Abstract
Retroviral integrase participates in two catalytic reactions, which require interactions with the two ends of the viral DNA in the 3′processing reaction, and with a targeted host DNA in the strand transfer reaction. The 3′-hydroxyl group of 2′-deoxyadenosine resulting from the specific removing of GT dinucleotide from the viral DNA in the processing reaction provides the attachment site for the host DNA in a transesterification reaction. We synthesized oligonucleotides (ONs) of various lengths that mimic the processed HIV-1 U5 terminus of the proviral long terminal repeat (LTR) and are ended by 2′-deoxyadenosine containing a 3′-O-phosphonomethyl group. The duplex stability of phosphonomethyl ONs was increased by covalent linkage of the modified strand with its complementary strand by a triethylene glycol loop (TEG). Modified ONs containing up to 10 bases inhibited in vitro the strand transfer reaction catalyzed by HIV-1 integrase at nanomolar concentrations.
Introduction
Integrase (IN) mediates the insertion of the viral genome into the host cell chromosome and is therefore essential for retroviral replication. Integrase binds to the ends of both viral long terminal repeats (LTR) and specifically cleaves the phosphodiestere bond between A and G bases within the sequence 5′-GCAGT. This specific cleavage is termed the 3′-end processing reaction and two nucleotides (GT) are removed from each cDNA 3′-end in this process. The recessed 3′-hydroxyl group of adenosine provides the site of attachment to the host DNA in the next joining (strand transfer) reaction Citation1–7. The completion of the integration process consists of single-stranded gap repair at the site of concerted insertion followed by the joining of the 5′-viral ends to the target DNA [Citation8]. Since integrase has no counterpart in the living cell and the inhibition of integrase does not interfere with normal cellular processes it has become an attractive target in the development of new anti-HIV drugs.
HIV-1 integrase is a 32 kDa protein composed of three domains [Citation9,Citation10]. The N-terminal domain (NTD; residues 1–49) contains a conserved zinc-finger-like (HHCC) motif, and its helical bundle is stabilized by the coordination of a single zinc atom [Citation11]. Mutations within the HHCC domain cause a loss of the 3′-end processing and strand transfer activities of HIV-1 IN but do not impair disintegration Citation9,Citation12–14. The catalytic core domain (CCD, residues 50–212) contains active site residues Asp-64, Asp-116, and Glu-152 (DDE triad), which are essential for all catalytic activities of integrase. The CCD alone can mediate only disintegration in vitro [Citation10,Citation15]. The C-terminal domain, which is also termed the DNA-binding domain (DBD; residues 213–288), is the least conserved domain and is not responsible for catalysis. Although the three-dimensional structures for each of the individual domains of HIV-1 IN [Citation11,Citation16–19] and for two-domain integrases consisting of the catalytic core and C-terminal domain (CCD-DBD) [Citation20,Citation21] or the core and N-terminal domains [Citation22] were solved, neither the structure of the entire integrase nor the complex of DNA with IN have been determined. Recently, the electron microscopy and the single-particle image analysis of an assembled complex of HIV-1 integrase with a DNA three-way junction substrates revealed a formation of a IN triangular base with a central channel, in which the targeted DNA might be tethered. The fourth lobe is in this model centered between two lobes and may bind the ends of the viral DNA [Citation23]. However, the detailed knowledge of the interactions of DNA with integrase is based only on functional studies and molecular modeling.
A number of compounds have been reported to inhibit the 3′-processing and/or the strand transfer activities of integrase. The most potent inhibitors discovered up to date belong to the group of diketoacid inhibitors, which selectively inhibit the second step of the integration process, a strand transfer reaction. These inhibitors bind to the catalytic site of IN Citation24–27. HIV integrase could accommodate also nucleotide analogs, which bind to the C-terminal domain around the residue Lys 244 [Citation28]. DNA aptamers composed from guanine-rich sequences, which adopt stable G-quadruplex structures, were shown to strongly inhibit HIV-1 integrase Citation29–31. Denaturing of a DNA quadruplex and forming a lipofectin-DNA complex enabled delivering of the aptamers into the cells [Citation32]. The G-quadruplex structures are flexible and can adopt different structures in dependence on conditions and therefore they might interact with the distinct binding sites in HIV IN Citation32–34. Phosphorothioate oligonucleotides were also tested as efficient inhibitors of the integrase 3′-processing [Citation35,Citation36].
