Abstract
A α-carbonic anhydrase (CA, EC 4.2.1.1) has been purified and characterized biochemically from the mollusk Mytilus galloprovincialis. As in most mollusks, this α-CA is involved in the biomineralization processes leading to the precipitation of calcium carbonate in the mussel shell. The new enzyme had a molecular weight of 50 kDa, which is roughly two times higher than that of a monomeric α-class enzyme. Thus, Mytilus galloprovincialis α-CA is either a dimer, or similar to the Tridacna gigas CA described earlier, may have two different CA domains in its polypeptide chain. The Mytilus galloprovincialis α-CA sequence contained the three His residues acting as zinc ligands and the gate-keeper residues present in all α-CAs (Glu106-Thr199), but had a Lys in position 64 and not a His as proton shuttling residue, being thus similar to the human isoform hCA III. This probably explains the relatively low catalytic activity of Mytilus galloprovincialis α-CA, with the following kinetic parameters for the CO2 hydration reaction: kcat = 4.1 × 105 s−1 and kcat/Km of 3.6 × 107 M−1 × s−1. The enzyme activity was poorly inhibited by the sulfonamide acetazolamide, with a KI of 380 nM. This study is one of the few describing in detail the biochemical characterization of a molluskan CA and may be useful for understanding in detail the phylogeny of these enzymes, their role in biocalcification processes and their potential use in the biomimetic capture of the CO2.
Introduction
Mollusk shell is composed of about 95% calcium carbonate (CaCO3) and 5% of organic componentsCitation1. The shell is formed by three elements secreted by the mantle (the dorsal body of the animal): a) a thin outer periostracum; b) a middle prismatic, which is a crystalline form of calcium carbonate; c) an inner calcareous (nacreous) layer appearing with a vividness texture or iridescent mother-of-pearl (nacre), depending on the speciesCitation2–4. Prismatic and nacreous layers are in the forms of calcium carbonate crystal; but, the two layers display a crystal polymorphism because the prismatic layer develops calcite and the nacreous layer forms aragoniteCitation5. The shell grows in circumference by the addition of material from the edge of the mantle, while grows in thickness by deposition from the general mantle surface. Calcium for shell growth is obtained from diet and/or from seawater, whereas carbonate is originated from the concentration of CO2/bicarbonate present in the tissue of the organismCitation6. The interconversion of CO2 and HCO3− is balanced naturally to maintain the equilibrium between dissolved inorganic carbon dioxide (CO2), carbonic acid (H2CO3), bicarbonate (HCO3−) and carbonate (CO32−)Citation7–9. The spontaneous hydration/dehydration reaction: CO2 + H2O ⇋ HCO3− + H+ is very slow at physiological pH. In this context, a family of metalloenzymes, named carbonic anhydrase (CA; EC 4.2.1.1) assumes a pivotal role in the biomineralization process of the mollusks and is essential for the shell developmentCitation7,Citation8,Citation10–16.
CAs, in fact, have the primary function to strongly accelerate the interconversion of CO2 and HCO3− playing an important role during the calcium carbonate formation in mollusks, such as acid–base regulation, calcification and mineralizationCitation17,Citation18. It has been reported in literature that intracellular or cytoplasmic membrane bound CAs are essential for cytoplasmic acid/base balance and for transport mechanisms of CO2 and carbonate ions; different CA isoforms has been identified in the calcifying epithelia of the mollusksCitation17,Citation19; CA activity has been reported in the extracellular skeletal matrix of the cnidarian exoskeleton, crustacean calcium storage concretions, fish otolith and molluskan shellCitation17,Citation19,Citation20; moreover, nacrein, the protein involved in the nacreous layer formation of shell and pearl, identified for the first time in the Japanese pearl oyster Pinctada fucata, function has a CA because it has a CA domain in its amino acid sequenceCitation21.
The CA superfamily includes seven distinct classes known as the α, β, γ, δ, ζ, η and θ. In addition to biomineralization, these enzymes are involved in many physiologic processes, such as photosynthesis, respiration, CO2 transport, as well as metabolism of xenobiotics (e.g., cyanate in Escherichia coli)Citation13–15,Citation22–31. Some of the catalytically active α- and θ-CAs can also catalyze the hydrolysis of esters, such as 4-nitrophenyl acetate (4-NpA)Citation32. However, no esterase activity was detected so far for enzymes belonging to the other five classes (β-, γ-, δ-, ζ- and η-CAs)Citation33. The α-, β-, δ-, η- and, perhaps θ-CAs use Zn(II) ions at the active site, the γ-CAs are probably Fe(II) enzymes (but they are active also with bound Zn(II) or Co(II) ions)Citation34–41, whereas the ζ-class CAs are cambialistic enzymes, active both with Cd(II) or Zn(II) bounded within the active site in order to perform the physiologic reaction catalysisCitation42–44. The metal ion from the enzyme active site is coordinated by three His residues in the α-, γ-, δ- and θ-classes, by one His and two Cys residues in β- and ζ-CAs or by two His and one Gln residues in η-class with the fourth ligand being a water molecule/hydroxide ion acting as nucleophile in the catalytic cycle of the enzymeCitation8,Citation12,Citation13,Citation45–47. In metazoans, CAs belong predominantly to the α-CA family, but recently β-CA family members have been identifiedCitation17. In the last years, CAs have acquired a great importance in biotechnological applications, such as in the achievement of an artificial respiration system, selective biosensors for metal ions, and in the carbon capture process (CCP)Citation48,Citation49. In the CCP context, a number of CO2 sequestration methods have been proposed in order to capture CO2 using different types of CA enzymatic bioreactorsCitation48,Citation50.
