 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Emergence of multidrug-resistant bacteria forces us to explore new therapeutic strategies, and proteins involved in key metabolic pathways are promising anti-bacterial targets. Bifunctional flavin-adenine dinucleotide (FAD) synthetases (FADS) are prokaryotic enzymes that synthesise the flavin mononucleotide (FMN) and FAD cofactors. The FADS from the human pathogen Streptococcus pneumoniae (SpnFADS)–causative agent of pneumonia in humans − shows relevant catalytic dissimilarities compared to other FADSs. Here, by integrating thermodynamic and kinetic data, we present a global description of the riboflavin kinase activity of SpnFADS, as well as of the inhibition mechanisms regulating this activity. Our data shed light on biophysical determinants that modulate species-specific conformational changes leading to catalytically competent conformations, as well as binding rates and affinities of substrates versus products. This knowledge paves the way for the development of tools − that taking advantage of the regulatory dissimilarities during FMN biosynthesis in different species − might be used in the discovery of specific anti-pneumococcal drugs.
Introduction
Streptococcus pneumoniae is the causative agent of human pneumonia diseaseCitation1, meningitis and bacteremia in children and adults. It is estimated that 1.6 millions of people, including 1 million of children under age five die every year of pneumonia diseaseCitation2,Citation3. The irruption during the last decades of multi-drug resistant pneumococci has revealed the need of finding new drugs, as well as novel drug targets. The bifunctional flavin-adenine dinucleotide (FAD) synthetase (FADS) from S. pneumoniae (SpnFADS) arises as a potential drug targetCitation4,Citation5, since it synthesises the essential cofactors flavin mononucleotide (FMN) and FADCitation6, involved in a plethora of vital processes as part of flavoproteins and flavoenzymesCitation7–9. As other bacterial FADSs, SpnFADS produces FMN and FAD through two sequential activities. First, a riboflavin kinase activity (RFK) at its C-terminus module phosphorylates riboflavin (RF) to FMN, and then the adenosine 5′-triphosphate (ATP):FMN:adenylyltransferase (FMNAT) activity of the enzyme N-terminus module transforms FMN into FADCitation10–12. Two characteristics stand out among the suitable properties of FADSs as drug targets. First, bacterial FADSs differ from the human proteins that synthesise FMN and particularly FAD. Thus, the eukaryotic FMNAT activity is catalysed by an enzyme with a completely different protein folding and chemistry relative to the N-terminus of bacterial FADSCitation13–16. Regarding FMN production, the monofunctional Homo sapiens RFK has been hardly characterised so far. Overall, it is structurally homologous to the RFK module of bacterial enzymes, but only structures containing bound ligands are availableCitation17,Citation18. Nonetheless, structural data predict differences in conformational changes to achieve the catalytic complexCitation19,Citation20, and the scarce biochemical information suggests differences in redox environmental requirements for maximal activityCitation11,Citation12. Second, the members of the prokaryotic FADSs family studied up to now differ catalytically among them, which might facilitate the design of new species-specific medicines. Structurally, when comparing SpnFADS with the member of the family so far best characterised—which is that from the organism Corynebacterium ammoniagenes (CaFADS)—it presents a very similar structure, with little differences in the position of some key loops. However, these two proteins only share the 26% of sequence homologyCitation6. Despite the overall structural similitude among prokaryotic FADSsCitation6,Citation12,Citation18, SpnFADS shows three main distinctive functional behaviors; (i) it mainly stabilises monomers—which are the functional formCitation6—or traces of dimers, during catalysis; (ii) its FMNAT activity requires reduced FMN as a substrate; and (iii) its RFK activity is not regulated by the RF substrateCitation6.
Here, we focus on the SpnFADS RFK activity, using pre-steady and steady-state biophysical techniques to describe this activity and the inhibition mechanism employed by this enzyme to regulate FMN synthesis. We take a close look at the thermodynamic and kinetic basis that determine the ligand binding order, the co-operativity between ligands and the inhibitory mechanism performed by the reaction products. Furthermore, we compare our results with those obtained for the FADS from C. ammoniagenes to identify key regulatory differences between both proteins.