Here, we investigate the effect of the modified phosphonate oligonucleotides of different lengths on binding to the HIV-1 integrase and on inhibition of the strand transfer reaction. We show that the modification of the 3′hydroxyl group by a 3′-O-phosphonomethyl group results in efficient inhibition of HIV-1 integrase at nanomolar concentration. However, a predominant force for the inhibition is the length of the modified DNA, which effects its proper binding to the integrase.
Materials and methods
Solid-phase synthesis of oligonucleotides
All modified oligonucleotides were synthesized by the “trityl off” phosphoramidite method from the 3′- to the 5′-end on a 0.5 μmol scale using a GeneSyn synthesizer. For the introduction of the triethylene glycol linker (TEG) we used 8-dimethoxytrityloxy-3,6-dioxaoctyl-(2-cyanoethyl)diisopropylphosphoramidite. Phosphonate unit A (see ) was attached to the 2-cyanoethanol type of solid support by the phosphotriester method [Citation37,Citation38] using an appropriate 3′-phosphonate monomer [Citation39].
Figure 1. Chemical structures of modified oligonucleotides with triethylenglycol loop and 3′-phosphonomethyl residue.
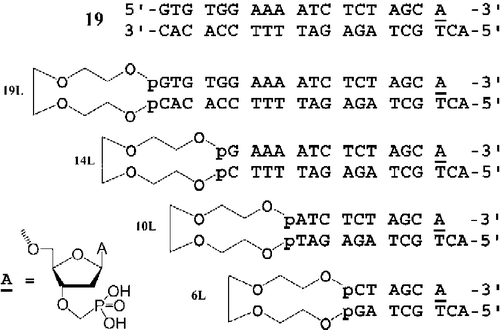
The synthesized oligonucleotides were deprotected [Citation37,Citation38] and purified by reverse phase HPLC followed by PAGE under denaturing conditions. The molecular weights of the purified oligonucleotides were confirmed by MALDI-TOF.
Purification of HIV-1 integrase
The plasmid (pRP 274) encoding HIV-1 integrase was kindly provided by Dr. Ronald Plasterk (Netherlands Cancer Institute, Amsterdam). Expression of integrase was accomplished by IPTG induction (0.4 mM) of the Escherichia coli strain BL 21(DE3) at OD600 = 1.0 at 37°C. The cells were harvested after 4 h and HIV-1 integrase was purified using a modification of the method described by Sherman and Fyfe. Pelleted cells were resuspended in 20 mM Hepes, pH 7.5, 1 mM EDTA, 5 mM DTT (buffer A) containing 200 mM NaCl and lysed by lysozyme (0.4 mg/mL). The cell lysate was sonicated several times in short bursts and then centrifuged for 30 min at 40,000 × g. The insoluble pellet containing integrase was suspended in Buffer A with 100 mM NaCl, homogenized with a Dounce homogenizer, and centrifuged (40,000 × g, 30 min). The washed pellet was resuspended in Buffer A with 1 M NaCl, stirred on ice for 1 h, and centrifuged (40,000 × g, 30 min). Integrase was precipitated from the solution with ammonium sulfate at 30% saturation. The precipitate was resuspended in Buffer A with 1 M NaCl, dialyzed against Buffer B (Buffer A containing 0.8 M ammonium sulfate and 200 mM NaCl), and clarified by centrifugation (10, 000 × g, 30 min). The integrase-containing supernatant was loaded onto a butyl-Sepharose 4B column equilibrated with buffer B. IN was eluted from the column with a linear gradient of Buffer B and Buffer A containing 80 mM NaCl, 300 mM ammonium sulfate, and 10% glycerol. Fractions containing IN were diluted with two volumes of Buffer A and 10% glycerol (Buffer C) and were loaded onto a heparin-Sepharose column equilibrated with Buffer C. IN was eluted from the column by a linear gradient of NaCl (from 0.2 M to 1 M) in Buffer C. Fractions containing integrase were dialyzed against Buffer A containing 1 M NaCl and 20% glycerol and were aliquoted and stored at − 70°C.