In the present study, we characterized and determined the kinetic constants of the CA purified from the mantle tissue of the bivalve Mediterranean mussel, Mytilus galloprovincialis. This CA has been purified by ammonium sulfate precipitation and ion-exchange chromatography followed by affinity chromatography. From the determination of its N-amino terminal sequence the purified metalloenzyme it has been assigned to the α-class of the CA superfamily. The kinetic study of the active α-CA in the mantle of the adult specimens of Mytilus galloprovincialis may provide further information on the physiological role and function of this metalloenzymes in the process of biomineralization of the bivalves. This is one of the few contributions, apart the characterization of the α-CA from Tridacna gigas, in which molluskan CAs are investigated in detail at the biochemical levelCitation51.
Materials and methods
Chemicals
All the chemicals were commercial products of the purest quality and purchased from Sigma. Immobilion-P membranes were from Perkin-Elmer (Waltham, MA).
Animals
Adult specimens of Mytilus galloprovincialis were collected in the proximity of Gulf of Naples. The mussels were maintained in seawater at a temperature of 4 °C. Bivalve mantels were quickly removed and frozen at −70 °C.
Enzyme purification
All the purification steps were carried out at a temperature of 4 °C. Approximately, 20 g of mussel mantles were homogenized in 50 ml of 20 mM Tris-HCl buffer pH 8.3 containing 10−3 M PMSF, 10−3 M benzamidine and 2 × 10−3 M EDTA. The homogenate was centrifuged twice for 30 min at 12,000 × g, and the resulting supernatant was centrifuged again for 45 min at 100,000 × g. This supernatant containing approximately 200 mg protein in a volume of 60 ml was subject to an ammonium sulfate precipitation. An amount of ammonium sulfate gradually was added to obtain saturations of 30%. After resting for 14 h at 4 °C, the sample was centrifuged at 12,000 × g at 4 °C for 30 min. The hydratase activity was detected in the supernatant fraction. This fraction was dialyzed against 20 mM Tris-HCl buffer pH 8.3. An amount of samples containing 5 mg of total protein was applied to a 4.6 × 50 mm anion-exchange Q column (BioSuite Q-PEEK, Waters Corporation) mounted on an Ultimate 3000 HPLC system (Dionex). The column was pre-equilibrated with 20 mM Tris-HCl, pH 7.5 (Buffer A). The CA was eluted by a linear gradient from 0% Buffer A to 100% Buffer B (20 mM Tris-HCl, pH 7.5, containing 0.5 M NaCl) with a flow rate set to 1 ml/min with a continuous monitoring of the absorbance at 280 and 220 nm; 1-ml fractions were collected. The fractions were tested for the CA presence using the protonography technique. A peak of CA activity was eluted at a concentration of NaCl of approximately 0.1 M. At this stage of the purification, the CA was 60% pure and the obtained recovery was of 1 mg of the native protein. The active fraction was pooled, dialyzed against 20 mM Tris-HCl buffer pH 7.5 and further purified by affinity chromatography on a p-aminomethylbenzenesulfonamide agarose resin (pAMBS; Sigma-Aldrich). 1 ml of pAMBS resin was applied to an empty econocolumn (BioRad) and equilibrated with 0.1 M Tris-HCl, pH 7.5 buffer containing 0.2 M K2SO4, 0.5 mM EDTA. The sample containing about 1 mg of total protein was loaded on the p-AMBS column equilibrated as aforementioned. Unbound proteins were removed by washing extensively with the same buffer. The bound CA was eluted using 0.4 M KSCN dissolved in 0.1 M Tris-HCl, pH 7.5 buffer. The CA-containing fractions were pooled, dialyzed and concentrated by ultrafiltration. The CA-containing sample was subject to protonography. At this stage of the purification, the CA isolated from M. galloprovincialis was 90% pure with a total concentration of 0.1 mg. Its N-amino terminal sequence was determined for assigning the CA at one of the seven classes of CA described in literature.
SDS-PAGE, electroblotting and sequencing
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was carried out according to LaemmliCitation52. Samples were dissolved in buffer with 5% b-mercaptoethanol. Gel was stained with Coomassie blue. Blotting from gel onto an Immobilion-P membrane was performed as described by MatsudairaCitation53. N-terminal sequencing was performed on the blotted protein by automated Edman degradationCitation54. Protein concentration was determined by Bio-Rad assay kit.
