleobase-amino acid pi-pi interactions and comparison to analogous DNA-protein pi-pi contacts. RNA. 2016 May;22(5):696–708.
- Wilson KA, Kung RW, D’Souza S, et al. Anatomy of noncovalent interactions between the nucleobases or ribose and pi-containing amino acids in RNA-protein complexes. Nucleic Acids Res. 2021 Feb 26;49(4):2213–2225.
- Kataoka K, Togawa H, Harada A, et al. Spontaneous formation of polyion complex micelles with narrow distribution from antisense oligonucleotide and cationic block copolymer in physiological saline. Macromolecules. 1996 Jan 1;29(26):8556–8557.
- Thompson N. Topics in fluorescence spectroscopy. Techniques. 1991;1:337.
- Ruponen M, Yla-Herttuala S, Urtti A. Interactions of polymeric and liposomal gene delivery systems with extracellular glycosaminoglycans: physicochemical and transfection studies. Biochim Biophys Acta. 1999 Jan 8;1415(2):331–341.
- Schmitt S, Nuhn L, Barz M, et al. Shining light on polymeric drug nanocarriers with fluorescence correlation spectroscopy. Macromol Rapid Commun. 2022 Feb;43(12):e2100892.
- Rodriguez PC, Quiceno DG, Zabaleta J, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004 Aug 15;64(16):5839–5849.
- Zhou Z, Murdoch WJ, Shen Y. Synthesis of an esterase-sensitive degradable polyester as facile drug carrier for cancer therapy. J Polym Sci A Polym Chem. 2016;54(4):507–515.
- Fernando IR, Ferris DP, Frasconi M, et al. Esterase- and pH-responsive poly(beta-amino ester)-capped mesoporous silica nanoparticles for drug delivery. Nanoscale. 2015 Apr 28;7(16):7178–7183.
- Uchida S, Kinoh H, Ishii T, et al. Systemic delivery of messenger RNA for the treatment of pancreatic cancer using polyplex nanomicelles with a cholesterol moiety. Biomaterials. 2016 Mar;82:221–228.
- Gan XJ, Liu SP, Liu ZF, et al. Fluorescence quenching method for the determination of carbazochrome sodium sulfonate with aromatic amino acids. Luminescence. 2013 May-Jun;28(3):265–269.
- Gan X, Liu S, Liu Z, et al. Determination of tetracaine hydrochloride by fluorescence quenching method with some aromatic amino acids as probes. J Fluoresc. 2012 Jan;22(1):129–135.
- Kauffman WB, Guha S, Wimley WC. Synthetic molecular evolution of hybrid cell penetrating peptides. Nat Commun. 2018 Jul 2;9(1):2568.
- Wang F, Hu K, Cheng Y. Structure-activity relationship of dendrimers engineered with twenty common amino acids in gene delivery. Acta Biomater. 2016 Jan;29:94–102.
- Creusat G, Zuber G. Self-assembling polyethylenimine derivatives mediate efficient siRNA delivery in mammalian cells. Chembiochem. 2008 Nov 24;9(17):2787–2789.