Methods
Cloning, expression, and purification of SpnFADS
SpnFADS was cloned, overexpressed, and purified as previously describedCitation6. In short, Escherichia coli Bl21 Star™ (DE3) cells were transformed with a pET-15b vector that contains the DNA sequence encoding SpnFADS. Transformed cells were grown and protein expression induced through isopropyl β-D-1-thiogalactopyranoside (IPTG) addition. Cells were harvested and broken by sonication. The supernatant was loaded into a His-Trap affinity column and the protein eluted applying a 10–500 mM imidazole gradient. The His6-Tag was removed and then the protein was loaded into HisTrap HP and GSTrap 4B connected columns. The unbound fraction was further purified by size exclusion chromatography. The protein purity was tested and pure protein aliquots were conserved at −80 °C.
Steady-state RFK activity
The SpnFADS RFK activity was measured at 25 °C in 500 µL of 20 mM 1,4-piperazinediethanesulfonic acid (PIPES) and 0.8 mM MgCl2, pH 7.0. Reaction samples contained different concentrations of RF (0.5–30 µM) and ATP (10–500 µM), as previously describedCitation21,Citation22. The inhibitory effect of the products of the reaction was analysed as previously describedCitation23. In short, the SpnFADS RFK activity was determined at increasing concentrations of FMN, varying the ATP concentration and keeping the RF constant (when studying the inhibitory effect of FMN), and at increasing concentrations of adenosine 5′-diphosphate (ADP), varying the RF concentration and keeping the ATP fixed (when studying the inhibitory effect of ADP). The flavin composition of the supernatant was determined using an Alliance high performance liquid chromatography (HPLC) system (Waters, Milford, MA, USA) equipped with a 2707 autosampler and a HSST3 column (4.6 × 50 mm, 3.5 mm; Waters) preceded by a precolumn (4.6 × 20 mm, 3.5 mm; Waters) as previously describedCitation21,Citation22. The FMN concentration was quantified using its standard curve. All the experiments were carried out in triplicate.
Michaelis–Menten (Km) and catalytic rate (kcat) constants were obtained by fitting the obtained data to the Michaelis equationCitation24. The inhibitory mechanism performed by the products of the RFK reaction − FMN and ADP − was analysed by evaluating their effects on the Km and kcat values, obtained by the individual fitting of data sets to the Michaelis–Menten model. Additionally, the data sets were globally fit utilising the Lineweaver–Burk equations for mixed inhibitionCitation25 (EquationEquation (1)(1)
(1) ).
(1)
(1)
where, [S] and [I] are the concentration of substrates and product inhibitor, respectively. Ki and K’i are the product inhibition constantsCitation25. Experiments were performed in triplicate. The estimated errors in kcat, Km, and Ki were within ±15% of their values.
Pre-steady-state kinetics
Kinetic experiments in the pre-steady state were registered as previously describedCitation23, using stopped-flow spectroscopy on an Applied Photophysics SX17. MV spectrophotometer, using the Xscan software (Applied Photophysics Ltd., Leatherhead, UK). Fast kinetic measurements were carried out as previously describedCitation23, at 25 °C in PIPES 20 mM pH 7.0, 0.8 mM MgCl2. About 0.2 µM SpnFADS was mixed with reaction samples that contained increasing concentrations of the flavin ligands (FLV, herein indicating RF or FMN), in the presence and in the absence of ADP or ATP (herein referred as ANP). Controls were measured in the same buffer but without MgCl2. All concentrations indicated here are the final ones in the reaction cell. The kinetic traces were registered until obtaining three reproducible traces.
Kinetic traces were fit to exponential equations (EquationEquation (2)(2)
(2) ), where each exponential term describes a different process. A linear correction term was added (EquationEquation (3)
(3)
(3) ) when a specific process was not finished within the measuring timeframe.
(2)
(2)
(3)
(3)
where, Ai and kobs,i are the amplitude and the observed kinetic constant for each process (i) that contributes to the overall time-dependent fluorescence change.
The processes whose kobs showed a linear dependency on the flavin concentration were fit to a one-step model that describes the kinetic equilibrium for the formation and the dissociation of enzyme-flavin complex (EquationEquation (4)(4)
(4) )
(4)
(4)
where, kon and koff are the complex formation and dissociation kinetic constants, respectively.
Experiments were performed in triplicate. The flavin photobleaching was analysed as previously describedCitation23 (not shown).