Carbonic anhydrase assay
CA activity assay was a medication of the procedure described by Capasso et al.Citation55. Briefly, the assay was performed at 0 °C using CO2 as substrate following the pH variation due to the catalyzed conversion of CO2 to bicarbonate. Bromothymol blue was used as the indicator of pH variation. The production of hydrogen ions during the CO2 hydration reaction lowers the pH of the solution until the color transition point of the dye is reached. The time required for the color change is inversely related to the quantity of CA present in the sample. Wilbur-Anderson units were calculated according to the following definition: One Wilbur-Anderson unit (WAU) of activity is defined as (T0 − T)/T, where T0 (uncatalyzed reaction) and T (catalyzed reaction) are recorded as the time (in seconds) required for the pH to drop from 8.3 to the transition point of the dye in a control buffer and in the presence of enzyme, respectively.
Esterase activity
Activity for p-nitrophenylacetate (p-NpA) hydrolysis was determined at 0 °C using a modification of the method proposed by Armstrong et al.Citation56. The reaction mixture contained 0.3 ml of freshly prepared 3 mM p-NpA and 0.7 ml of 15 mM Tris sulfate buffer, pH 7.6. An aliquot of enzyme solution was added, and the catalyzed reaction was monitored reading the increase in A348 nm for 5 min. The catalyzed reactions were corrected for the non-enzymatic reaction. One enzyme unit was defined as the amount capable of producing an OD348 nm = 0.03 in 5 min.
Kinetic and inhibition assays
An applied photophysics stopped-flow instrument has been used for assaying the CA catalyzed CO2 hydration activity. Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 20 mM Tris (pH 8.3) as buffer, and 20 mM NaClO4 (for maintaining constant the ionic strength), following the initial rates of the CA-catalyzed CO2 hydration reaction for a period of 10–100 s. The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters (by Lineweaver–Burk plots) and inhibition constants. For each inhibitor at least six traces of the initial 5–10% of the reaction have been used for determining the initial velocity. The uncatalyzed rates were determined in the same manner and subtracted from the total observed rates. Stock solutions of inhibitor (10–100 mM) were prepared in distilled–deionized water and dilutions up to 0.01 mM were done thereafter with the assay buffer. Inhibitor and enzyme solutions were preincubated together for 15 min at room temperature prior to assay, in order to allow for the formation of the E–I complex or for the eventual active site mediated hydrolysis of the inhibitor. The inhibition constants were obtained by non-linear least-squares methods using PRISM 3 and the Cheng–Prusoff equation, as reported earlier, and represent the mean from at least three different determinations.
Protonography
Protonography is a simple and inexpensive method, similar to zymography, which allowed the detection of CA activity on the polyacrylamide gel following the formation of H+ ions produced by hydratase activity of CAsCitation57–59. Samples were run on SDS-PAGE at a concentration of about 1 μg per well. SDS-Page was carried out as described in the section “SDS-PAGE”, with the exception that samples were mixed with Laemmli loading buffer without 2-mercaptoethanol and without boiling the samples, in order to prevent protein denaturation induced by heating. The gel was run at 180 V until the dye front ran off the gelCitation57–59. Following the electrophoresis, the gel was treated with Triton X-100 at 2.5% and kept under stirring for one hour to remove the SDS. The gel was subjected to a washing step of 20 min with 100 mM Tris-HCl, pH 8.3, containing 10% isopropanol. It was washed two times for 10 min. Finally, the gel was incubated for 30 min at 4 °C under stirring with 0.1% bromothymol blue (BTB, the pH indicator) in 100 mM Tris-HCl pH 8.3. To detect the hydratase activity, the gel was immersed in distilled water saturated with CO2 prepared by bubbling CO2 into 200 ml of distilled water for about 3 h. The localized decrease of the pH value, due to the presence of the enzymatic activity of CAs, was detected through the formation of yellow band due to the change of color of the BTB from blue (alkaline pH) to yellow (acidic pH)Citation29,Citation57–61.
Results and discussion
Purification of the native form of CA
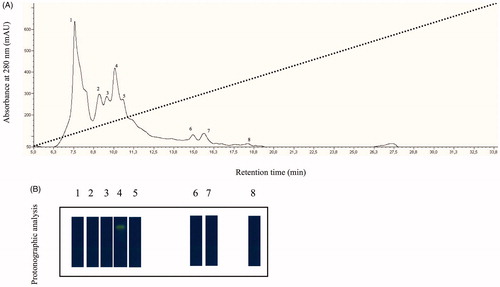
The native CA was isolated and purified to homogeneity at 4 °C from the mantles of about 30 mussels belonging to the species Mytilus galloprovincialis, the Mediterranean mussel. Most of the CA activity was recovered in the soluble fraction of cell extract after ultracentrifugation as described in “Materials and methods” section. Contaminants proteins, such as actin and chitinase, were precipitated by addition of solid ammonium sulfate (30% saturation). Following the ammonium sulfate precipitation, the hydratase activity was detected in the supernatant fraction, which was fractioned by anion-exchange chromatography as showed in . The elution profile from anion-exchange chromatography showed the presence of at least 8 peaks with a significant absorption at 280 nm (, panel A). All the peaks were analyzed by protonographic technique, which allowed the detection of CA activity on the polyacrylamide gel following the formation of H+ ions produced by hydratase activity of CAs. The protonogram was stained with bromothymol blue, an indicator monitoring the pH variation. This dye appears blue in its deprotonated form, while its color changes to yellow in the protonated form. The production of hydrogen ions during the CO2 hydration lowers the pH of the solution until the transition point of the dye is reached (pH 6.8) and the yellow color appears on the gel (, panel B). As shown in , the protonograms clearly showed a hydratase activity only associated to the peak number 4 eluted at a concentration of NaCl of approximately 0.1 M and with a retention time of about 10 min. At this stage of the purification, the CA was 60% pure as in agreement with the SDS-PAGE analysis of the native enzyme (data not shown). The hydratase-containing peak was further purified by pAMBS affinity chromatography.
