Abstract
Increasing knowledge of the relationship between cancer and dysregulated polyamine catabolism suggests interfering with aberrant polyamine metabolism for anticancer therapy that will have considerable clinical promise. SMO (spermine oxidase) plays an essential role in regulating the polyamines homeostasis. Therefore, development of SMO inhibitors has increasingly attracted much attention. Previously, we successfully purified and characterised SMO. Here, we presented an in silico drug discovery pipeline by combining pharmacophore modelling and molecular docking for the virtual screening of SMO inhibitors. In vitro evaluation showed that N-(3-{[3-(dimethylamino)propyl]amino}propyl)-8-quinolinecarboxamide (SI-4650) inhibited SMO enzyme activity, increased substrate spermine content and reduced product spermidine content, indicating that SI-4650 can interfere with polyamine metabolism. Furthermore, SI-4650 treatment suppressed cell proliferation and migration. Mechanistically, SI-4650 caused cell cycle arrest, induced cell apoptosis, and promoted autophagy. These results demonstrated the properties of interfering with polyamine metabolism of SI-4650 as a SMO inhibitor and the potential for cancer treatment.
1. Introduction
Natural polyamines putrescine (Put), spermidine (Spd) and spermine (Spm), are ubiquitous low-molecular-mass organic polycations in eukaryotesCitation1. These molecules can bind to the negatively-charged biological macromolecules such as DNA, RNA, proteins, and phospholipids by electrostatic interactions. Importantly, these molecules are involved in many fundamental processes including stabilisation of chromatin structure, regulating gene expression, maintaining the stability of membrane phospholipids, protection from oxidative damage, cell proliferation, differentiation, and apoptosisCitation2–6.
Polyamines are basic regulators of cell growth and survival, and the precise regulation of polyamine metabolism is essential for normal life activitiesCitation7. Numerous studies have shown that polyamine metabolic disorders are closely associated with many diseases, including cancerCitation8, inflammationCitation9, atherosclerosisCitation10, strokeCitation11, renal failureCitation12, and diabetesCitation13. Excessive accumulation or depletion of intracellular polyamines (especially Spd) can have deleterious effects on mammalian cells and can lead to cell deathCitation14. In cancer, polyamine metabolism is frequently dysregulated, overall indicating that the tumour progression is highly dependent on the elevated polyamines levelsCitation15. Previous studies have found that high levels of polyamines can promote tumour growth, invasion and metastasis, while reducing polyamine content can inhibit the proliferation of tumour cellsCitation16,Citation17. Therefore, the polyamine metabolic pathway has provided rational drug targets for intervention in cancer.
Polyamine catabolic enzymes include spermidine/spermine N1-acetyltransferase (SSAT), N1-acetylpolyamine oxidase (APAO), and spermine oxidase (SMO). Our previous work has shown that SMO is an FAD-dependent enzyme and can be rapidly induced from nearly undetectable to very high levelsCitation18. Furthermore, SMO is found both in the cytoplasm and in the nucleus and catalyses Spm into the aldehyde 3-aminopropanal, H2O2, and SpdCitation19–21. As an inducible flavoenzyme, expression level of SMO is frequently upregulated in response to various diseases including tumours, infection and inflammationCitation22–26. In infected gastric mucosal epithelial cells, the bacterial toxin CagA can up-regulate the expression of SMO, resulting in increased ROS production and DNA damage, and long-term chronic accumulation of these lesions will eventually lead to gastric cancerCitation27–29. Additionally, significant up-regulation of SMO activity was found in invasive prostate cancerCitation5, neuroblastoma cellsCitation30, and liver cancer cellsCitation31,Citation32. These data have strongly implied that inhibition of SMO is a rational drug target for cancer therapy.
In view of the fact that SMO abnormal expression is implicated in the development of cancer, small molecule inhibitors of SMO will have important potential application value in the prevention and treatment of cancer. Many potent inhibitors of SMO have been described, but by far the most widely reported are MDL 72527N1,N4-(bis(2,3-butadienyl)-1,4-butanediamine) and oligamine analogue. MDL 72527 is an irreversible inhibitor of APAO, and it was subsequently proved to be a rather weak competitive SMO inhibitorCitation33. In our previous study, we have confirmed that cis-3,8,13,18,23,28,33,38,43,48-decaazapentacontene-25 can inhibit SMO activityCitation34. Although these inhibitors exhibited antitumor activity, the poor efficacy and side effects limited the clinical application for cancer treatmentCitation35,Citation36. More recently, a spermine analogue, 1,12-diamino-2,11-bis(methylidene)-4,9-diazadodecane, was synthesised as an enzyme-activated inhibitor of SMO, but the related biological data were not providedCitation37. Despite these progresses, there are problems with efficacy and side effects, necessitating novel potent SMO inhibitors for anti-tumour therapy.
Although it is clear that SMO is a rational target for drug design, the detailed structure information is limited by the fact that no crystal structure of SMO currently exists. However, the molecular modelling structure of Spm and SMO has demonstrated the structure features and could be used as a powerful tool to search for new inhibitorsCitation38–40. In this investigation, we took advantage of the structural information of SMO-Spm complex and performed structure-based virtual screening to screen promising hit compounds against SMO. By combining the structure-based pharmacophore modelling and molecular docking method, we identified SI-4650 as an inhibitor of SMO that notably inhibited SMO activity and reduced the polyamine content in a lung cancer model. Furthermore, we also investigated the effects of SI-4650 on the cell proliferation, migration, cell cycle, apoptosis, and autophagy.