Isothermal titration calorimetry
We performed isothermal titration calorimetry (ITC) assays to elucidate both the ligand binding order and the thermodynamic inhibition produced by the binding of the FLV and ANP ligands. Titrations were performed on an AutoITC200 (MicroCal, Malvern, UK) thermostated at 20 °C, as previously describedCitation23. Approximately 25 µM SpnFADS contained in a 200 µL-cell was titrated with solutions of 180 µM RF, 250 µM FMN, or 350 µM ATP or ADP. Additionally, titrations with ANP ligands into mixtures that contain the protein pre-bound to FLV ligands were performed, as well as titrations with FLV ligands into pre-bound SpnFADS:ANP complexes. The titrations were conducted through 19 stepwise injections of 2 µL of the titrating ligand to the calorimetric cell, as previously describedCitation21,Citation26. Both the ligands and the protein were dissolved in 20 mM PIPES, pH 7.0, either in presence of 0.8 mM MgCl2 or in absence of this cation, and degassed before the titration. The enthalpy change (ΔH), the association constant (Ka), and the binding stoichiometry (N) were obtained through non-linear least squares regression of the data using a homemade fitting routine corresponding to a model for one or two independent binding sites, implemented in Origin 7.0 (OriginLab, Northampton, MA, USA), as previously describedCitation21,Citation26. The Gibbs free energy (ΔG), the entropic contribution (−TΔS), and the dissociation constant (Kd) were obtained using well-known thermodynamic equations.
Cooperativity constants (α) between ANP and FLV ligands were obtained as previously describedCitation23,Citation27,Citation28. Particular titrations with ANP ligands into mixtures of the protein and 100 µM FLV were fit to a homemade fitting routine corresponding to a model that considers the influence of the FLV ligand in the protein binding affinity for the ANP ligandCitation27,Citation28.
Experiments were performed in triplicate. The errors considered in the measured parameters (±15% in Kd and Ka values, ±0.3 kcal mol−1 in ΔG, ΔH, and −TΔS and ±20% in α) were assumed to be larger than the standard deviation between replicates and the numerical error after fitting analysis.
Results and discussion
The products of the reaction inhibit the RFK activity of SpnFADS
Feedback inhibition is a frequent strategy to regulate enzymes involved in key metabolic pathwaysCitation29,Citation30. Some bacteria regulate FMN synthesis through the inhibition of the RFK activity of their bifunctional FADSs by the reaction products–FMN and ADP–and/or the RF substrateCitation19,Citation23, although the inhibition level triggered differs between organismsCitation6,Citation23. Here, our Michaelis–Menten plots of the SpnFADS RFK activity, for both reaction substrates–RF and ATP–at increasing concentrations of the FMN and ADP products, reveal that kcat decreases while KmATP and KmFMN increase (. These data point out to mixed or uncompetitive inhibition mechanisms. Therefore, to discern the inhibition mechanism, we carried out Lineweaver–Burk representations (), which reveal that both products act as mixed inhibitors (. Thus, FMN and ADP bind to the free enzyme as well as to the SpnFADS:ATP and SpnFADS:RF complexes, respectively. However, their affinities for the free protein and for the complex considerably differ (), being KiADP and KiFMN 6.4 and 5.5 times smaller than K´iADP and K´iFMN, respectively. That indicates that both reaction products bind preferentially to the free enzyme. Also, the considerably smaller inhibition constants for FMN compared to ADP (KiADP/KiFMN = 100) reveals the flavin product as a much more potent inhibitor ().
Figure 1. Inhibition of the SpnFADS RFK reaction by FMN and ADP products. Michaelis–Menten plots as a function of (A) ATP and (C) RF substrates at different concentrations of the ADP and FMN products. Lineaweaver–Burk representation at different (B) ADP and (D) FMN concentrations with global fitting to the equation for mixed inhibition.
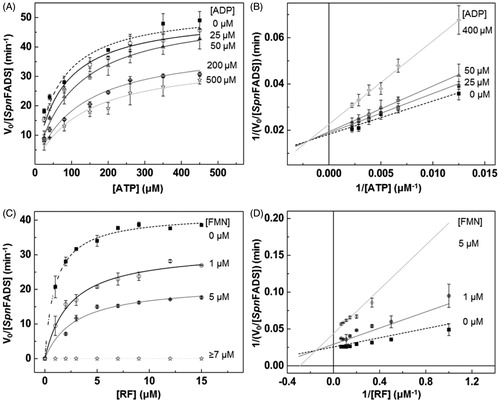
Table 1. Steady-state kinetic parameters for the SpnFADS RFK activity as calculated by global fitting to the Lineweaver–Burk equation.