Figure 1. (A) Elution profile from the anion-exchange chromatography column of the supernatant obtained from the ammonium sulfate precipitation. Dot line represents the linear gradient from 0 to 0.5 NaCl; (B) Protonographic analysis carried out on the peaks eluted from the column. The yellow band denotes the hydratase activity due to the native CA purified by the mantles of the Mediterranean mussels, M. galloprovincialis.
SDS-Page and protonography
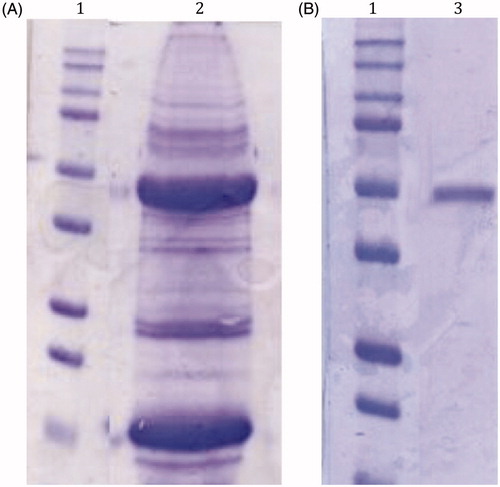
In is shown the SDS-PAGEs obtained analyzing the sample after the tissue homogenization of the mussels (panel A) and the column affinity chromatography (panel B). The native CA was purified to an apparent homogeneity, as indicated by a single protein band after SDS-PAGE (, panel B, lane 3). The molecular weight estimated by SDS-PAGE was 50.0 kDA under both reducing and non-reducing conditions. Generally, a subunit molecular mass of about 26.0 kDa was calculated on the basis of the amino acid sequence translated from the α- or β-CAs usually found in metazoans, although the Tridacna gigas CA was observed to possess a MW of 70 kDA, being a dual glycoprotein with two different CA domain, one at the amino-, the other at the carboxy-terminal parts, of the proteinCitation51. So, with the information available at this moment, we cannot rule out that the M. galloprovincialis CA may be a homodimer, or as the T. gigas enzyme, a glycoprotein with two different CA domains.
Figure 2. SDS-Page of the native CA purified from the mantles of M. galloprovincialis. Panel A, lane 2: cell extract protein from the mussel mantles before the purification; Panel B, lane 3: purified CA from pAMBS affinity column. Panel A and B, lane 1: molecular markers. Starting from the top: 250 kDa, 150 kDa, 100 kDa, 75 kDa, 50 kDa, 37 kDa, 25 kDa, 20 kDa, 15 kDa and 10 kDa.
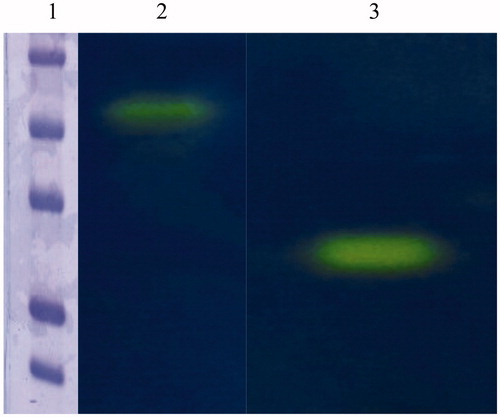
The hydratase activity of the purified mussel CA was confirmed by protonography. showed the protonogram obtained with the native mussel CA. The commercial bovine CA (bCA) was used as positive control. The yellow band corresponded to the CA position on the gel. The SDS-PAGE and protonogram showed clearly that the native mussel CA had a dimeric arrangement, suggesting that this enzyme may acts as a physiological dimer. This is very intriguing because generally α-CAs are monomeric enzymes. But, in the last years, it has been discovered dimeric arrangements for several members of the α-class, such as SspCA, from Sulfurihydrogenibium yellowstonense YO3AOP1Citation39; NgCA, from Neisseria gonorrhoeaeCitation62; CrCA, from Chlamydomonas reinhardtiiCitation63; AoCA, from Aspergillus oryzaeCitation64; and three human isoforms, indicated with the acronyms hCA VI, hCA IX and hCA XIICitation65–67. Moreover, CAs (α-type) from mammalian sources catalyze the reversible hydrolysis of esters. Thus, with p-NpA as substrate, the presence of esterase activity was investigated in the native mussel enzyme. The mussel CA showed an esterase specific activity 100 times smaller than that of the commercially available bovine bCA II.
