ABSTRACT
Background: Nuclear pore membrane protein 121 (POM121) plays a crucial role in nucleocytoplasmic transport, but its significance in tumorigenesis and the progression of colorectal cancer (CRC) remains unknown. The aim of this study was to evaluate the relationship between POM121 and CRC.
Methods: POM121 expression in colorectal tissues was analyzed at both the gene and protein levels. We investigated the connection between POM121 expression and clinicopathological features, as well as overall survival. A gene set enrichment analysis (GSEA) was performed, and a protein-protein interaction (PPI) network was constructed to determine the mechanism of POM121 in CRC.
Results: CRC tissues displayed a striking increase in POM121 expression compared with colonitis and pericarcinomatous mucosa tissues (66.61% vs 24.36% vs 24.11%, respectively, p < 0.0167). POM121 overexpression was significantly associated with lymph node metastasis, distant metastasis, TNM stage, venous invasion, perineural invasion, preoperative CEA and CA19-9 levels, and Ki67 expression. CRC patients with high POM121 levels tended to have poor overall survival rates. POM121 may participate in the regulation of the cell cycle and DNA repair in CRC.
Conclusions: Our results suggest that POM121 has the potential to serve as a novel prognostic biomarker in CRC patients.
1. Introduction
Colorectal cancer (CRC)is the third most frequent malignancy worldwide, with 1.4 million new cases and 700,000 cancer-related deaths every year [Citation1].Chemotherapy and surgery are efficient methods for primary CRC, and the 5-year survival rate of patients in early stages is close to 90% [Citation2].Unfortunately, early CRC shows occult clinical symptoms, and more than half of patients are ignored until an advanced stage is entered [Citation3].It has been reported that the 5-year survival rate is only 10% or less in CRC patients with distant metastases [Citation4].Compared with colonoscopy screening, the fecal occult blood test is more commonly applied in CRC screening, but the latter inevitably results in a large number of false-positives and subsequent limitations [Citation5,Citation6].Therefore, it is necessary to identify new sensitive biomarkers that can be applied to detect early, noninvasive CRC.
Nuclear pore membrane protein 121 (POM121) is an important part of the nuclear pore complex (NPC) with wide distribution in vertebrates [Citation7].POM121 has been identified to participate in nuclear envelope assembly and nuclear transport and acts as an NPC anchor [Citation8–Citation10]. Previous studies have focused on the role of POM121 in NPC formation, but increasing evidence has confirmed the versatility of POM121 in other domains. Saito et al. found that N-terminally truncated POM121inhibits HIV-1 replication [Citation11], and Guo et al. demonstrated the role of full-length POM121inefficientHIV-1 nuclear import [Citation12], indicating that POM121 is involved in HIV infection. A recent study showed that POM121 exerts a potent inhibitory effect on macrophage inflammation via the NF-κB signaling pathway [Citation13].POM121 is also reported to be involved in acute lymphoblastic leukemia development by fusing with PAX5 [Citation14,Citation15]and promoting the proliferation and therapeutic resistance of prostate cancer by increasing E2F1, MYC, and AR nuclear import [Citation16].More research is essential to explore the function and mechanism of POM121 in tumorigenesis and progression.
The aim of this study was to determine the role of POM121 in CRC. POM121 expression in CRC and colorectal tissues was detected and compared. The association among POM121 expression, clinicopathologic features and the prognosis of CRC patients was analyzed. We also preliminarily discuss the mechanism by which POM121 plays its pivotal roles in CRC by bioinformatics analysis.
2. Methods
2.1. Tissue samples and clinical data
The research was performed based on the Declaration of Helsinki and acquired the approval of the Ethics Committee of Nanjing Hospital Affiliated to Nanjing Medical University. All patients enrolled in the study received a biopsy or surgery at the Nanjing Hospital Affiliated to Nanjing Medical University between December 2012 and May 2016 and provided written informed consent. No patient underwent chemotherapy or radiotherapy before surgery. A total of 771 colorectal samples were detected, including 581 carcinoma tissues, 112 pericarcinomatous tissues and 78 colonitis tissues. The following clinicopathologic information of 581CRC patients and the 29 fresh colorectal cancer samples (completely randomized) was also assessed: sex, age, tumor location, histological type, differentiation, tumor size, lymph node metastasis, distant metastasis, TNM stage, venous invasion, perineural invasion, preoperative carcinoembryonic antigen (CEA) level, preoperative CA19-9 level, and Ki67 expression.