Equivalent data obtained for the so far best characterised FADS–that belonging to C. ammoniagenes (CaFADS)–show that ADP and FMN are, respectively, competitive and uncompetitive inhibitors of its RFK activityCitation23. The FMN product appears as a considerable more potent inhibitor of the RFK activity of CaFADS than of SpnFADS, as the KmRF/KiFMN values of 7.1 and 0.8 for CaFADS and SpnFADS, respectively, show. This, together with the fact that RF strongly inhibits the CaFADS RFK activity but not the SpnFADS oneCitation6, points to species-specific inhibition and activity modulation mechanisms. Physiologically, other preferred regulatory strategies, such as the use of reduced FMN during the FMNAT activity, and the different oligomeric assemblies established along both activities of these enzymesCitation6,Citation31,Citation32, might be also behind the distinct requirements for the RFK activity regulation.
The binding of the substrates of the RFK reaction is the fastest and most favored process for SpnFADS
Then we used stopped-flow spectrophotometryCitation23,Citation33 to kinetically identify individual steps during the SpnFADS RFK reaction. Here, we took advantage of two aspects; (i) RF and FMN have the same fluorescence spectra and yieldsCitation34 and (ii) under oxidising conditions flavins only bind and get transformed at the RFK module of SpnFADSCitation6. Consequently, although RF would be transformed into FMN, we would only observe fluorescence changes derived from flavin binding, flavin dissociation, and/or conformational changes at the RFK module.
When mixing SpnFADS with RF or FMN (herein FLV) ligands, we only observed flavin photobleaching, similarly to that reported for CaFADSCitation23. This suggests that SpnFADS is not able either to bind oxidised flavins or to internalise their isoalloxazine ring. On the contrary, fast and intense exponential flavin fluorescence variations were observed when mixing SpnFADS simultaneously with ANP (ATP or ADP) and FLV ligands. All samples showed fast initial fluorescence decays, but mixtures containing ATP also presented subsequent fluorescence increases (). As recently reported for CaFADSCitation23, we relate the initial fluorescence decay to FLV binding and internalisation through a conformational change of the loop-L4c, that closes the flavin binding site when ANP is previously bound (Supplementary Figure SP1)Citation20. The succeeding fluorescence raise can be similarly related to an ATP-induced conformational change that re-opens the flavin binding site, making the isoalloxazine ring accessible again to the solventCitation23 (, schemes). kobs1 and kobs2 represent, respectively, the observed rates for these processes. The linear dependence of kobs1 on FLV concentrations () allows us calculating flavin association and dissociation rate constants (kon and koff, respectively) and the derived dissociation constants (Kd). The binding of the RFK substrates–RF and ATP–is the fastest process (), and also shows the largest fluorescence decay amplitude (). Hence, contrary to that reported for CaFADSCitation23, the SpnFADS RFK site preferably binds the RF and ATP substrates over other combinations of ligands, being the binding of the FMN product in presence of the ATP substrate the least favored combination. In addition, kobs2 values show that the conformational rearrangement for flavin release is considerably faster when RF is the flavin initially bound (). Moreover, kobs2 shows a biphasic behavior negatively affected by the FMN concentration, indicating that accumulation of FMN hinders its release. Noticeable, kobs1 and kobs2 for processes that involve RF and ATP are in the kcat range, revealing that the reaction steps represented by these parameters are relevant for catalysis. Nevertheless, as the kobs1/kobs2 ratio indicates, the binding and the internalisation of the reaction substrates is 4 times faster than the subsequent conformational change that releases flavins to the solvent, being this last process the SpnFADS RFK reaction bottleneck.
Figure 2. Pre-steady-state stopped-flow kinetics for RF and FMN binding to SpnFADS in the presence of adenine nucleotides. (A) Kinetic traces for the flavin fluorescence evolution upon mixing the protein with ANP-FLV combinations. (B) Example of fittings of kinetic traces. Evolution of (C) kobs1 and (D) kobs2 on FLV concentration, with schemes representing the corresponding processes in the insets.
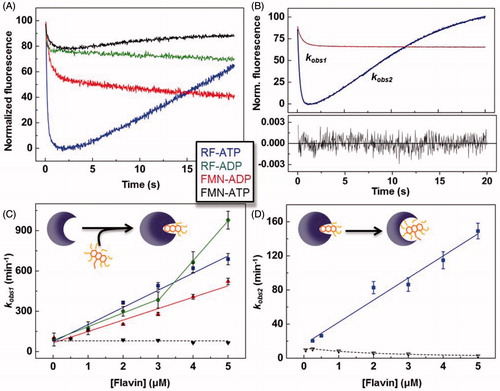
Table 2. Pre-steady-state kinetic parameters for flavins binding and dissociation to SpnFADS in the presence of adenine nucleotides.