Assay for HIV-1 IN activity and inhibition studies
For activity measurements, HIV-1 integrase at the final concentration of 220 nM was incubated with a 5′-end 32P-labeled linear oligonucleotide substrate (S) with the sequence 5′-TGTGGAAAAATCTCTAGCA-3′ annealed to the complementary 21-mer ON containing at its 5′ end GT dinucleotide at concentrations ranging from 3–200 nM in a reaction buffer (20 mM Mops, pH 7.2, 50 mM NaCl, 50 μM EDTA, 10 mM mercaptoethanol, 10% glycerol (w/v), 7.5 mM MnCl2, 0.1 mg/mL BSA) at 37°C for 1–60 min. For inhibitor tests, HIV-1 integrase was preincubated with inhibitors in various molar ratios at 37°C for 15 min. Subsequently, 30 nM labeled substrate was added and the reaction continued for an additional 30 min. The reaction was stopped by adding an equal volume of Maxam–Gilbert loading buffer (98% deionized formamide, 10 mM EDTA, 0.025% xylene cyanol, and 0.025% bromophenol blue). Samples were heated at 100°C for 3 min and portions (5 μL) of the samples were resolved by electrophoresis on a denaturing 15% polyacrylamide gel (7 M urea, 0.09 M Tris borate, pH 8.3, 2 mM EDTA). Gels were dried and subjected to autoradiography or analyzed using a Molecular Dynamics Phosphor Imager. The scanned gels were quantitated with ImageQuant 3.1 software (Molecular Dynamics).
UV-cross linking experiments
The HIV integrase (220 nM) was incubated with 50 nM modified ONs labeled at the 5′-end with 32P at 25°C for 15 min. Then the reaction mixture was spotted onto parafilm and irradiated at 254 nm for 10 min at room temperature. Then, the mixtures were recovered in Eppendorf tubes, containing an equal volume of SDS-PAGE sample buffer and were heated at 90°C for 3 min. To analyze the photoadducts, the samples were loaded onto a 12% SDS-PAGE. After electrophoresis, the SDS-PAGE gel was silver-stained and dried on Whatman 3 MM paper. Autoradiography was performed by exposing the gel to X-ray film.
Results
The length dependent inhibition of the strand transfer reaction by covalently linked LTR mimics with the 3′-O-phosphonomethyl group
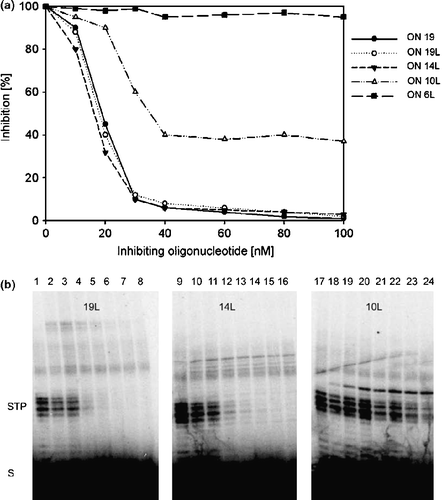
In this study we used a series of phosphonomethyl ONs of different length, which mimick the processed HIV U5 LTR sequence and contain an isopolar methylphosphonate residue attached to the 3′-hydroxyl of 2′-deoxyadenosine moiety at the 3′end for inhibition of the strand transfer activity of HIV-1 IN. To increase the stability of the shorter ON inhibitors, we covalently linked the 5′-end of the phosphonate modified oligonucleotide to the 3′-end of the complementary strand by a triethylene glycol spacer (TEG). We prepared 19-, 14-, 10-, and 6-mer oligonucleotides (ONs) linked with TEG (ON 19L, ON 14L, ON 10L, and ON 6L) (for structures see ) and for comparison we used the 3′-O-phosphonomethyl 19mer annealed to the complementary strand (without TEG). The in vitro inhibition of the strand transfer activity of HIV-1 IN by ds phosphonate 19-mer (ON 19), ON 19L, and ON 14L was almost identical. These compounds inhibited HIV-1 IN with IC50 about 20 nM. The inhibition by 10L ON was significantly lower and 6L ON was totally ineffective in inhibiting HIV IN (). We also tested the inhibition of HIV-1 IN by ss phosphonate 19-mer. The efficiency of this compound was about one order of magnitude lower (data not shown). The autoradiograms of the strand transfer inhibition showed that the TEG compounds inhibited formation of the lower molecular strand transfer products (STP) in dose-dependent manner (). The identical inhibition profiles were obtained under both arrangements, i.e. when HIV IN was preincubated with tested compounds and when the competitor was added to the reaction mixture simultaneously with the DNA substrate.