2.2. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) analysis
To compare POM121gene expression in CRC and normal tissues, total RNA was extracted from29 pairs of cancerous and tumor-adjacent normal tissues using a TRIzol Plus RNA Purification Kit (#12183555, Invitrogen, USA). cDNA was reverse transcribed with a SuperScript Choice System for cDNA Synthesis (#18090019, Invitrogen, USA) according to the manufacturer’s instructions. qRT-PCR analysis was performed using Power SYBR Green PCR Master Mix (#4367659, Applied Biosystems, USA) on a Pharmaceutical Analytics QuantStudio 5 Real-Time PCR System. The primer sequences used in the experiments were as follows: human POM121 forward, 5ʹ-CGTTTGCCTTCAACGTGAGC-3ʹ and reverse, 5ʹ-AAAAGTGTTGCCGAAAGGTGC-3ʹ; and GAPDH forward, 5ʹ-CTGGGCTACACTGAGCACC-3ʹ and reverse, 5ʹ-AAGTGGTCGTTGAGGGCAATG-3ʹ. The GAPDH gene was regarded as the housekeeper gene to normalize POM121 mRNA expression, and 2−ΔΔCt was calculated to determine the relative quantification of the POM121 gene. All analyzes were repeated at least three times.
2.3. Tissue Microarray (TMA) Construction
TMAs were constructed using a TMA Grand Master (3DHISTECH, Hungary). Core tissue biopsies 1.5 mm in diameter were acquired from donor tissues embedded in paraffin and were then embedded into a ‘recipient’ block. The recipient block was cut into sections with a thickness of 3 μm and placed onto polylysine-coated glass slides. A total of 14TMAsweremanufactured, including 10carcinoma TMAs (581 carcinoma samples), 2 normal colorectal TMAs (112 pericarcinomatous samples) and 2colonitis TMAs (78 colonitis samples).
2.4. Immunohistochemistry (IHC)analysis
The TMA sections were depauperated with xylene, rehydrated with graded alcohols, and then heated in 0.01 mM citrate buffer (pH 6.0) for 10 min to retrieve antigens. The slides were incubated in H2O2 (3%) for 15 min to block endogenous peroxidase activity. POM121 was detected with rabbit polyclonal antibody (1:100, # PA5-83681, Invitrogen, USA) and a goat anti-rabbit IgG secondary antibody (1:5000, #31460, Invitrogen, USA). Reactions were detected using an UltraVisionQuanto Detection System HRP DAB (#TL-125-QHD, Thermo, USA). All sections were judged and scored by two pathologists blinded to each other and to the clinical information. According to the staining intensity, cells in each sample were scored as 0 (no staining), 1 (weak staining), 2 (moderate staining), or 3 (strong staining). The intensity percentage score was calculated as 100 × the product of the percentage of cells with the same intensity and the relevant intensity score. Four intensity percentage scores were added together to yield a final staining score that ranged from 0 (no staining) to 300 (all cells with strong staining). The X-tile software program was used to identify significant cutoff points according to the overall survival of CRC patients. For POM121, the cutoff value was selected as 130: samples with a score between 0 and 130 were considered low or no expression, while samples with a score greater than 130 were considered high expression.
2.5. Gene set enrichment analysis (GSEA) and protein-protein interaction (PPI) network
GSEA is a common bioinformatic method to interpret and analyze microarray and other similar data, speculating relevant pathways within which regulated genes are significantly enriched. We downloaded the RNAseq data of CRC patients from the NCBI-GEO database(available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE25071) and conducted the analysis using GSEA v3.0 software. The samples were separated into a high and a low group (critical value = median POM121 level). A total of 1000 permutations were used to calculate the p values. A meaningful gene set was defined as a p-value < 0.05 and an FDR < 0.25. We explored the proteins interacting with POM121 at https://string-db.org/cgi/input.pl and visualized them in a PPI network with Cytoscape v3.6.0 software.