2. Materials and methods
2.1. Chemical reagents and cell culture
SI-4650 (N-(3-{[3-(dimethylamino)propyl]amino}propyl)-8-quinolinecarboxamide) was purchased from SPECS (Zoetermeer, The Netherlands), DMSO, 3-(4,5-dimethylthiazol)-2,5-diphenyltetrazolium bromide (MTT), Spermine, horseradish peroxidase, luminol, 1,7-Diaminoheptane were purchased from Sigma (St. Louis, MO), foetal calf serum was purchased from Gibco (Grand Island, NY), Turbofect transfection reagent, DNA marker and protein marker were purchased from Thermo Fisher Scientific (Waltham, MA), Transwell chamber was purchased from Corning (Corning, NY), FITC Annexin V Apoptosis Detection Kit was purchased from BD Biosciences Inc. (San Jose, CA). The primary antibodies were all obtained from Santa Cruz Biotechnologies: p62(catalog no. 18420-1-AP), p21(catalog no. 103551-AP), CyclinB1(catalog no. 55004-1-AP), LC3(catalog no. 12741 s), Bax(catalog no. 50599-2-Ig), Bcl-2(catalog no. 12789-1-AP). All other chemicals used in the experiments were commercial products of reagent grade.
The A549 cell line was obtained from the Cell Bank of China (Wuhan, China). Cells were cultured in RPMI-1640 supplemented with 10% foetal calf serum, 5% CO2 at 37 °C.
2.2. Virtual screening
As a first step in our pipeline, we carried out a structure-based pharmacophore modelling based on the complex structure of SMO-Spm. The structure based other than the ligand-based pharmacophore modelling was performed since the SMO inhibitors reported so far have only little structural diversity. The structure of SMO in complex with Spm (PMDB entry: PM0076222.pdb) was used as the molecular template for our investigation. The interactions between ligand and receptor were analysed by using the Receptor-Ligand Interactions protocol in Discovery Studio 3.1 (Accelrys, San Diego, CA). The pharmacophore model was constructed based on the critical interactions identified by Receptor-Ligand Interactions protocol and other reported literatures.
The obtained pharmacophore model was used as a three-dimensional query for retrieving potent SMO inhibitors from the chemical database SPECS (500,000 molecules). All the compounds of the abovementioned database passed the Lipinski “rule of five” for drug likeness, ensuring fewer false positives and higher quality results. Subsequently, a further docking study was carried out for the remaining compounds. Each compound was docked into the active site of the protein using the Autodock software. Compounds were ranked according to the scoring function incorporated in Autodock. On the basis of the ranking order and energy score, selected compounds were purchased and shifted to the subsequent biochemical assays.
2.3. Cell proliferation assay
The in vitro bioactivities of the 28 selected hit compounds were first examined by MTT assay against human lung cancer A549 cell line in which SMO is overexpressed. Briefly, A549 cells were seeded in 96-well plates at a density of 1 × l03/well. After 24 h incubation, the culture medium was changed to 1640 medium containing 28 small molecule drugs with the same final concentrations (100 µM). After 48 h incubation, 200 µL MTT (0.2 g/L) was added to each well and cultured for another 4 h. Then supernatant was removed and 150 µL DMSO was added. It was shaken for 5 min for the crystal dissolution. When the precipitate is fully dissolved, the absorbance at a wavelength 490 nm was measured with a Full wavelength microplate reader.
The antiproliferation effects of SI-4650 at different concentration (5 µmol/L, 10 µmol/L, 20 µmol/L, 40 µmol/L, 80 µmol/L, 160 µmol/L) and time (24, 48, and 72 h) against A549 cells were also evaluated by MTT assay.
2.4. Expression and purification of SMO and APAO
SMO and APAO was expressed and purified as described in our previous workCitation34 and Bianchi et al.Citation39. Briefly, the plasmid was used to transform the BL21(DE3) strain of Escherichia coli (Novagen) and transformed cells were selected on LB agar with 50 µg/mL ampicillin. The expression of protein was induced in LB medium by the addition of 1 mM IPTG for 4 h at 37 °C. Cell lysates were prepared under denaturing conditions with 8 M urea and protein was purified from the lysate by Ni-NTA resin according to the manufacturer’s protocol. The resulting denatured protein was renatured in buffers containing decreasing concentrations of urea (5 M urea, 4 h; 2.5 M urea, 4 h; 1 M urea, 12 h; and 0 M urea, 12 h) and 50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and 0.2 µM flavin adenine dinucleotide (FAD).
2.5. In vitro enzyme inhibition assay
The activity of the purified SMO/APAO was evaluated by chemiluminesence analysis according to our previous work. Briefly, Luminol was prepared as a 100 mM stock solution in DMSO and diluted to 100 µM with H2O, immediately prior to use. Purified SMO/APAO was assayed in a 100 mM glycine buffer, pH 8.0, 50 µL luminol, 20 µg horseradish peroxidase, and the polyamine substrate as indicated. These selected compounds with different concentrations (from 0 to 3 mmol/L) and other regents with the exception of the polyamine substrate were combined and incubated for 2 min at 37 °C, then the tube was transferred to the luminometer, substrate was added, and the resulting chemiluminescence was integrated over 20 s. The integral values are calibrated against standards containing known concentrations of H2O2 and the activities are expressed as pmols H2O2/mg protein/min.
2.6. Detection and quantification of cellular polyamines
The cellular polyamine content was measured using the HPLC method. Briefly, A549 cells were treated with SI-4650 (80 µmol/L) for 48 h, then the cell culture medium was removed. Cells were collected to a new Eppendorf tube and washed with 1.0 mL of PBS (pH 7.4) by centrifugation at 800 rpm at 4 °C for 4 min and discarded the supernatant fluid, then 800 µL cell lysate was added to the tube. After 40 min, the tube was centrifuged at 12,000 rpm for 15 min and the supernatant fluid was transferred into a new 4.0 mL Eppendorf tube. Cell lysate with the same protein content and 20 µL 1,7-diamino-heptane (1 mmol/L) as an internal standard were added into the tube and mixed thoroughly. The mixture was alkalinised by adding 2 mol mL−1 NaOH solution, followed by 10 µL benzoyl chloride. After standing for 20 min under water bath at 40 °C, reaction was terminated by adding the saturated sodium chloride solution. Polyamine derivatives were extracted into diethyl ether, followed by evaporating to dryness. The residue was redissolved in 1.0 mL methanol and filtered using 0.22 µm microporous membrane filtration. Protein was determined by BCA assay.