Figure 3. Protonogram obtained using the native CA from Mytilus galloprovincialis and the commercial bovine CA (bCA). Lane 2: mussel CA protonogram showing a band (yellow color) with an apparent molecular weight of 50 kDa; Lane 1: bovine CA is present in a monomeric state corresponding at a molecular weight of about 26 kDa; Lane 1: molecular markers. Starting from the top: 75 kDa, 50 kDa, 37 kDa, 25 kDa and 20 kDa.
Primary structure analysis
The N-amino terminal sequence of the electroblotted enzyme was found to be SWGYGNDNGP. Using the Basic Local Alignment Search Tool (BLAST) and using as query sequence the N-amino terminal obtained as described above, the purified native CA completely matched (query cover 100%) the N-amino terminal of the α-CA previously identified in the genome of the Mediterranean mussel, Mytilus galloprovincialis (). The M. galloprovincialis CA is a polypeptide chain formed by 255 amino acid residues. In , the M. galloprovincialis CA sequence was aligned with Homo sapiens CA isoforms (hCA I and hCA II) sequences. It may be observed that similar to the other investigated α-CA, the mussel metalloenzyme has the conserved three His ligands, which coordinate the Zn(II) ion crucial for catalysis (His94, 96 and 119, hCA I numbering system). Interesting to note that the proton shuttle residue (His64) is conserved in the human isoforms, but is missing in the mussel enzyme, being replaced by a Lys residue. This is also the situation for the human isoform hCA III, which has Lys64 instead of His64, and also shows a reduced catalytic activity compared with isoforms hCA I and IICitation68. This residue assists the rate determining step of the catalytic cycle transferring a proton from the water coordinated to the Zn(II) ion to the environment, with formation of zinc hydroxide representing the nucleophilic species of the enzyme. A Lys in position 64 is less efficient than a His for assisting this process, which explains why the molluskan enzyme has a lower catalytic activity compared with hCA I and IICitation68. M. galloprovincialis enzyme also has the gate-keeping residues (Glu106 and Thr199), which orientate the substrate for catalysis, and are also involved in the binding of inhibitors.
Figure 4. Amino acid sequence of CA from Mytilus galloprovincialis deposited in the protein data bank. In bold, the N-amino terminal sequence obtained from the electroblotted mussel CA. The asterisk indicates amino acid identity.

Figure 5. Amino acid multialignment obtained using the Mytilus galloprovincialis CA and Homo sapiens CA isoforms (hCA I and hCA II). The zinc ligands (His94, 96 and 119) and the gate-keeper residues (Glu106 and Thr199) are conserved in the mussel and mammalian sequences. The proton shuttle residue (His64) is missing in the M. galloprovincialis enzyme. hCA I numbering system was used. The asterisk (*) indicates identity at all aligned positions; the symbol (:) relates to conserved substitutions, while (.) means that semi-conserved substitutions are observed. Multialignment was performed with the program Muscle, version 3.1.

Enzyme kinetics
Using the stopped-flow technique, the kinetic parameters were determined for the native α-CA using CO2 as a substrateCitation69. The activity of the mussel enzyme was compared with that of other α-CAs such as Homo sapiens isoforms (hCA I and hCA II), Stylophora pistillata isoforms (STPCA and STPCA-2) ()Citation18,Citation70,Citation71. As shown in , the mussel α-CA had a kcat = 4.1 × 105 s−1 with a kcat/Km ratio 1.38 times lower with respect to that of the hCA I, which possessed a kcat of 2.0 × 105 s−1. Moreover, the kinetic constants of the native mussel CA are very similar to those obtained for the coral isoform STPC and only slightly lower than the isoform STPCA-2. This is very intriguing since mussel α-CA respect to the two coral isoforms, lacked of a His64. This residue is involved in the transfer of proton from the water coordinated to the Zn(II) ion to the environment with the function to accelerate the rate of the catalytic cycle. STPCA-2 from scleractinian coral Stylophora pistillata is considered highly active and has an intracellular localization in the oral endoderm and in the aboral tissue of the coral. Interesting to note that the native mussel CA is not well affected by acetazolamide inhibition showing a KI of 380 mM (). The intracellular localization of the native mussel CA and its kinetic similarity with STPCA-2, could allow that the Mytilus galloprovincialis α-CA is involved in pH regulation and/or in the delivery of bicarbonate helping the mussel during the formation of the shell.
Table 1. Kinetic parameters for the CO2 hydration reaction catalyzed by the purified native mussel CA, the Homo sapiens CA isoforms (hCA I and hCA II) and coral CA isoforms (STPCA and STPCA-2). Acetazolamide (AAZ) inhibition data are also shown.