Table 3. Cooperativity coefficients for the binding of the different combinations of FLV and ANP ligands to SpnFADS in Mg2+ absence.
Considering our results in the context of those reported for the RFK cycle of CaFADS, some key facts are worthy to be highlighted. Both proteins require ANP nucleotides to bind/internalise flavin ligands, and they show the same overall individual processes along the reaction. Additionally, both FADSs are able to bind all the possible combinations of ANP-FLV ligands, although differences in the relative binding rates and magnitude of the associated spectroscopic changes must be behind the lower inhibition levels shown by SpnFADS. Thus, while binding of the substrates of the RFK reaction—that is RF and ATP—is the fastest and most favored process for SpnFADS (), it is the slowest one for CaFADSCitation23. This fact together with a 10-times higher KiADP, and a 9-times higher KmRF/Ki FMN ratio for CaFADS than for SpnFADS, explains the larger inhibition by the reaction products observed for CaFADS ()Citation23. Comparison of the kobs2 behavior for both enzymes ()Citation23—kobs2 linearly depends on the RF concentration for SpnFADS while it shows a biphasic behavior for CaFADS—shows that during the CaFADS RFK reaction, high RF concentrations inhibit the ATP-induced conformational change for flavin release, while SpnFADS does not present such inhibition. This might be a determinant of the strong inhibition by RF that CaFADS suffersCitation21,Citation23, which is absent in SpnFADS.
Thermodynamics explain the modest inhibition of the SpnFADS RFK activity
We next used ITC to determine whether the identified kinetic processes were relevant to reach the thermodynamic equilibrium. We titrated with the substrates and the products of the RFK reaction, both free SpnFADS and its binary mixtures with either ANP or FLV ligands. Since Mg2+ is necessary for the RFK reaction to occur, we performed all titrations with and without this cation, to determine its role on ligands binding. However, titrations that involve both reaction substrates only were executed without Mg2+, since the reaction heat would mask the binding enthalpy ().
Figure 3. Gibbs free energy diagrams for the interaction of SpnFADS with its ligands. Titrations performed (A) without MgCl2, and (B) with 0.8 mM MgCl2. Numbers indicate the ΔG value in kcal mol−1. The arrows thickness is proportional to the protein fraction able to bind the ligand. NM denotes paths where the reaction heat would mask the binding enthalpy.
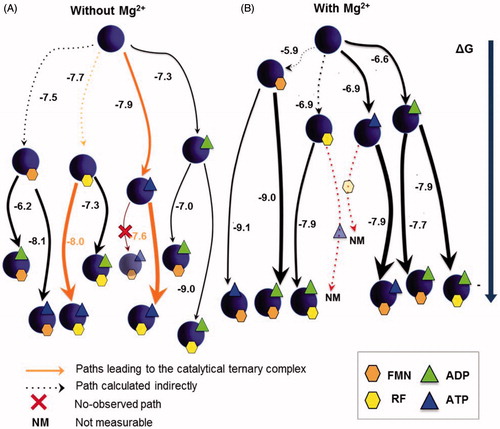
illustrates all the possible binding pathways occurring in the interaction landscape of the SpnFADS RFK module with the substrates and products. As reported for CaFADSCitation23, Mg2+ is key in the formation of SpnFADS:ANP:FLV complexes ( and Supplementary Table SP1): (i) by increasing the protein fraction prone to interact with a specific ligand, and consequently, the probability of the pathway, and (ii) by favoring the formation of ternary complexes from the binary ones (compare arrows thickness and ΔG values in ). The stabilisation of these ternary complexes by Mg2+ is mainly a consequence of the less unfavorable binding entropy (Supplementary Table SP1), which suggests different conformations in ternary complexes with and without MgCl2. It is worthy to highlight that although FLV binding to the free protein is not directly observed by ITC or stopped-flow spectrophotometry, flavins—particularly when Mg2+ is present—highly modify the protein affinity for ANP ligands (, Supplementary Table SP1). This cooperative effect—also observed with CaFADSCitation23—evinces that RF and FMN act as slow-binding ligandsCitation35. However, although their binding is too slow to be measured within the experimental time, it can be indirectly estimatedCitation27,Citation28,Citation36.