2.6. Statistical analysis
All statistical analyzes were performed with SPSS 19.0 statistical software package (SPSS, Inc., USA). Comparisons between two groups were performed by Student’s t-test, and the relationship between POM121 expression and clinical features was evaluated by the χ2 test. The Kaplan-Meier method was utilized to generate survival curves, and the log-rank test was performed to compare the differences in overall survival (OS). To determine independent prognostic markers for CRC patients, univariate and multivariate analyzes were performed using the Cox regression model. Statistical significance was set as a two-tailed p-value < 0.05.
3. Results
3.1. Expression of POM121 mRNA in colorectal tissues
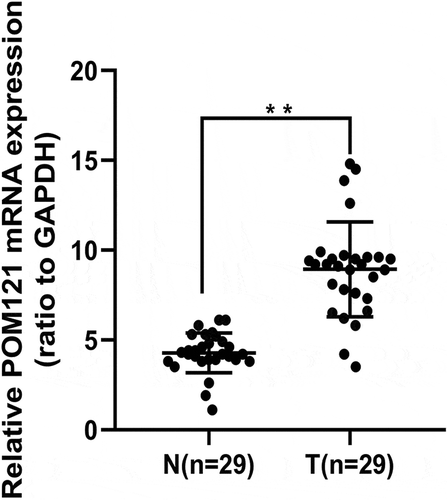
To investigate the expression of POM121 mRNA, qRT-PCR was performed in 29 pairs of fresh CRC tissues and matched noncancerous tissues. As shown in , POM121 mRNA in CRC tissues was significantly higher than that in normal colorectal tissues (p < 0.001).
3.2. Expression of the POM121 protein in colorectal tissues
Distinct POM121 staining was performed in CRC, colonitis and pericarcinomatous tissues. The POM121 protein, which is located in the nuclear membrane, was upregulated in CRC tissues (). Only a fraction of colonitis (24.36%, 19/78) and pericarcinomatous (24.11%, 27/112) tissues showed high POM121 expression, whereas high expression of thePOM121proteinwas detected in 66.61%of CRC tissues (387/581)(p < 0.001, ).
Figure 2. IHC staining for POM121 expression in colorectal samples. (a),(b)Positive IHC staining of POM121 in well-differentiated adenocarcinoma. (c), (d)Positive IHC staining of POM121 in moderately differentiated adenocarcinoma. (e), (f)Positive IHC staining of POM121 in poorly-differentiated adenocarcinoma.(g), (h) Negative IHC staining of POM121 in normal colorectal tissue. (Original images×4, scale bars500 μm; enlarged images×40, scale bars 50 μm).
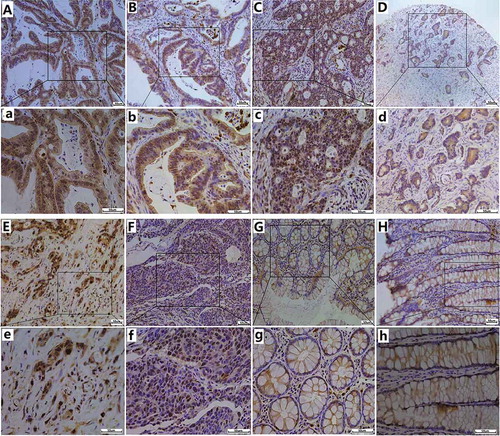
Table 1. POM121 expression in colorectal tissues.
3.3. Association between POM121 expression and clinicopathologic parameters in CRC patients
We also collected clinical information from 581 CRC patients and analyzed the relationship between POM121 expression and clinicopathologic characteristics (). The results showed that high POM121 expression was significantly associated with advanced lymph node metastasis(χ2 = 13.2883, P = 0.001), distant metastasis(χ2 = 8.987, P = 0.003), TNM stage(χ2 = 10.289, P = 0.016), positive venous invasion(χ2 = 3.877, P = 0.049), perineural invasion(χ2 = 40.581, P = 0.000), high preoperative CEA(χ2 = 26.916, P = 0.000)and CA19-9(χ2 = 14.010, P = 0.001) levels, and positive Ki67 expression(χ2 = 32.348, P = 0.000) ().This evidence indicates that POM121 expression is involved in the regulation of cell invasion and metastasis in CRC. However, there was no significant association between gender, age, location, histological type, differentiation, and tumor size.