Conclusions
A α-CA has been purified and characterized biochemically from the mollusk Mytilus galloprovincialis. As in most mollusks, this α-CA seems to be involved in the biomineralization processes leading to the precipitation of calcium carbonate in the mussel shell. The new enzyme has a molecular weight of 50 kDa, which is roughly two times higher than that of a monomeric α-class enzyme. Thus, Mytilus galloprovincialis α-CA is either a dimer, or similar to the Tridacna gigas CA described earlier, may have two different CA domains in its polypeptide chain. The Mytilus galloprovincialis α-CA sequence contains the three His residues acting as zinc ligands and the gate-keeper residues (Glu106-Thr199) present in all α-CAs, but had a Lys in position 64 and not a His as proton shuttling residue, being thus similar to the human isoform hCA III. This probably explains the relatively low catalytic activity of Mytilus galloprovincialis α-CA, with the following kinetic parameters for the CO2 hydration reaction: kcat= 4.1 × 105 s−1 and kcat/Km of 3.6 × 107 M−1 × s−1. The enzyme activity was poorly inhibited by the sulfonamide acetazolamide, with a KI of 380 nM. This study is one of the few describing in detail the biochemical characterization of a molluskan CA and may be useful for understanding in better detail the phylogeny of these enzymes and their role in biocalcification processes. In addition, the mussel CA is an attractive candidate for its potential use in carbon dioxide sequestration. The recombinant DNA technology will make possible the ingegnerization of the mussel CA and production of large quantities of the metalloenzyme with the aim to use the M. galloprovincialis CA, either free or immobilized, in the CO2 biomimetic capture process.
Disclosure statement
The authors report no conflicts of interest.
Funding
This research was financed by the grant “SMART GENERATION – Sistemi e tecnologie sostenibili per la generazione di energia – PON03PE_00157_1, OR3 – Bio-sistemi di cattura ed utilizzazione della CO2” funded by MIUR.
References
- Marin F, Le Roy N, Marie B. The formation and mineralization of mollusk shell. Front Biosci (Schol Ed) 2012;4:1099–125.
- Marie B, Luquet G, Bedouet L, et al. Nacre calcification in the freshwater mussel Unio pictorum: carbonic anhydrase activity and purification of a 95 kDa calcium-binding glycoprotein. Chembiochem 2008;9:2515–23.
- Barthelat F. Growing a synthetic mollusk shell. Science 2016;354:32–33.
- Olson IC, Kozdon R, Valley JW, Gilbert PU. Mollusk shell nacre ultrastructure correlates with environmental temperature and pressure. J Am Chem Soc 2012;134:7351–58.
- Arivalagan J, Yarra T, Marie B, et al. Insights from the shell proteome: biomineralization to adaptation. Mol Biol Evol 2016;34:66–77.
- Venn AA, Tambutte E, Holcomb M, et al. Impact of seawater acidification on pH at the tissue-skeleton interface and calcification in reef corals. Proc Natl Acad Sci U S A 2013;110:1634–39.
- Capasso C, Supuran CT. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin Ther Targets 2015;19:1689–704.
- Capasso C, Supuran CT. An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria? J Enzyme Inhib Med Chem 2015;30:325–32.
- Zoccola D, Innocenti A, Bertucci A, et al. Coral carbonic anhydrases: regulation by ocean acidification. Mar Drugs 2016;14:109. doi:10.3390/md14060109.
- Capasso C, Supuran CT. Anti-infective carbonic anhydrase inhibitors: a patent and literature review. Expert Opin Ther Pat 2013;23:693–704.
- Capasso C, Supuran CT. Sulfa and trimethoprim-like drugs – antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J Enzyme Inhib Med Chem 2014;29:379–87.
- Capasso C, Supuran CT. An overview of the selectivity and efficiency of the bacterial carbonic anhydrase inhibitors. Curr Med Chem 2015;22:2130–39.
- Capasso C, Supuran CT. An overview of the carbonic anhydrases from two pathogens of the oral cavity: Streptococcus mutans and Porphyromonas gingivalis. Curr Top Med Chem 2016;16:2359–68.
- Ozensoy Guler O, Capasso C, Supuran CT. A magnificent enzyme superfamily: carbonic anhydrases, their purification and characterization. J Enzyme Inhib Med Chem 2016;31:689–94.
- Supuran CT, Capasso C. The η-class carbonic anhydrases as drug targets for antimalarial agents. Expert Opin Ther Targets 2015;19:551–63.
- Xiang L, Kong W, Su JT, et al. Amorphous calcium carbonate precipitation by cellular biomineralization in mantle cell cultures of Pinctada fucata. PLoS One 2014;9:e113150.
- Bertucci A, Moya A, Tambutte S, et al. Carbonic anhydrases in anthozoan corals – a review. Bioorg Med Chem 2013;21:1437–50.
- Bertucci A, Tambutte S, Supuran CT, et al. A new coral carbonic anhydrase in Stylophora pistillata. Mar Biotechnol (NY) 2011;13:992–1002.
- Le Goff C, Ganot P, Zoccola D, et al. Carbonic anhydrases in cnidarians: novel perspectives from the Octocorallian Corallium rubrum. PLoS One 2016;11:e0160368.
- Zoccola D, Ganot P, Bertucci A, et al. Bicarbonate transporters in corals point towards a key step in the evolution of cnidarian calcification. Sci Rep 2015;5:9983.