Titrations without MgCl2 () allow establishing an interaction diagram that includes “pseudo-reactive” pathways with the reaction substrates (SpnFADS:ATP:RF). Comparison of this diagram with that obtained for CaFADSCitation23 reveals important differences, which might contribute to the regulatory dissimilarities displayed between both proteins. Thus, (i) two alternative pathways lead to the SpnFADS:ATP:RF complexes, while CaFADS presents only one pathwayCitation23 and (ii) SpnFADS:ATP:RF complexes are almost the most stable ones—as their lowest position in the ΔG diagram indicates—and also the most probable—thickest orange arrows—. This situation is completely different for CaFADS, where all the other non-productive CaFADS:ANP:FLV complexes are favored against CaFADS:ATP:RFCitation23.
We can extract two main conclusions from these results. First, both proteins achieve the catalytic FADS:RF:ATP complex through different mechanisms; SpnFADS seems to follow a random sequential binding of the RFK substrates, while CaFADS requires its concerted fitCitation23. Second, as the RFK reaction progresses and the products accumulate, pathways different from the reactive one are favored in the case of CaFADS, which reduces the efficiency of the overall catalysisCitation23. For its part, the pathway that leads to SpnFADS:ATP:RF is the most favored, at least until reaching a high [products]/[substrates] ratio.
Conformational differences explain the dissimilar regulation of prokaryotic RFK cycles
Structurally, the different mechanisms by which RF and ATP get allocated in the RFK active site might be related to the simpler conformational changes required by SpnFADS. shows the relative conformation of the catalytic PTAN motif in SpnFADS regarding CaFADS () and CaFADS:FMN:ADP structures (). Remarkably, the conformation of this motif in SpnFADS resembles that of CaFADS in a ternary complex with the products of the RFK reaction. So, the ligand-induced conformational change of this motif, which is crucial for the CaFADS RFK cycleCitation20, is not necessary for SpnFADS. This allows flexibility in the allocation order of substrates in the SpnFADS RFK active site.
Figure 4. The C-terminal modules of CaFADS and SpnFADS. (A) Superposition of the RFK modules of SpnFADS (PDB 3OP1) and CaFADS (PDB 2X0K). Zoom into the conformation of the PTAN motif of SpnFADS and the corresponding residues in (B) CaFADS and (C) CaFADS:ADP:FMN (PDB 5A89).
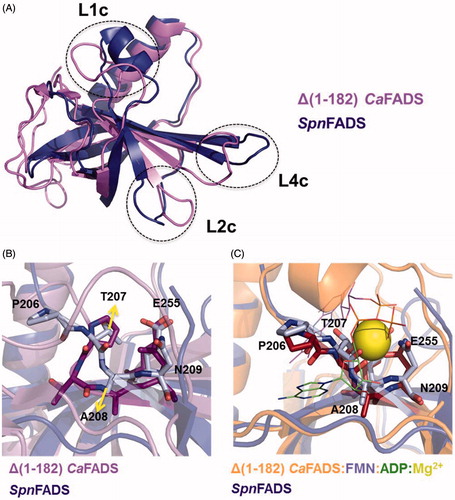
also reveals highly different ΔG values for titrations of either free SpnFADS or of its binary complexes with the same ligand, as well as that flavins are able to bind the enzyme in the presence of ANP ligands but not in its absence. These effects can be explained considering binding cooperative effects between ANP and FLV ligands (), and slow binding of flavins to free SpnFADS. Therefore, we carried out a co-operativity study that again came out with fundamental differences regarding heterotropic cooperativity in the CaFADS RFK module. Our data show that for SpnFADS the RF and ATP substrates always show positive cooperativity (). On the contrary, in the case of CaFADS low RF concentrations facilitate ATP binding, while increasing RF amounts hinder itCitation23. This different cooperative behavior of the substrates explains the inhibition by the RF substrate that CaFADS shows but not SpnFADS. Structurally, such effect might be also related to the different conformation of the PTAN motif. In CaFADS, the occupation of the ANP binding site by RF —when it is in excess— might prevent the ligand-induced conformational change of this motif, which is necessary for ATP binding ()Citation23. This structural rearrangement is not necessary for SpnFADS (), and therefore, the excess of RF does not hinder ATP binding.