Table 2. POM121 expression level and CRC patients’ clinicopathological characteristics.
3.4. Connection between POM121 expression and the prognosis of CRC patients
Prognostic factors in CRC were determined using univariate and multivariate logistic regression analyzes. POM121 expression (HR = 1.005; 95% CI, 1.004–1.007; P = 0.000)was confirmed as a prognostic factor in univariate analyzes, as was age (HR = 1.406; 95% CI, 1.120–1.766; P = 0.003), distant metastasis(HR = 5.367; 95% CI, 4.142–6.905; P = 0.000), TNM stage(HR = 1.136; 95% CI, 1.006–1.283; P = 0.040), venous invasion(HR = 1.242; 95% CI, 1.083–1.423; P = 0.002), and Ki67expression (HR = 1.240; 95% CI, 1.128–1.363; P = 0.000). In multivariate analyzes, high POM121 expression(HR = 1.005; 95% CI, 1.003–1.006; P = 0.000), distant metastasis(HR = 5.472; 95% CI, 4.416–7.191; P = 0.000), and TNM stage (HR = 1.136; 95% CI, 1.006–1.283; P = 0.040) were significantly associated with poor OS (). Kaplan-Meier survival analysis confirmed that CRC patients with high POM121 expression had a significantly shorter survival than those with low POM121 expression ().
Figure 3. Survival curves of CRC patients using Kaplan-Meier plots. (a) Overall survival rates in patients with low POM121 expression(blue line)and high POM121 expression (green line).(b) Overall survival rates of patients in TNM 0 + I(blue line), TNMII(green line), TNM III (gray line)and TNM IV (purple line).(c) Overall survival rates in patients with positive venous invasion (blue line) and negative venous invasion (green line). (d) Overall survival curves by distant metastases: M0 (blue line) and M1 (green line).
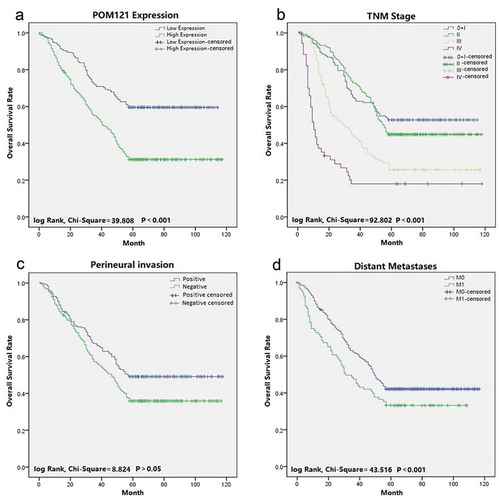
Table 3. Univariate and multivariable analysis of prognostic factors for overall survival in CRC.
3.5. Main enriched KEGG pathways in CRC tissues with high POM121 expression and the proteins interacting withPOM121
A GSEA is performed to analyze tissue microarrays based on groups or pathways rather than individual genes and can identify more subtle changes in gene expression [Citation17].In this study, the RNAseq data of CRC patients with different levels of POM121 expression were compared, and the results showed that the genes that varied significantly in patients with high POM121 expression were enriched in the cell cycle, spliceosome, basal transcription factors, DNA replication, mismatch repair and base excision repair pathways (), ). We constructed a putative protein interaction network to investigate the mechanism of action of POM121 in CRC. MYC, CCNA1, and other proteins were found to have a close relation with POM121 ().
Figure 4. The main enriched KEGG pathways and the protein interaction network. (a)Gene expression levels in patients with high and low POM121expressionwere compared through GSEA. Gene sets involved in the cell cycle, spliceosome, basal transcription factors, DNA replication, mismatch repair and base excision repair were enriched in the POM121-high expression phonotype. (b) MYC, CCNA1, and44 other proteins were confirmed to interact with POM121. (c) MYC andCCNA1 showed a significant relationship with POM121 expression in CRC.