HPLC analytical were performed according to the following procedures. Derivative polyamines were separated on a luna C18 column (5 µm, 150 mm × 4.6 mm) held at 30 °C. The column was eluted with a gradient mixture of acetonitrile (phase A) and water (phase B) at the flow rate of 1 mL min−1. The detection wavelength is 254 nm.
2.7. Transwell
Briefly, A549 cells were seeded in six-well plates at a density of 1 × 105 cells/well in 1640 medium. After 24 h, the growth medium was then replaced with the same medium but containing SI-4650 (80 µmol/L final concentration). Following 48 h, 5 × 104 cells were plated into the upper chamber of a 24-well transwell in serum free 1640 and exposed to 20% FBS as inducers of migration (0.8 mL, lower chamber). After the cells were incubated for 18 h in a 5% CO2 humidified incubator at 37 °C, the basement membranes were washed with PBS, and the non-migrated cells on the top surface of the thin basement membranes were scraped off with cotton swabs. The migrated cells adhering to the lower surface were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet solution and counted under a microscope (100×) in five random and visual fields per well.
2.8. Transmission electron microscopy
In brief, SI-4650 (80 µmol/L) treated A549 cells were collected after 48 h and washed with PBS. The supernatant was discarded, and 2.5% glutaraldehyde was added to the tube to fix the cells overnight. Then the solution was removed and pellet then washed by PBS and postfixed in 1% (w/v) osmium tetroxide for another 1.5 h and washed twice with PBS. The sample was subjected to continuous serial dehydration with ethanol (50–100%), followed by dehydrated with acetone. Following embedding, ultrathin sections (80 nm) were prepared using a microtome and each section was mounted on a copper grid. Ultrathin sections were stained with 2.5% (w/v) uranyl acetate (22 min) and 0.3% (w/v) lead citrate (22 min). The autophagosomes were observed under a transmission electron microscope and imaged.
2.9. Cell cycle analysis using flow cytometry
The effect of SI-4650 on cell cycle distribution was determined by flow cytometry. Briefly, A549 cells were seeded in 6-well plates at a density of 1 × 105 cells/well for 24 h. After 24 h, the growth medium was then replaced with the same medium but containing SI-4650 (80 µmol/L final concentration). Following 48 h and 72 h, cells were suspended, washed with PBS, centrifuged, and fixed in 75% ethanol (v/v) at 4 °C overnight. Then the cells were resuspended in 1 mL of PBS containing 50 µg/mL ribonuclease A and 0.5 mg/mL PI. Cells were incubated in the dark for 30 min at room temperature. The cell cycle analysis was performed by using a flow cytometer.
2.10. Quantification of cellular apoptosis using flow cytometry
Apoptotic cells were quantitated using the FITC Annexin V Apoptosis Detection Kit. After cells were treated with SI-4650 (80 µmol/L final concentration) for 48 h as described previously. Cells were harvested and washed with cold PBS, and then cells were resuspended in 100 µL 1× binding buffer. Subsequently, 5 µL of FITC Annexin V and 5 µL of PI were added. The cells were gently mixed and incubated in the dark for 15 min at 25 °C. 400 µL of 1× binding buffer was added to each sample before flow cytometry analysis.
2.11. Quantification of cellular autophagy
A549 cells were seeded in eight-well plates at a density of 2 × 104 cells/well for 24 h. According to the manufacturer's instructions, cells were transfected with pEGFP-LC3 plasmid for 24 h and then treated with SI-4650 at a final concentration 80 µmol/L for 48 h. GFP-LC3II-positive punctate pattern was observed under confocal microscope equipped with oil immersion lens (40×) with 405- and 488 nm excitation lasers. Numbers of autophagosomes were counted by using the Image J programme.
2.12. Western blotting analysis
The expression level of cellular proteins of interest was determined using Western blotting assays. Cells were harvested and lysed in radioimmune precipitation assay (RIPA) buffer. Protein concentration was quantified by using a BCA protein assay kit. An equal amount of protein sample was dissolved by 12% SDS-PAGE and electrotransferred from gel to PVDF membrane. After blocking in 5% non-fat milk for 2 h, the membranes were immunoblotted overnight at 4 °C with primary antibodies against LC3I, LC3II, Cyclin-B1, p21, Bcl-2, p62, and Bax, followed by incubation with respective secondary anti-mouse or anti-rabbit antibody for 1 h at room temperature. The membrane was detected by the enhanced chemiluminescence (ECL) detection system. Protein level was normalised to the matching densitometric value of the internal control β-actin.
2.13. Statistical analysis
Statistical analysis was performed using SPSS version 18.0 software. Data are presented as the mean ± standard deviation (SD), and the normality was analysed by K-S and Q-Q plot. One-way analysis of variance with Tuckey’s post-hoc test was used for multiple comparisons. Values of p < 0.05 were considered statistically significant.
3. Results
3.1. Identification potent SMO inhibitors based on virtual screening
In this study, structure-based pharmacophore modelling and molecular docking were employed to identify promising SMO inhibitors. The possible interactions formed between Spm and SMO were identified by the receptor-ligand interactions module. As shown in , residues HIS58, GLN176, GLU200, SER503, GLN176, and THR504 can form hydrogen bonds with Spm respectively, and residues THR202 and TYR458 can interact with Spm by van der waals interactions. It is worth noting that the hydrogen bonds formed by residues HIS58, GLN176, GLU200, SER503, and Spm are stronger than others. In order to obtain small molecules with structure diversity, we only chose two hydrogen bonds involved in GLU200 and SER503 to construct the pharmacophore features. According to the previous studies, GLU192 and SER194 are the major determinants for SMO substrate specificityCitation38. Thus, we added a hydrogen bond acceptor feature based on GLU192 by manually. Finally, a pharmacophore model containing three hydrogen bond acceptors (H1, H2, and H3) features was obtained. shows the associated residues of these pharmacophore features. The hydrogen bond acceptor features H1 and H2 stem from the backbone carboxyl oxygen of SER503 and GLU192, respectively, and H3 originates from the hydroxyl oxygen in the side chain of GLU200.