- Miyamoto H, Miyoshi F, Kohno J. The carbonic anhydrase domain protein nacrein is expressed in the epithelial cells of the mantle and acts as a negative regulator in calcification in the mollusc Pinctada fucata. Zoolog Sci 2005;22:311–15.
- Vullo D, Del Prete S, Osman SM, et al. Burkholderia pseudomallei gamma-carbonic anhydrase is strongly activated by amino acids and amines. Bioorg Med Chem Lett 2017;27:77–80.
- Vullo D, Del Prete S, De Luca V, et al. Anion inhibition studies of the beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem Lett 2016;26:1406–10.
- Vullo D, Del Prete S, Capasso C, Supuran CT. Carbonic anhydrase activators: activation of the β-carbonic anhydrase from Malassezia globosa with amines and amino acids. Bioorg Med Chem Lett 2016;26:1381–5.
- Supuran CT, Capasso C. New light on bacterial carbonic anhydrases phylogeny based on the analysis of signal peptide sequences. J Enzyme Inhib Med Chem 2016;31:1254–60.
- Melis C, Meleddu R, Angeli A, et al. Isatin: a privileged scaffold for the design of carbonic anhydrase inhibitors. J Enzyme Inhib Med Chem 2017;32:68–73.
- Del Prete S, Vullo DD, Fonzo P, et al. Sulfonamide inhibition profile of the gamma-carbonic anhydrase identified in the genome of the pathogenic bacterium Burkholderia pseudomallei the etiological agent responsible of melioidosis. Bioorg Med Chem Lett 2017;27:490–5.
- Del Prete S, Vullo D, De Luca V, et al. Anion inhibition profiles of alpha-, beta- and gamma-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem 2016;24:3413–17.
- Del Prete S, De Luca V, Vullo D, et al. A new procedure for the cloning, expression and purification of the beta-carbonic anhydrase from the pathogenic yeast Malassezia globosa, an anti-dandruff drug target. J Enzyme Inhib Med Chem 2016;31:1156–61.
- De Luca V, Vullo D, Del Prete S, et al. Cloning, characterization and anion inhibition studies of a gamma-carbonic anhydrase from the Antarctic bacterium Colwellia psychrerythraea. Bioorg Med Chem 2016;24:835–40.
- Del Prete S, Vullo D, De Luca V, et al. Biochemical characterization of recombinant beta-carbonic anhydrase (PgiCAb) identified in the genome of the oral pathogenic bacterium Porphyromonas gingivalis. J Enzyme Inhib Med Chem 2015;30:366–70.
- Del Prete S, De Luca V, Scozzafava A, et al. Biochemical properties of a new alpha-carbonic anhydrase from the human pathogenic bacterium, Vibrio cholerae. J Enzyme Inhib Med Chem 2014;29:23–27.
- Del Prete S, Vullo D, De Luca V, et al. Biochemical characterization of the delta-carbonic anhydrase from the marine diatom Thalassiosira weissflogii, TweCA. J Enzyme Inhib Med Chem 2014;29:906–11.
- Pinard MA, Lotlikar SR, Boone CD, et al. Structure and inhibition studies of a type II beta-carbonic anhydrase psCA3 from Pseudomonas aeruginosa. Bioorg Med Chem 2015;23:4831–38.
- Ferraroni M, Del Prete S, Vullo D, et al. Crystal structure and kinetic studies of a tetrameric type II β-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Acta Crystallogr D Biol Crystallogr 2015;71:2449–56.
- De Simone G, Monti SM, Alterio V, et al. Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense. Bioorg Med Chem Lett 2015;25:2002–6.
- Zolnowska B, Slawinski J, Pogorzelska A, et al. Carbonic anhydrase inhibitors. Synthesis, and molecular structure of novel series N-substituted N′-(2-arylmethylthio-4-chloro-5-methylbenzenesulfonyl)guanidines and their inhibition of human cytosolic isozymes I and II and the transmembrane tumor-associated isozymes IX and XII. Eur J Med Chem 2014;71:135–47.
- De Luca L, Ferro S, Damiano FM, et al. Structure-based screening for the discovery of new carbonic anhydrase VII inhibitors. Eur J Med Chem 2014;71:105–11.
- Di Fiore A, Capasso C, De Luca V, et al. X-ray structure of the first ‘extremo-alpha-carbonic anhydrase’, a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr D Biol Crystallogr 2013;69:1150–9.
- Supuran CT. Structure-based drug discovery of carbonic anhydrase inhibitors. J Enzyme Inhib Med Chem 2012;27:759–72.
- Supuran CT. Carbonic anhydrases – an overview. Curr Pharm Des 2008;14:603–14.
- Supuran CT. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin Drug Discov 2017;12:61–88.
- Bhatt A, Mahon BP, Cruzeiro VW, et al. Structure-activity relationships of benzenesulfonamide-based inhibitors towards carbonic anhydrase isoform specificity. Chembiochem 2017;17:213–22.
- Alterio V, Langella E, Viparelli F, et al. Structural and inhibition insights into carbonic anhydrase CDCA1 from the marine diatom Thalassiosira weissflogii. Biochimie 2012;94:1232–41.