Figure 2. a: The inhibition of the HIV IN strand transfer reaction by TEG-linked oligonucleotides. Curves correspond to inhibition profiles of ON 19L, ON 14L, ON 6L, ON 10L and ds ON 19. HIV-1 integrase (220 nM) was preincubated with inhibitors in various molar ratios at 37°C for 15 min. Results are shown as relative activity, 100% corresponds to HIV-1 IN activity in the absence of inhibiting oligonucleotide. b: Autoradiograms illustrating the effect of ON 19L, ON 14L, and ON 10L on HIV-1 IN strand transfer reaction. Lanes 1, 9, 17: the strand transfer activities of HIV-1IN without the inhibitor; Lanes 2–8: 220 nM HIV IN in the presence of 10, 20, 40, 60, 80, 100 and 150 nM ON 19L; lanes 10–16: 220 nM HIV IN in the presence of 10, 20, 40, 60, 80, 100 and 150 nM ON 14L; lanes 18–24: 220 nM HIV IN in the presence of 10, 20, 40, 60, 80, 100 and 150 nM ON 10L.
The triethylene glycol spacer stabilizes the duplex form of modified oligonucleotides
Shortening of oligonucleotide structures can be accompanied by an impaired ability to form duplexes. In order to achieve better stability of the duplex DNA structure, we chemically linked modified and complementary ONs with three triethylene glycol moieties. The experimentally determined melting temperatures of TEG-linked oligonucleotides are summarized in . Introduction of the flexible loop significantly increased the Tm of all ON structures, indicating a stabilization of ds ON forms. It is of interest that the melting temperature of ON 6L is 5°C higher than that of ds ON 19. However, the transition profiles of TEG-ON melting curves are slightly different from those for typical duplexes and display different kinetics of the pairing process (not shown).
Table I. The melting temperatures for ON 19 and TEG-linked oligonucleotides. Temperature melting curves of modified ONs were measured on a Varian Cary 100 Bio spectrophotometer (THERMAL program) in 50 mM TRIS-HCl pH 7.2 with 1 mM EDTA and 100 mM NaCl, at a total strand concentration 1 μM.
To investigate requirement of the pre-hybridization process on oligonucleotide inhibition efficiencies, we compared the inhibition profiles of TEG ONs' prepared by heating to 70°C and slow cooling to room temperature, with those obtained for ONs 19L–10L added directly to the reaction without the pre-heating step. We obtained similar inhibition profiles for both arrangements (data not shown), confirming that the triethylene glycol loop is sufficient for stabilization of the double stranded form of DNA.