Figure 1. The virtual screening of SMO inhibitors. (A) The complex structure of Spm and SMO. The circle represents the binding site and the Spm is shown in pink. (B) The detailed interactions of SMO with Spm. Pink circles represent residues involved in hydrogen bonds interactions. Green circles represent residues involved in van der Waals interactions. Blue dashed arrow represents hydrogen bond. (C) The pharmacophore model developed based on the structure of Spm and SMO. The pharmacophore features are colour coded with pink for hydrogen bond features (H1, H2, and H3). Mapping of Spm with the pharmacophore model. (D) The chemical structure of SI-4650.
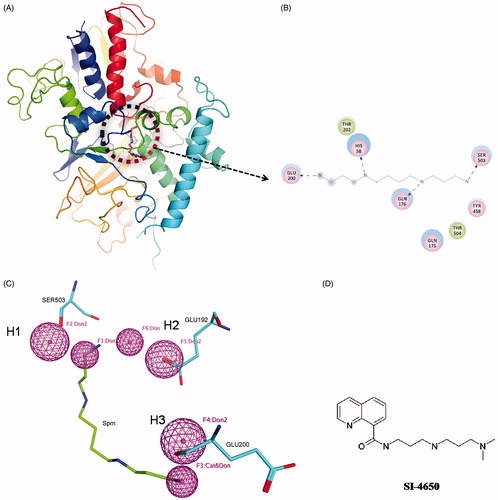
Based on the above pharmacophore model, a total of 120 potent inhibitors were finally selected. Subsequently, the 120 compounds were docked into the SMO protein pocket confirmed by Spm using the Autodock programme. On the basis of the ranking order and energy score (<–4.0 kcal/mol), 28 structurally diverse compounds (Supporting Information Table S1) were purchased and shifted to the subsequent biochemical assays. We then tested the effects of these compounds on inhibition activity against A549 cell line at 100 µmol/L (Supporting Information Figure S1). Among these compounds, SI-4650 exhibited the strongest inhibitory effect and was thus selected for the subsequent investigations. The chemical structures of SI-4650 is presented in .
3.2. SI-4650 inhibits polyamine metabolism enzyme activity and interferes with polyamine metabolism
To explore the inhibitory activity of SI-4650 on SMO and APAO enzyme in vitro, recombinant human SMO and APAO protein was expressed and the enzymatic activity was evaluated as described in our previous studyCitation34. SI-4650 showed inhibitory effect against SMO with IC50 value of 381.8 ± 0.17 µM (). The Ki value was obtained by Cheng-Prusoff method and the value is calculated to be 289 µM. As for APAO (), the IC50 value is 35.4 ± 0.26 µM and Ki value is 32.3 µM.
Figure 2. The effects of SI-4650 on SMO and APAO activity and cellular polyamine content. The chemiluminesence method indicating the inhibition effects of SI-4650 on SMO (A) and APAO (C). Purified protein was incubated in the presence of SI-4650 with different concentration (from 0 to 3 mM) and 3 mM substrate. The inhibition rate of SMO (B) and APAO (D) by SI-4650. The data are shown as means ± SD. Compared with control *p < 0.05, **p < 0.01. (E) The effect of SI-4650 on cellular polyamine content determined by HPLC. A549 cells were treated with DMSO or 80 μmol/L SI-4650 for 48 h before being collected for the indicated polyamine analyses. The values are presented as the means ± SD from three independent experiments (Student’s t-test). *p < 0.05, **p < 0.01, ns, not significant.
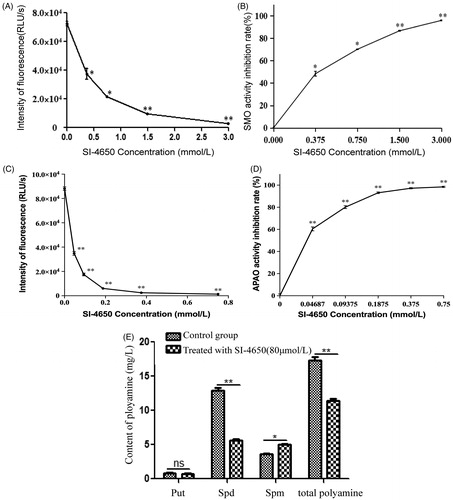
Measuring the kinase activity and cell viability of SI-4650 in vitro revealed that SI-4650 exhibited significant inhibitory activities. We then further tested the effect of SI-4650 on the intracellular polyamine content. As expected, SI-4650 treatment decreased the total intracellular polyamine content in A549 cells, while the Put level was not significantly different in each sample (). Furthermore, the A549 cells incubated with 80 µmol/L SI-4650 for 48 h exhibited higher Spm level than control cells. In contrast, SI-4650 treatment specifically inhibited Spd production. Collectively, these results suggested that SI-4650 can inhibit SMO and APAO activity and interfere with intracellular polyamine metabolism in A549 cells.
3.3. Structural bases for inhibition enzyme activity of SI-4650
In order to analyse the potential inhibition properties of SI-4650, a comparative structural analysis of the binding modes formed by SMO with Spm and SI-4650 has been performed. showed the predicted interactions occurring between inhibitor chemical groups and SMO active site residues. As shown in , molecule docking results revealed that SI-4650 occupied the binding site of the substrate Spm and appeared as a similar conformation. Obviously, SI-4650 is not able to establish Spm-like interactions with residues in the active site, it makes potential contacts including hydrogen bond interaction, cation-Pi interaction (terminal amino group and TYR458) and hydrophobic interactions. In comparison with hydrogen bond formed by Spm, this cation-Pi interaction displayed similar role in the binding process. In particular, GLU192 and SER194 formed hydrophobic interactions with SI-4650, which may increase the binding affinity.