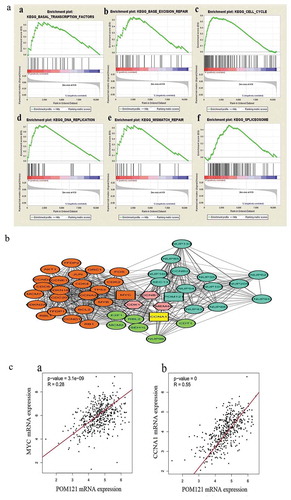
Table 4. Enriched pathways for differential expression POM121 in CRC.
4. Discussion
Nucleoporins (Nups) are the chief components of nuclear pore complexes (NPCs) which can regulate cellular signaling between cytoplasm and nucleus [Citation18]. Except transport-independent functions, Nups also participate in cancer formation and progression. Tpr is the first validiert Nup contributing to the mechanism of oncogenesis, and decreased in human colorectal tumors [Citation19]. Nup98 expression decreased in murine and human hepatocellular carcinomas and acts as a potential tumor suppressor through regulatingp53 target genes [Citation20]. Nup88 overexpressed in a large number of tumors, such as CRC, endometrial cancer, breast cancer [Citation21–Citation23]. Nup62 is highly expressed in elevated in squamous cell carcinomas and controls cell fate through regulation of p63 nuclear transport [Citation24]. In addition, Nup93, Nup188, Nup205, Nup358 and other Nups also play a role in colon cancer cells [Citation25,Citation26]. Different Nups show different expression level in cancers, but no doubt that some of them affect tumorigenesis and progression. POM121 is reported to overexpress in lethal prostate cancer, but little is known its role in CRC.
In our study, POM121 mRNA and protein expression was examined using qRT-PCR and IHC in gastric tissues. The POM121 mRNA level in CRC tissues was significantly higher than that in tumor-adjacent tissues. Interestingly, POM121 mRNA did not have a strictly linear relation with protein expression, and insufficient samples and spatial-temporal disparities may account for this phenomenon. To confirm the upregulation of POM121 in CRC tissues, TMAs that comprised 771 samples of colorectal tissues were constructed. Similar toPOM121mRNA, high levels of the POM121 protein were detected more often in CRC tissues (66.61%) than in pericarcinomatous (24.11%) and colonitis mucosa (24.36%) tissues. POM121 overexpression was associated with advanced lymph node metastasis, distant metastasis, TNM stage, positive venous invasion, perineural invasion, high preoperative CEA and CA19-9 levels, and positive Ki67 expression, indicating that POM121 participates in the proliferation, migration and invasion of CRC. Moreover, CRC patients with high POM121 expression had poor overall survival rates. This finding suggests that POM121 may serve as a new prognostic indicator for CRC patients.
According to the GSEA, CRC tissues displaying increased expression of the POM121overexpressed gene groups that are involved in the cell cycle, spliceosome, basal transcription factors, DNA replication, mismatch repair and base excision repair. The loss of checkpoint control of the cell cycle is the basis of genetic instability, and dysregulation of the cell cycle can result in the abnormal proliferation of cancer cells [Citation27]. Alternative splicing plays an important role in gene regulation, and aberrant splicing can activate oncogenes or devitalize tumor suppressors [Citation28]. POM121 may also affect the invasion and metastasis of CRC by regulating spliceosome-associated factors [Citation29].The regulatory function of POM121 in transcription factors allows it to disturb the homeostasis of cellular signaling crucial for carcinogenesis [Citation30].Mismatch repair proteins are reported to initiate DNA hypermethylation alteration and tumorigenesis [Citation31]. Base excision repair is a key pathway that removes damaged DNA bases, with the potential to drive carcinogenesis [Citation32]. POM121 showed close relationships with DNA replication, mismatch repair and base excision repair, suggesting that POM121 may promote CRC oncogenesis by modulating DNA replication and repair.
We constructed a presumptive protein network to further explore the functional mechanism of POM121 and found that MYC, CCNA1, and their downstream/upstream proteins had a major impact on POM121 function. MYC amplifies a selective gene to regulate the cell cycle, promoting cancer cell growth and proliferation [Citation33]. MYC also controls the transcription of the components of the spliceosome and accelerates malignant transformation [Citation34].TP53, BCL2, AKT1 and other proteins interacting with MYC can affect cancer proliferation via the cell cycle and apoptosis [Citation35–Citation37]. CCNA1is a cell cycle regulatory factor and participates in cell death induced by DNA damage, which is closely linked with mismatch repair and base excision repair [Citation38]. CDT1 contributes to the cyclin/CDK pathway and minichromosome maintenance to regulate DNA replication [Citation39]. The results obtained from the PPI network confirmed the enriched pathways selected in the GSEA to some degree.