Binding of modified ONs to HIV integrase
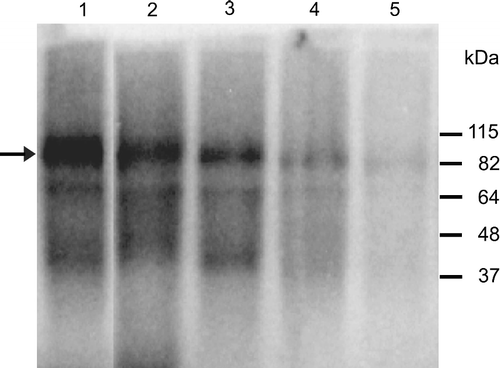
To prove binding of truncated TEG-linked ON inhibitors to HIV-1 IN, the labeled ONs were subjected to UV cross-linking experiments. The results are shown in . All bands on the autoradiogram correspond to the dimeric form of integrase with a bound radiolabelled ON. We did not detect any cross-linked product corresponding to the monomer or higher multimer of HIV IN with all tested ONs. The intensities of the bands corresponding to cross-linked complexes of ON 19L and ds ON 19 with HIV IN were roughly equal (data not shown), indicating that the triethylene glycol spacer does not influence their binding affinities to HIV IN. ON 14L was bound to integrase at approximately the same level, since the intensity of the band was similar to that of ON 19L. In agreement with the previous inhibition experiments, we detected band of the weaker intensity corresponding to the complex of HIV IN-ON 10L and no cross-linked product was observed for ON 6L. These results confirmed that inhibition efficiency of truncated modified ONs is strictly dependent on their proper binding to the integrase molecule.
Figure 3. UV-cross linking of HIV IN and ON 19 (lane 1), ON 19L (lane 2), ON 14L (lane 3); ON 10L (lane 4) and ON 6L (lane 5). The reaction mixture contained 220 nM HIV integrase and 50 nM 5′-end labelled oligonucleotide. The arrow indicates the position of the cross-linked complex of HIV integrase-DNA.
Discussion
In this study we tested truncated phosphonate oligonucleotides for their inhibition of HIV-1 IN strand transfer activity and for binding to the molecule of HIV-1 integrase. These compounds of various lengths mimicked the processed HIV U5 LTR sequence and contained a 3′-O-phosphonomethyl group. Their duplex stability was increased by covalent linkage of the U5-(+) 5′-end and the U5-( − ) 3′-end with a triethylene glycol spacer. The integrase ds oligonucleotide substrate analogs with a modified 3′ hydroxyl of the terminal 2′-deoxyadenosine represent promising inhibitors of the strand transfer reaction, since the 3′hydroxyl group is the attacking nucleophile in the joining reaction with a target DNA in the reaction catalyzed by the integrase. We previously showed that ds modified 21- and 19-mer oligonucleotides inhibited the HIV-1 IN in the nanomolar concentration range [Citation40]. In this work, we have made an effort to prepare sufficiently potent, stable, and short oligonucleotide inhibitors of the strand transfer reaction. We demonstrated that the covalent linkage of the modified ON strand with the complementary strand by the triethylenglycole loop increased the duplex stability of modified ONs, since the melting temperatures of all TEG ONs were higher than those of ONs without TEG. The results obtained from inhibition study with TEG ONs demonstrated that truncation of modified ONs up to 14 bases did not significantly change their inhibition activities. The IC50 values for these derivatives were about 20 nM. The shortening of oligonucleotides to 10 bases significantly increased the IC50 value and further truncation up to 6 bases abolished the inhibition activity. The binding UV cross-linking experiments of modified ONs to HIV-1 IN confirmed that critical contacts between IN and the LTR-derived oligonucleotide are primarily mediated by the conservative deoxyadenosine and 9 adjacent bases, and show that bases in positions 7–13 from the conserved adenosine are important for optimal fitting of DNA into the integrase molecule. Further modification of oligonucleotide backbone of truncated phosphonate oligonucleotide might contribute to a design of oligonucleotide inhibitor with potential cellular stability.
Acknowledgements
This work was supported by the “Centre for New Antivirals and Antineoplastics” 1M6138896301 of the programme of Czech Ministry of Education, by research projects Z 40550506, by grant No. 203/05/P557, 203/05/0827of the Czech Science Foundation, and by a grant No. IAA 4055304 of the Grant agency of the Academy of Sciences of the Czech Republic.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- PO Brown, B Bowerman, HE Varmus, and JM Bishop. (1989). Retroviral integration-structure of the initial covalent product and its precursor, and a role for the viral in protein. Proc Natl Acad Sci USA 86:2525–2529.
- T Fujiwara, and K Mizuuchi. (1988). Retroviral DNA integration-structure of an integration intermediate. Cell 54:497–504.
- M Katzman, RA Katz, AM Skalka, and J Leis. (1989). The avian retroviral integration protein cleaves the terminal sequences of linear viral-DNA at the in vivo sites of integration. J Virol 63:5319–5327.