Figure 3. The binding mode analysis of SI-4650. The binding modes of SI-4650 in SMO (A) and APAO (C). Pink represents substrate and cyan represents SI-4650. The detailed interactions of SI-4650 with SMO (B) and APAO (D). Blue dashed arrow represents hydrogen bond. Orange line with symbols represent cation-Pi interaction.
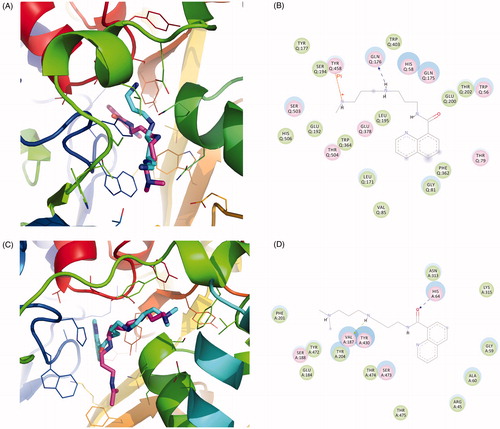
In addition, the binding mode of SI-4650 and APAO was also analysed by molecular docking. As shown in , SI-4650 partially occupied the binding site of the substrate and adopted a similar conformation as that observed in the SMO and SI-4650 complex. It is noted that the terminal amino group lost the cation-Pi interaction and the oxygen of carbonyl formed a stronger hydrogen bond with HIS64, respectively.
3.4. SI-4650 inhibits cell proliferation and migration
To examine the biological effect in greater detail, we tested the inhibitory effect of SI-4650 against A549 cell line at different concentration (5 µmol/L, 10 µmol/L, 20 µmol/L, 40 µmol/L, 80 µmol/L, 160 µmol/L) for 24, 48, and 72 h. As presented in , SI-4650 inhibited proliferation of A549 cells in a dose- and time-dependent manner. Furthermore, 80 µmol/L SI-4650 reduced the cell viability by approximately 50%. These results indicated that SI-4650 displayed anti-proliferation effect and blockage of high polyamine content-independent cell growth via its SMO inhibitory effect.
Figure 4. The effects of SI-4650 on cell proliferation and migration in A549 cells. (A) A549 cells were treated with 0, 5, 10, 20, 40, and 80 and 160 µmol/L SI-4650 for 24, 48, and 72 h. Cell proliferation was then performed by using the MTT assay. The data are shown as means ± SD. (B) A549 cells were starved for 24 h and then treated with SI-4650 at 80 μM for 24 h. The numbers of migrated cells were counted under an inverted fluorescence microscope in five randomly selected visual fields per well within the same area. Scale bar: 100 μm. (C) Percent migration inhibition rate relative to control treated A549 cells. The values are presented as the means ± SD from three independent experiments (Student’s t-test), ***p < 0.001.
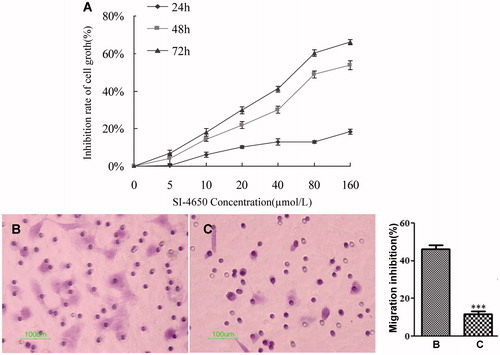
To evaluate the effect of SI-4650 on A549 cell migration, cells were treated with 80 µmol/L SI-4650 for 48 h. As shown in , the numbers of migrated cells that adhered to the lower surface of the Transwell chambers were counted under an inverted microscope in five randomly chosen visual fields per well within the same area. Compared with the control group, the migration of the A549 cells was significantly suppressed (p < 0.001) and the inhibition rate was 76.47%. These results suggested that treatment of SI-4650 can decrease the A549 cells migration in vitro.
3.5. SI-4650 induces cell cycle arrest in A549 cells
To investigate whether SI-4650 affects the cell cycle in A549 cells, we assessed the cell cycle distribution of A549 cells treated with SI-4650 by flow cytometry. We treated A549 cells with SI-4650 at 80 µM for 48 and 72 h, and then we measured the cell cycle distribution ( and ). Compared to the control cells, the percentage of A549 cells in S/G2 phase was increased from 16.68% at basal level to 23.7% and 27.06% after 48 and 72 h treatment with 80 µM SI-4650, respectively. In contrast, the number of cells in G0/G1 phase declined to 73.86% after 72 h treatment of SI-4650. These results suggested that SI-4650 can arrest the cells at S/G2 transition.
Figure 5. The effects of SI-4650 on cell cycle distribution and the expression levels of cyclin B1 and p21 in A549 cells. The A549 cells treated with DMSO (A), SI-4650 at 80 µM for 48 h (B), SI-4650 at 80 µM for 72 h (C) were fixed, stained by PI, and analysed by flow cytometry. (D) Protein expression levels were measured by Western blot analysis. The A549 cells were treated with SI-4650 at 80 µM for 48 h, and β-actin was used as the internal control.