Collectively, our results shed light on the kinetic and thermodynamic basis behind the substrates and products inhibition regulatory differences in the RFK cycles of SpnFADS and CaFADS. These essential enzymes differ in the substrates binding order, as well as in the inhibitory potency of the products and their action mechanism. The lesser inhibition level presented by SpnFADS—regarding CaFADS—is because of the binding of the reaction substrates is the fastest and thermodynamically most favored process, regardless of products accumulation. The structural origin of such divergences is the dissimilar conformation that the key PTAN motif shows in both FADSs, which points to species-specific conformational changes during the RFK activity. This fact, together with other differential regulatory strategies, such as the use of reduced substrates, provide us with a broader frame in which we could work in the development of new anti-microbials specific for S. pneumoniae. That way, it might be feasible to envisage drugs binding to a particular SpnFADS conformation, being unable to interact–due to the structural constraints and functional differences–with other FADSs.
Conclusions
In conclusion, here we present a complete description of the RFK catalytic cycle of SpnFADS, integrating thermodynamic and kinetic data, both in the pre-steady and in the steady-state, obtained using different biophysical and biochemical tools. We consider our research highly relevant in a double way. On one hand, combining physico-chemical tools commonly used in the biophysical characterisation of proteins, we have designed a strategy to go beyond the information directly extracted from this kind of techniques to offer a model that contributes to explain the dynamics during the SpnFADS RFK catalytic cycle. On the other hand, the results derived from our study are also relevant for the scientific community. Thus, we have found important differences between the RFK catalytic cycles of two structurally similar essential proteins, i.e. CaFADS and SpnFADS, shedding light on the thermodynamic and kinetic determinants that led to these inhibitory differences. Taking into account the pathogenic character of S. pneumoniae, the essentiality of its FADS and the last tendencies in discovering species-specific drugs, which reduce the resistances emergence, the knowledge here presented might facilitate the development of drugs able to bind a specific conformation of SpnFADS, having no effect on other members of the FADS family.
IENZ_1461857_Supplementary_Material.pdf
Download PDF (333.9 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Drijkoningen JJC, Rohde GGU. Pneumococcal infection in adults: burden of disease. Clin Microbiol Infect off Publ Eur Soc Clin Microbiol Infect Dis 2014;20:45–51.
- Lynch JP, Zhanel GG. Streptococcus pneumoniae: epidemiology, risk factors, and strategies for prevention. Semin Respir Crit Care Med 2009;30:189–209.
- World Health Organization (WHO). Causes of child mortality. Global Health Observatory; 2015. Available from: http://www.who.int/gho/child_health/mortality/causes/en/
- Serrano A, Ferreira P, Martínez-Júlvez M, Medina M. The prokaryotic FAD synthetase family: a potential drug target. Curr Pharm Des 2013;19:2637–48.
- Sebastián M, Anoz-Carbonell E, Gracia B, et al. Discovery of antimicrobial compounds targeting bacterial type FAD synthetases. J Enzyme Inhib Med Chem 2018;33:241–54.
- Sebastián M, Lira-Navarrete E, Serrano A, et al. The FAD synthetase from the human pathogen Streptococcus pneumoniae: a bifunctional enzyme exhibiting activity-dependent redox requirements. Sci Rep 2017;7:7609.
- Parsons HG, Dias VC. Intramitochondrial fatty acid metabolism: riboflavin deficiency and energy production. Biochem Cell Biol Biochim Biol Cell 1991;69:490–7.
- Myllykallio H, Lipowski G, Leduc D, et al. An alternative flavin-dependent mechanism for thymidylate synthesis. Science 2002;297:105–7.
- Gross E, Kastner DB, Kaiser CA, Fass D. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell 2004;117:601–10.
- Efimov I, Kuusk V, Zhang X, McIntire WS. Proposed steady-state kinetic mechanism for Corynebacterium ammoniagenes FAD synthetase produced by Escherichia coli. Biochemistry (Mosc) 1998;37:9716–23.
- Wang W, Kim R, Yokota H, Kim S-H. Crystal structure of flavin binding to FAD synthetase of Thermotoga maritima. Proteins 2005;58:246–8.
- Matern A, Pedrolli D, Groszhennig S, et al. Uptake and metabolism of antibiotics roseoflavin and 8-demethyl-8-aminoriboflavin in riboflavin-auxotrophic Listeria monocytogenes. J Bacteriol 2016;198:3233–43.
- Barile M, Passarella S, Bertoldi A, Quagliariello E. Flavin adenine dinucleotide synthesis in isolated rat liver mitochondria caused by imported flavin mononucleotide. Arch Biochem Biophys 1993;305:442–7.
- Barile M, Brizio C, Valenti D, et al. The riboflavin/FAD cycle in rat liver mitochondria. Eur J Biochem 2000;267:4888–900.