5. Conclusions
In summary, this study suggests that POM121 is overexpressed in CRC tissues and that high POM121 expression is associated with a poor prognosis. Therefore, the POM121 cloud acts as a novel prognostic biomarker and therapeutic target for CRC patients.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Contributors
Lizhou Jia and Tengqi Wang designed the study; Lizhou Jia, Tengqi Wang and Haibin Sun acquired the data and drafted the article; Lizhou Jia, Wei Zhao, Yinshengboer Bao, Riletu En and Yongjing Tian analyzed and interpreted the data; Lizhou Jia revised the article critically for important intellectual content. All the authors approved the version to be published.
Consent for publication
Consent for publication was obtained from all authors.
Declaration of interest
The authors have declared that no competing financial interests exist.
Ethical statement
This study was performed in accordance with medical ethical standards and was approved by the Human Research Ethics Committee of Nanjing Hospital Affiliated to Nanjing Medical University. Written informed consents were obtained from all study participants.
Acknowledgments
We thank all patients enrolled in the study. We also thank the American Journal Experts, for editing the English text of a draft of this manuscript.
Additional information
Funding
References
- Bundgaard-Nielsen C, Baandrup UT, Nielsen LP, et al. The presence of bacteria varies between colorectal adenocarcinomas, precursor lesions and non-malignant tissue. BMC Cancer. 2019;19:399.
- Coppede F, Lopomo A, Spisni R, et al. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World J Gastroenterol. 2014;20:943–956.
- Geng F, Wang Z, Yin H, et al. Molecular targeted drugs and treatment of colorectal cancer: recent progress and future perspectives. Cancer Biother Radiopharm. 2017;32:149–160.
- Oconnell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new american joint committee on cancer sixth edition staging. J Natl Cancer Inst. 2005;96:1420–1425.
- Jin J. Screening for colorectal cancer. JAMA. 2016;315:2635.
- Burch J, Soaresweiser K, John DJBS, et al. Diagnostic accuracy of faecal occult blood tests used in screening for colorectal cancer: a systematic review. J Med Screen. 2007;14:132–137.
- Shaulov L, Gruber R, Cohen I, et al. A dominant-negative form of POM121 binds chromatin and disrupts the two separate modes of nuclear pore assembly. J Cell Sci. 2011;124:3822–3834.
- Antonin W, Franz C, Haselmann U, et al. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell. 2005;17:83–92.
- Mitchell JM, Mansfeld J, Capitanio J, et al. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol. 2010;191:505–521.
- Sevil Y, Rachel SM, Birgit K, et al. NLS-mediated NPC functions of the nucleoporin Pom121. FEBS Lett. 2010;584:3292–3298.
- Saito H, Takeuchi H, Masuda T, et al. N-terminally truncated POM121C inhibits HIV-1 replication. PLoS One. 2017;12:e0182434.
- Guo J, Liu X, Wu C, et al. The transmembrane nucleoporin Pom121 ensures efficient HIV-1 pre-integration complex nuclear import. Virology. 2018;521:169–174.
- Ge W, Yue Y, Xiong S. POM121 inhibits the macrophage inflammatory response by impacting NF-kappaB P65 nuclear accumulation. Exp Cell Res. 2019;377:17–23.
- Fortschegger K, Anderl S, Denk D, et al. Functional heterogeneity of PAX5 chimeras reveals insight for leukemia development. Mol Cancer Res. 2014;12:595–606.
- Denk D, Bradtke J, Konig M, et al. PAX5 fusion genes in t(7;9)(q11.2;p13) leukemia: a case report and review of the literature. Mol Cytogenet. 2014;7:13.
- Rodriguez-Bravo V, Pippa R, Song WM, et al. Nuclear pores promote lethal prostate cancer by increasing POM121-driven E2F1, MYC, and AR nuclear import. Cell. 2018;174:1200–1215 e1220.