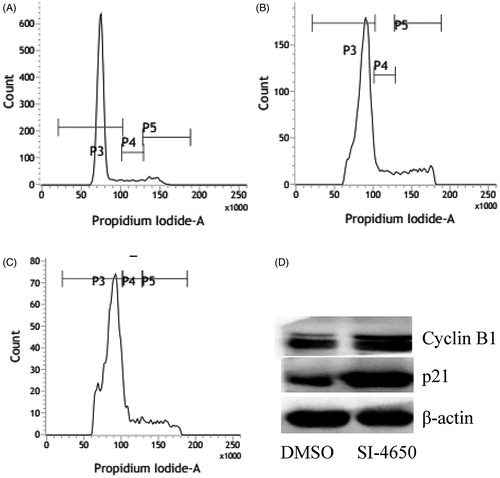
Table 1. Cell cycle distribution of the A549 cells detected by FCM (%).
To explore the mechanism for SI-4650-induced cell cycle arrest, we evaluated the expression levels of cyclin B1 and p21 in A549 cells using western blotting assay. As presented in , treatment of A549 cells with SI-4650 at 80 µM for 48 h significantly increased the level of cyclin B1 and p21. These results indicated that SI-4650 induces S/G2 arrest via up-regulating the expression of cyclin B1 and p21 in A549 cells.
3.6. SI-4650 induces apotosis in A549 cells
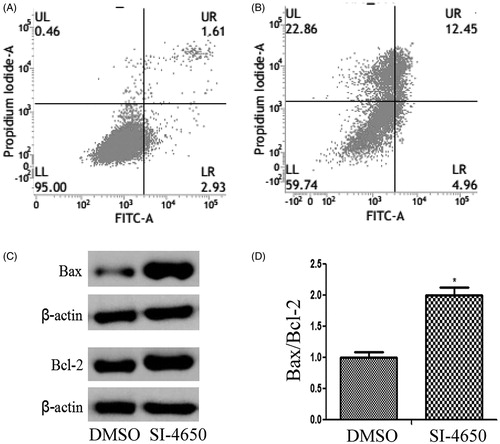
It is well known that cell apoptosis plays a pivotal role in the occurrence and development of tumours. In the present study, the apoptotic cells were determined by Annexin V-Propidium Iodide (PI) staining by flow cytometry. While A549 cells were treated with SI-4650 at 80 µM for 48 h, the total percentage of apoptotic cells was increased from 4.54% to 17.41%. This demonstrated that SI-4650 had an obvious inductive effect on the apoptosis of A549 cells as compared to negative control (p < 0.01). Importantly, the percentage of necrotic cells was also increased from 0.46% to 22.86%. These results indicated that in addition to apoptotic pathway, SI-4650 may induce other forms of cell death of A549 cells.
To probe the mechanisms responsible for SI-4650-induced apoptosis in A549 cells, we evaluated the expression levels of pro-apoptotic protein Bax and the anti-apoptotic protein Bcl-2 in A549 cells treated with SI-4650 at 80 µM for 48 h. As presented in , treatment of A549 cells with SI-4650 significantly increased the expression levels of Bax and Bcl-2 compared to the control group. However, incubation with SI-4650 resulted in a significant increase in Bax/Bcl-2 ratio in A549 cell line. These results indicated that SI-4650 may induce cell apotosis via up-regulating the protein expression levels of Bcl-2 and Bax.
Figure 6. The effects of SI-4650 on cell apoptosis and the expression levels of Bcl-2 and Bax. (A) Flow cytometry of A549 cells treated with DMSO (A) or SI-4650 (B) at 80 µM for 48 h and stained with a mixture of FITC-annexin V and PI. (C) Protein expression levels were measured by Western blot analysis. The A549 cells were treated with SI-4650 at 80 µM for 48 h, and β-actin was used as the internal control. (D) The relative ratio of Bax and Bcl-2. *p < 0.05.
3.7. Promotion of cell autophagy induced by SI-4650 in A549 cells
In order to determine the effect of SI-4650 treatment on cell autophagy, the protein expression levels of LC3I, LC3II, and p62 were evaluated by the western blot analysis. As presented in , compared with the control group, the LC3II protein expression levels were increased after treated with SI-4650 at 80 µmol/L for 48 h. On the contrary, SI-4650 treatment obviously decreased the expression level of p62 in the A549 cells. These results indicated that SI-4650 treatment can promote cell autophagy in A549 cells.
Figure 7. SI-4650 induced Autophagy. (A) A549 cells were treated for 48 h with SI-4650 (80 μM), along with DMSO as control. Levels of LC3 and p62 were analysed by western blotting. (B) A549 cells incubated without or with 80 μM SI-4650 for 48 h were imaged by transmission electron microscopy. (C) The effects of SI-4650 on A549 cells with GFP-LC3. A549 cells were transfected with pEGFP-LC3 plasmid and then treated with SI-4650 (80 μM) or DMSO for 48 h. Autophagy spot aggregation in A549 cells with GFP-LC3 was pictured by laser scanning confocal microscopy. Scale bar, 100 μm.
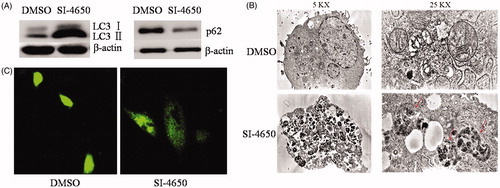
The induction of autophagy by SI-4650 was further determined by the observation of the formation of autophagosomes using a transmission electron microscope in A549 cells. As shown in , following 80 µmol/L SI-4650 treatment for 48 h, the number of intracellular autophagosomes and vacuoles increased compared with that of the control group (no treatment). These results were also evidenced by the observation of punctate GFP- LC3II distribution under confocal microscopy. Obviously, SI-4650 induced prominent autophagy as indicated by enhanced GFP-LC3II puncta (). These results demonstrated that SI-4650 can promote the accumulation of GFP-LC3II protein to autophagosomes, thereby promoting autophagy in A549 cells.