- FD Bushman, T Fujiwara, and R Craigie. (1990). Retroviral DNA integration directed by HIV integration protein in vitro. Science 249:1555–1558.
- R Craigie, T Fujiwara, and F Bushman. (1990). The IN protein of Moloney murine leukemia-virus processes the viral-DNA ends and accomplishes their integration in vitro. Cell 62:829–837.
- RA Katz, G Merkel, J Kulkosky, J Leis, and AM Skalka. (1990). The avian retroviral in protein is both necessary and sufficient for integrative recombination in vitro. Cell 63:87–95.
- FD Bushman, and R Craigie. (1991). Activities of human-immunodeficiency-virus (HIV) integration protein in vitro-specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA 88:1339–1343.
- B Van Maele, and Z Debyser. (2005). HIV-1 integrated: An interplay between HIV-1 integrase, cellular and viral proteins. AIDS Rev 7:26–43.
- A Engelman, and R Craigie. (1992). Identification of conserved amino-acid-residues critical for human-immunodeficiency-virus type-1 integrase function-in vitro. J Virol 66:6361–6369.
- FD Bushman, A Engelman, I Palmer, P Wingfield, and R Craigie. (1993). Domains of the integrase protein of human-immunodeficiency-virus type-1 responsible for polynucleotidyl transfer and zinc-binding. Proc Natl Acad Sci USA 90:3428–3432.
- ML Cai, RL Zheng, M Caffrey, R Craigie, GM Clore, and AM Gronenborn. (1997). Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol 4:567–577.
- AD Leavitt, L Shiue, and HE Varmus. (1993). Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem 268:2113–2119.
- KA Vincent, V Ellison, SA Chow, and PO Brown. (1993). Characterization of human-immunodeficiency-virus type-1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol 67:425–437.
- DC van Gent, C Vink, AAMO Groeneger, and RHA Plasterk. (1993). Complementation between HIV integrase proteins mutated in different domains. EMBO J 12:3261–3267.
- C Vink, AAMO Groeneger, and RHA Plasterk. (1993). Identification of the catalytic and DNA-binding region of the human-immunodeficiency-virus type-I integrase protein. Nucleic Acids Res 21:1419–1425.
- F Dyda, AB Hickman, TM Jenkins, A Engelman, R Craigie, and DR Davies. (1994). Crystal structure of the catalytic domain of HIV-1 integrase: Similarity to other polynucleotidyl transferases. Science 266:1981–1986.
- Y Goldgur, F Dyda, AB Hickman, TM Jenkins, R Craigie, and DR Davies. (1998). Three new structures of the core domain of HIV-1 integrase: An active site that binds magnesium. Proc Natl Acad Sci USA 95:9150–9154.
- APAM Eijkelenboom, RAP Lutzke, R Boelens, RHA Plasterk, R Kaptein, and K Hard. (1995). The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol 2:807–810.
- PJ Lodi, JA Ernst, J Kuszewski, AB Hickman, A Engelman, R Craigie, GM Clore, and AM Gronenborn. (1995). Solution structure of the DNA-binding domain of HIV-1 integrase. Biochemistry 34:9826–9833.
- JCH Chen, J Krucinski, LJW Miercke, JS Finer-Moore, AH Tang, AD Leavitt, and RM Stroud. (2000). Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: A model for viral DNA binding. Proc Natl Acad Sci USA 97:8233–8238.
- ZG Chen, YW Yan, S Munshi, Y Li, J Zugay-Murphy, B Xu, M Witmer, P Felock, A Wolfe, V Sardana, EA Emini, D Hazuda, and LC Kuo. (2000). X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50–293)-An initial glance of the viral DNA binding platform. J Mol Biol 296:521–533.
- JY Wang, H Ling, W Yang, and R Craigie. (2001). Structure of a two-domain fragment of HIV-1 integrase: Implications for domain organization in the intact protein. EMBO J. 20:7333–7343.
- G Ren, K Gao, FD Bushman, and M Yeager. (2007). Single-particle image reconstruction of a tetramer of HIV integrase bound to DNA 3. J Mol Biol 366:286–294.