- Ning QY, Wu JZ, Zang N, et al. Key pathways involved in prostate cancer based on gene set enrichment analysis and meta analysis. Genet Mol Res. 2011;10:3856–3887.
- Raices M, D’Angelo MA. Nuclear pore complexes and regulation of gene expression. Curr Opin Cell Biol. 2017;46:26–32.
- Alfonso P, Canamero M, Fernandezcarbonie F, et al. Proteome analysis of membrane fractions in colorectal carcinomas by using 2D-DIGE saturation labeling. J Proteome Res. 2008;7:4247–4255.
- Singer S, Zhao R, Barsotti A, et al. Nuclear pore component Nup98 is a potential tumor suppressor and regulates posttranscriptional expression of select p53 target genes. Mol Cell. 2012;48:799–810.
- Zhang Z, Zhao Z, Jiang L, et al. Nup88 expression in normal mucosa, adenoma, primary adenocarcinoma and lymph node metastasis in the colorectum. Tumor Biol. 2007;28:93–99.
- Li Y, Zhang X, Ge J, et al. Can Nup88 expression be associated with atypical endometrial hyperplasia and endometrial cancer? A preliminary study. Pathol Res Pract. 2016;212:274–278.
- Agudo D, Gomezesquer F, Martinezarribas F, et al. Nup88 mRNA overexpression is associated with high aggressiveness of breast cancer. Int J Cancer. 2004;109:717–720.
- Hazawa M, Lin D, Kobayashi A, et al. ROCK‐dependent phosphorylation of NUP62 regulates p63 nuclear transport and squamous cell carcinoma proliferation. EMBO Rep. 2018;19:73–88.
- Labade AS, Karmodiya K, Sengupta K. HOXA repression is mediated by nucleoporin Nup93 assisted by its interactors Nup188 and Nup205. Epigenetics Chromatin. 2016;9:54.
- Vecchione L, Gambino V, Raaijmakers JA, et al. A vulnerability of a subset of colon cancers with potential clinical utility. Cell. 2016;165:317–330.
- Williams GH, Stoeber K. The cell cycle and cancer. J Pathol. 2012;226:352–364.
- Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene. 2015;34:1–14.
- Li Y, Guo H, Jin C, et al. Spliceosome-associated factor CTNNBL1 promotes proliferation and invasion in ovarian cancer. Exp Cell Res. 2017;357:124–134.
- Lim KS, Wong RW. Targeting Nucleoporin POM121-Importin β Axis in Prostate Cancer. Cell Chem Biol. 2018;25:1056–1058.
- Maiuri AR, Peng M, Sriramkumar S, et al. Mismatch repair proteins initiate epigenetic alterations during inflammation-driven tumorigenesis. Cancer Res. 2017;77:3467–3478.
- Marsden CG, Dragon J, Wallace SS, et al. Base excision repair variants in cancer. Methods Enzymol. 2017;591:119–157.
- Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849:506–516.
- Anczukow O, Krainer AR. The spliceosome, a potential Achilles heel of MYC-driven tumors. Genome Med. 2015;7:107.
- Alidousty C, Baar T, Martelotto LG, et al. Genetic instability and recurrent MYC amplification in ALK‐translocated NSCLC: a central role of TP53 mutations. J Pathol. 2018;246:67–76.
- Sur S, Nakanishi H, Steele R, et al. Depletion of PCAT-1 in head and neck cancer cells inhibits tumor growth and induces apoptosis by modulating c-Myc-AKT1-p38 MAPK signalling pathways. BMC Cancer. 2019;19:354.
- Ennishi D, Mottok A, Benneriah S, et al. Genetic profiling of MYC and BCL2 in diffuse large B-cell lymphoma determines cell of origin-specific clinical impact. Blood. 2017;129:2760–2770.
- Woo SH, Seo S, An S, et al. Implications of caspase-dependent proteolytic cleavage of cyclin A1 in DNA damage-induced cell death. Biochem Biophys Res Commun. 2014;453:438–442.
- Pozo PN, Matson JP, Cole Y, et al. Cdt1 variants reveal unanticipated aspects of interactions with cyclin/CDK and MCM important for normal genome replication. Mol Biol Cell. 2018;29:2989–3002.