4. Discussion
SMO plays pivotal roles in polyamine metabolism and is indispensable to cancer cell growth and survival. Therefore, it represents an attractive molecular target for human disease treatmentCitation41,Citation42. Obviously, small molecule inhibitors of SMO have potential as chemical tools for studying the biological function of SMO and cancer treatment. During the past decades, several attempts have been made to identify SMO inhibitors, but novel method and inhibitors are still urgently neededCitation35,Citation36. In this study, we developed a virtual screening pipeline and SI-4650 was screened out and identified as an inhibitor of SMO. Further study confirmed that SI-4650 can inhibit the activity of SMO and has anti-tumour effect, showing its potential for basic SMO and pharmaceutical research.
Pharmacophore-based virtual screening is a valuable tool in the drug discovery process and can be employed for a variety of tasksCitation43. However, pharmacophore model has not been developed for SMO inhibitors screening during the past decades. In light of the limited structural type of SMO inhibitors, we constructed a pharmacophore model based on the interactions between Spm and SMO38–40. This pharmacophore model containing three hydrogen bond acceptors features originated from SER503, GLU192, and GLU200. Based on the above pharmacophore model and molecular docking, SI-4650 was screened out. Subsequently, biochemical experiments indicated that this virtual screening strategy is efficient for identifying SMO inhibitor. Despite more data is still needed to validate and optimise the screening pipeline, these results presented here suggested a possibly useful approach to discover new inhibitors that is complementary to the strategies of designing SMO inhibitors.
In order to further confirm that SI-4650 can inhibit the activity of SMO, we tested the effects of SI-4650 on the enzymatic activity and intracellular polyamine content. As expected, the enzymatic activity of SMO was inhibited by SI-4650 at molecular level. Consistently, further study showed that SI-4650 treatment in A549 cells could decrease the intracellular Spd content and increase Spm content, while the Put level was not significantly different. It is well known that SMO can catalyse Spm into the aldehyde 3-aminopropanal, H2O2, and SpdCitation21–23. These results suggested that SI-4650 can inhibit the activity of SMO, preventing the conversion from Spm to Spd.
As the other member of PA oxidase family, APAO is capable of oxidising the N1-acetylated forms of spermine or spermidine to generate spermidine or putrescine. Currently, specific inhibitors either for SMO or APAO are still very challengeable. It is noted that SI-4650 can also inhibit the enzyme activity of APAO with a lower Ki value than SMO. Docking analysis results have shown that SI-4650 formed a stronger hydrogen bond interaction with HIS64 and the location is more favourable for the binding process. These structure characteristics are expected to explain the different binding affinity of SI-4650 for SMO and APAO. Albeit the specificity is still need to be improved, SI-4650 can be used as a lead compound to develop more specific SMO inhibitor.
The polyamine metabolism regulation is very complicated and many enzymes are involved in this process. Based on our results, we can infer that SI-4650 inhibits the SMO and APAO enzyme activities and reduces the total polyamine content. However, we can’t conclude that the decrease of polyamine levels is only triggered by SMO and APAO inhibition. Therefore, further investigations are still needed to validate whether SI-4650 can affect other enzymes of polyamine metabolism.
To investigate the antitumor activity of SI-4650, we tested the effects of SI-4650 on cell proliferation, migration and cell cycle distribution. From the data presented in this work, it must be concluded that SI-4650 can inhibit cell proliferation and migration, and induce S/G2 arrest in A549 cell line. It is well known that the cell cycle is regulated by cyclins, CDKs, and cyclin-dependent kinases. In particular, cyclin B protein is involved in the regulation of the progression of G2/M phase and p21, a cyclin-dependent kinase inhibitor, is involved in G1/S transition thereby inducing cell cycle arrestCitation44. In this study, we observed that SI-4650 treatment significantly increased the level of cyclin B1 and p21 in A549 cells. These results indicated that upregulation of cyclin B1, and p21 expression by SI-4650 may lead to the S/G2 phase arrest in A549 cells.
Apoptosis is a very tightly programmed cell death with distinct biochemical and genetic pathways. In this study, induced apoptosis of cells was observed in A549 cells on SI-4650 exposure. To explore the precise mechanism of apoptosis induced by SI-4650 in A549 cells, the Bcl-2 family proteins (Bax and Bcl-2) were examined. The Bax/Bcl-2 ratio has a close relationship with the extent of apoptosisCitation45. Our results showed that Bax and Bcl-2 were up-regulated at the same time, while the Bax/Bcl-2 ratio was significantly elevated in the A549 cells, indicating the increase of apoptosis. It is well known that Bcl-2 family proteins play an important role in mitochondrial apoptosis pathwayCitation46. Thus, SI-4650 may induce the apotosis in A549 cells via mitochondrial apoptosis pathway.
Autophagy can act as an alternative cell death pathway to apoptosisCitation47. In this study, transmission electron microscope images and confocal microscopy results showed that SI-4650 treatment increased the formation of intracellular autophagosomes in A549 cells. LC3, including LC3I(18 kDa) and LC3II(16 kDa), is associated with the formation of the autophagic vacuole. The amount of LC3II correlates with the extent of autophagosome formation and is considered as a specific molecular marker for autophagosomesCitation48. p62 is an autophagy adaptor protein and p62 reduction may activate autophagyCitation49. The induction of autophagy by SI-4650 was further evidenced by the increased LC3II and p62 reduction. Taken together, our results suggested that SI-4650 treatment can lead to apoptosis as well as autophagy in A549 cells.