- Torchetti EM, Bonomi F, Galluccio M, et al. Human FAD synthase (isoform 2): a component of the machinery that delivers FAD to apo-flavoproteins. Febs J 2011;278:4434–49.
- Leone P, Galluccio M, Barbiroli A, et al. Bacterial production, characterization and protein modeling of a novel monofuctional isoform of FAD synthase in humans: an emergency protein? Mol Basel Switz 2018;23:116.
- Karthikeyan S, Zhou Q, Mseeh F, et al. Crystal structure of human riboflavin kinase reveals a beta barrel fold and a novel active site arch. Struct Lond Engl 1993 2003;11:265–73.
- Frago S, Martínez-Júlvez M, Serrano A, Medina M. Structural analysis of FAD synthetase from Corynebacterium ammoniagenes. BMC Microbiol 2008;8:160.
- Mack M, Grill S. Riboflavin analogs and inhibitors of riboflavin biosynthesis. Appl Microbiol Biotechnol 2006;71:265–75.
- Herguedas B, Lans I, Sebastián M, et al. Structural insights into the synthesis of FMN in prokaryotic organisms. Acta Crystallogr D Biol Crystallogr 2015;71:2526–42.
- Serrano A, Frago S, Herguedas B, et al. Key residues at the riboflavin kinase catalytic site of the bifunctional riboflavin kinase/FMN adenylyltransferase from Corynebacterium ammoniagenes. Cell Biochem Biophys 2013;65:57–68.
- Serrano A, Frago S, Velázquez-Campoy A, Medina M. Role of key residues at the flavin mononucleotide (FMN): adenylyltransferase catalytic site of the bifunctional riboflavin kinase/flavin adenine dinucleotide (FAD) synthetase from Corynebacterium ammoniagenes. Int J Mol Sci 2012;13:14492–517.
- Sebastián M, Serrano A, Velázquez-Campoy A, Medina M. Kinetics and thermodynamics of the protein-ligand interactions in the riboflavin kinase activity of the FAD synthetase from Corynebacterium ammoniagenes. Sci Rep 2017;7:7281.
- Michaelis L, Menten ML. Die Kinetik der Invertinwerkung. Biochem Z 1933;49:333.
- Yamada Y, Merrill AH, McCormick DB. Probable reaction mechanisms of flavokinase and FAD synthetase from rat liver. Arch Biochem Biophys 1990;278:125–30.
- Frago S, Velázquez-Campoy A, Medina M. The puzzle of ligand binding to Corynebacterium ammoniagenes FAD synthetase. J Biol Chem 2009;284:6610–9.
- Velazquez-Campoy A, Goñi G, Peregrina JR, Medina M. Exact analysis of heterotropic interactions in proteins: characterization of cooperative ligand binding by isothermal titration calorimetry. Biophys J 2006;91:1887–904.
- Martínez-Júlvez M, Medina M, Velázquez-Campoy A. Binding thermodynamics of ferredoxin:NADP + reductase: two different protein substrates and one energetics. Biophys J 2009;96:4966–75.
- Weber G, Convery HJ, Lea MA, Stamm NB. Feedback inhibition of key glycolytic enzymes in liver: action of free fatty acids. Science 1966;154:1357–60.
- Fukao T, Tanabe M, Terauchi Y, et al. PI3K-mediated negative feedback regulation of IL-12 production in DCs. Nat Immunol 2002;3:875–81.
- Herguedas B, Martínez-Júlvez M, Frago S, et al. Oligomeric state in the crystal structure of modular FAD synthetase provides insights into its sequential catalysis in prokaryotes. J Mol Biol 2010;400:218–30.
- Marcuello C, Arilla-Luna S, Medina M, Lostao A. Detection of a quaternary organization into dimer of trimers of Corynebacterium ammoniagenes FAD synthetase at the single-molecule level and at the in cell level. Biochim Biophys Acta 2013;1834:665–76.
- Weber G. Fluorescence of riboflavin and flavin-adenine dinucleotide. Biochem J 1950;47:114–21.
- Sun M, Moore TA, Song PS. Molecular luminescence studies of flavins. I. The excited states of flavins. J Am Chem Soc 1972;94:1730–40.
- Bollen YJM, Westphal AH, Lindhoud S, et al. Distant residues mediate picomolar binding affinity of a protein cofactor. Nat Commun 2012;3:1010.
- Martinez-Julvez M, Abian O, Vega S, et al. Studying the allosteric energy cycle by isothermal titration calorimetry. Methods Mol Biol Clifton NJ 2012;796:53–70.