- R Dayam, J Deng, and N Neamati. (2006). HIV-1 integrase inhibitors: 2003-2004 update. Med Res Rev 26:271–309.
- Y Pommier, AA Johnson, and C Marchand. (2005). Integrase inhibitors to treat HIV/AIDS. Nat Rev Drug Discov 4:236–248.
- M Witvrouw, B Van Maele, J Vercammen, A Hantson, Y Engelborghs, E De Clercq, C Pannecouque, and Z Debyser. (2004). Novel inhibitors of HIV-1 integration. Curr Drug Metab 5:291–304.
- DJ Hazuda, P Felock, M Witmer, A Wolfe, K Stillmock, JA Grobler, A Espeseth, L Gabryelski, W Schleif, C Blau, and MD Miller. (2000). Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646–650.
- KL Williams, YJ Zhang, N Shkriabai, RG Karki, MC Nicklaus, N Kotrikadze, S Hess, SFJ Le Grice, R Craigie, VK Pathak, and M Kvaratskhelia. (2005). Mass spectrometric analysis of the HIV-1 integrase-pyridoxal 5′-phosphate complex reveals a new binding site for a nucleotide inhibitor. J Biol Chem 280:7949–7955.
- N Jing, and ME Hogan. (1998). Structure-activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J Biol Chem 273:34992–34999.
- N Jing, E De Clercq, RF Rando, L Pallansch, C Lackman-Smith, S Lee, and ME Hogan. (2000). Stability-activity relationships of a family of G-tetrad forming oligonucleotides as potent HIV inhibitors-A basis for anti-HIV drug design. J Biol Chem 275:3421–3430.
- VR de Soultrait, PY Lozach, R Altmeyer, L Tarrago-Litvak, S Litvak, and ML Andreola. (2002). DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J Mol Biol 324:195–203.
- N Jing, W Xiong, Y Guan, L Pallansch, and S Wang. (2002). Potassium-dependent folding: A key to intracellular delivery of G-quartet oligonucleotides as HIV inhibitors. Biochemistry 41:5397–5403.
- N Jing, C Marchand, J Liu, R Mitra, ME Hogan, and Y Pommier. (2000). Mechanism of inhibition of HIV-1 integrase by G-tetrad-forming oligonucleotides in vitro. J Mol Biol 275:21460–21467.
- AT Phan, V Kuryavyi, JB Ma, A Faure, ML Andreola, and DJ Patel. (2005). An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase. Proc Natl Acad Sci USA 102:634–639.
- Y Tamura, M Yoshida, J Suzuki, T Hiratou, N Miyano-Kurosaki, K Takai, and H Takaku. (2000). Properties of quadruplex oligonucleotides with anti-HIV-1 activity. Nucleic Acids Symp Ser 44:181–182.
- E Tramontano, P La Colla, and YC Cheng. (1998). Biochemical characterization of the HIV-1 integrase 3′-processing activity and its inhibition by phosphorothioate oligonucleotides. Biochemistry 37:7237–7243.
- D Rejman, J Snášel, R Liboska, Z Točík, O Pačes, S Kraliková, M Rinnová, P Kois, and I Rosenberg. (2001). Oligonucleotides with isopolar phosphonate internucleotide linkage: A new perspective for antisense compounds?. Nucleosides Nucleotides Nucleic Acids 20:819–823.
- T Szabo, A Kers, and J Stawinski. (1995). A new approach to the synthesis of the 5′-deoxy-5′-methylphosphonate linked thymidine oligonucleotide analogs. Nucleic Acids Res 23:893–900.
- D Rejman, M Masojídková, and I Rosenberg. (2004). Nucleosidyl-O-methylphosphonates: A pool of monomers for modified oligonucleotides. Nucleosides Nucleotides Nucleic Acids 23:1683–1705.
- J Snášel, D Rejman, R Liboska, Z Točík, T Ruml, I Rosenberg, and I Pichová. (2001). Inhibition of HIV-1 integrase by modified oligonucleotides derived from U5′ LTR. Eur J Biochem 268:980–986.