- Supuran CT. Structure and function of carbonic anhydrases. Biochem J 2016;473:2023–32.
- Buzas GM, Supuran CT. The history and rationale of using carbonic anhydrase inhibitors in the treatment of peptic ulcers. In memoriam Ioan Puscas (1932-2015). J Enzyme Inhib Med Chem 2016;31:527–33.
- Carta F, Supuran CT, Scozzafava A. Sulfonamides and their isosters as carbonic anhydrase inhibitors. Future Med Chem 2014;6:1149–65.
- Migliardini F, De Luca V, Carginale V, et al. Biomimetic CO2 capture using a highly thermostable bacterial alpha-carbonic anhydrase immobilized on a polyurethane foam. J Enzyme Inhib Med Chem 2014;29:146–50.
- Boone CD, Habibzadegan A, Gill S, McKenna R. Carbonic anhydrases and their biotechnological applications. Biomolecules 2013;3:553–62.
- Russo ME, Olivieri G, Capasso C, et al. Kinetic study of a novel thermo-stable alpha-carbonic anhydrase for biomimetic CO2 capture. Enzyme Microb Technol 2013;53:271–7.
- Leggat W, Dixon R, Saleh S, Yellowlees D. A novel carbonic anhydrase from the giant clam Tridacna gigas contains two carbonic anhydrase domains. FEBS J 2005;272:3297–305.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–85.
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem 1987;262:10035–38.
- Thomsen J, Bucher D, Brunfeldt K, Olesen H. An improved procedure for automated Edman degradation used for determination of the N-terminal amino acid sequence of human transcobalamin I and human intrinsic factor. Eur J Biochem 1976;69:87–96.
- Capasso C, De Luca V, Carginale V, et al. Biochemical properties of a novel and highly thermostable bacterial alpha-carbonic anhydrase from Sulfurihydrogenibium yellowstonense YO3AOP1. J Enzyme Inhib Med Chem 2012;27:892–97.
- Armstrong JM, Myers DV, Verpoorte JA, Edsall JT. Purification and properties of human erythrocyte carbonic anhydrases. J Biol Chem 1966;241:5137–49.
- Del Prete S, De Luca V, Iandolo E, et al. Protonography, a powerful tool for analyzing the activity and the oligomeric state of the gamma-carbonic anhydrase identified in the genome of Porphyromonas gingivalis. Bioorg Med Chem 2015;23:3747–50.
- Del Prete S, De Luca V, Supuran CT, Capasso C. Protonography, a technique applicable for the analysis of eta-carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:920–24.
- De Luca V, Del Prete S, Supuran CT, Capasso C. Protonography, a new technique for the analysis of carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:277–82.
- Del Prete S, De Luca V, De Simone G, et al. Cloning, expression and purification of the complete domain of the η-carbonic anhydrase from Plasmodium falciparum. J Enzyme Inhib Med Chem 2016;31:54–59.
- Del Prete S, Vullo D, De Luca V, et al. Comparison of the sulfonamide inhibition profiles of the alpha-, beta- and gamma-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem Lett 2016;26:1941–46.
- Huang S, Xue Y, Sauer-Eriksson E, et al. Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide. J Mol Biol 1998;283:301–10.
- Suzuki K, Yang SY, Shimizu S, et al. The unique structure of carbonic anhydrase alphaCA1 from Chlamydomonas reinhardtii. Acta Crystallogr D Biol Crystallogr 2011;67:894–901.
- Cuesta-Seijo JA, Borchert MS, Navarro-Poulsen JC, et al. Structure of a dimeric fungal α-type carbonic anhydrase. FEBS Lett 2011;585:1042–8.
- Pilka ES, Kochan G, Oppermann U, Yue WW. Crystal structure of the secretory isozyme of mammalian carbonic anhydrases CA VI: implications for biological assembly and inhibitor development. Biochem Biophys Res Commun 2012;419:485–89.
- Alterio V, Hilvo M, Di Fiore A, et al. Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX. Proc Natl Acad Sci U S A 2009;106:16233–38.
- Whittington DA, Grubb JH, Waheed A, et al. Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane-associated isozymes. J Biol Chem 2004;279:7223–28.
- Vullo D, Nishimori I, Scozzafava A, Supuran CT. Carbonic anhydrase activators: activation of the human cytosolic isozyme III and membrane-associated isoform IV with amino acids and amines. Bioorg Med Chem Lett 2008;18:4303–7.
- Khalifah RG. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biol Chem 1971;246:2561–73.
- Bertucci A, Innocenti A, Scozzafava A, et al. Carbonic anhydrase inhibitors. Inhibition studies with anions and sulfonamides of a new cytosolic enzyme from the scleractinian coral Stylophora pistillata. Bioorg Med Chem Lett 2011;21:710–14.
- Moya A, Tambutte S, Bertucci A, et al. Carbonic anhydrase in the scleractinian coral Stylophora pistillata: characterization, localization, and role in biomineralization. J Biol Chem 2008;283:25475–84.