Despite the potential utility of SI-4650, we note that this compound would need to be optimised to produce a SMO inhibitor with greater potency and specificity. Analysis of the binding mode of SI-4650 highlighted interesting features, which can shed light on the improvement direction. Due to the similar fold and conservation of key residues, the overall shape of the active site of SMO and APAO is very similar. Albeit SI-4650 adopted similar conformation in both complex structures, the binding site is different: in the SMO complex, SI-4650 binds to the left side of active pocket; whereas in the APAO complex, SI-4650 binds to the right side of active pocket. From the structural point, this difference was dominated by the hydrogen bond formed by terminal amino group and amide group. These observations led to the hypothesis that replacing amide group with hydrophobic group may reduce the affinity of SI-4650 and APAO, making energetically unfavourable the binding of SI-4650 to APAO and increasing the specificity toward SMO. Moreover, adding electron-withdrawing group to the terminal amino group would help form stronger cation-Pi interaction, thus facilitating the directing and binding of SI-4650 to the SMO. Notably, the structural details showed that despite SI-4650 can form new interactions with SER192 and GLU194, which is the determinant factors for the specificity, the type of interaction is hydrophobic rather than polar. Therefore, additional polar group for terminal amino group would increase the binding affinity and specificity. In addition, the water solubility of SI-4650 is low, making it unsuitable to use in some contexts. Based on these structural details, we can design some new active molecules with lower Ki values, and specific for SMO. To these ends further optimisation of the SI-4650 and in vivo evaluation of the effectiveness of SI-4650 are still needed in future work.
5. Conclusion
In summary, based on the developed virtual screening pipeline, we have identified computationally SI-4650 that function as a novel inhibitor of SMO. Experimentally, we have confirmed that SI-4650 can inhibit SMO enzyme activity and reduce the polyamine content in A549 cells. Furthermore, SI-4650 can inhibit the proliferation and migration, and induce cell cycle S/G2 arrest, apoptosis and autophagy in A549 cells. Collectively, these results suggested that SI-4650 can interfere with polyamine metabolism as a SMO inhibitor and has the potential for cancer treatment.
Supplemental Material
Download PDF (369.9 KB)Acknowledgements
The authors are thankful to State Key Laboratory of Natural and Biomimetic Drugs, Peking University, Beijing, China, for performing the virtual screening.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Pegg AE, Casero RA, Jr. Current status of the polyamine research field. Methods Mol Biol 2011;720:3–35.
- Amendola R, Cervelli M, Fratini E, et al. Spermine metabolism and anticancer therapy. Curr Cancer Drug Targets 2009;9:118–30.
- Igarashi K, Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochem Biophys Res Commun 2000;271:559–64.
- Bachrach U. Naturally occurring polyamines: interaction with macromolecules. Curr Protein Pept Sci 2005;6:559–66.
- Battaglia V, DeStefano Shields C, Murray-Stewart T, Casero RA, Jr. Polyamine catabolism in carcinogenesis: potential targets for chemotherapy and chemoprevention. Amino Acids 2014;46:511–19.
- Schipper RG, Penning LC, Verhofstad AA. Involvement of polyamines in apoptosis. Facts and controversies: effectors or protectors? Semin Cancer Biol 2000;10:55–68.
- Casero RA, Jr, Murray Stewart T, Pegg AE. Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer 2018;18:681–95.
- Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov 2007;6:373–90.
- Babbar N, Murray-Stewart T, Casero RA, Jr. Inflammation and polyamine catabolism: the good, the bad and the ugly. Biochem Soc Trans 2007;35:300–4.
- Park MH, Igarashi K. Polyamines and their metabolites as diagnostic markers of human diseases. Biomol Ther (Seoul) 2013;21:1–9.
- Igarashi K, Kashiwagi K. Protein-conjugated acrolein as a biochemical marker of brain infarction. Mol Nutr Food Res 2011;55:1332–41.
- Igarashi K, Ueda S, Yoshida K, Kashiwagi K. Polyamines in renal failure. Amino Acids 2006;31:477–83.
- Kramer DL, Diegelman P, Jell J, et al. Polyamine acetylation modulates polyamine metabolic flux, a prelude to broader metabolic consequences. J Biol Chem 2008;283:4241–51.
- Pegg AE. Functions of polyamines in mammals. J Biol Chem 2016;291:14904–12.
- Khomutov MA, Weisell J, Hyvonen M, et al. Hydroxylamine derivatives for regulation of spermine and spermidine metabolism. Biochemistry (Mosc) 2013;78:1431–46.
- Kee K, Vujcic S, Merali S, et al. Metabolic and antiproliferative consequences of activated polyamine catabolism in LNCaP prostate carcinoma cells. J Biol Chem 2004;279:27050–8.
- Nowotarski SL, Woster PM, Casero RA, Jr. Polyamines and cancer: implications for chemotherapy and chemoprevention. Expert Rev Mol Med 2013;15:e3.
- Wang Y, Hacker A, Murray-Stewart T, et al. Induction of human spermine oxidase SMO(PAOh1) is regulated at the levels of new mRNA synthesis, mRNA stabilization and newly synthesized protein. Biochem J 2005;386:543–7.
- Wang Y, Devereux W, Woster PM, et al. Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res 2001;61:5370–3.
- Cervelli M, Salvi D, Polticelli F, et al. Structure-function relationships in the evolutionary framework of spermine oxidase. J Mol Evol 2013;76:365–70.
- Murray-Stewart T, Wang Y, Goodwin A, et al. Nuclear localization of human spermine oxidase isoforms - possible implications in drug response and disease etiology. FEBS J 2008;275:2795–806.
- Cervelli M, Amendola R, Polticelli F, Mariottini P. Spermine oxidase: ten years after. Amino Acids 2012;42:441–50.
- Xu H, Chaturvedi R, Cheng Y, et al. Spermine oxidation induced by Helicobacter pylori results in apoptosis and DNA damage: implications for gastric carcinogenesis. Cancer Res 2004;64:8521–5.
- Hong SK, Chaturvedi R, Piazuelo MB, et al. Increased expression and cellular localization of spermine oxidase in ulcerative colitis and relationship to disease activity. Inflamm Bowel Dis 2010;16:1557–66.
- Goodwin AC, Jadallah S, Toubaji A, et al. Increased spermine oxidase expression in human prostate cancer and prostatic intraepithelial neoplasia tissues. Prostate 2008;68:766–72.
- Smirnova OA, Keinanen TA, Ivanova ON, et al. Hepatitis C virus alters metabolism of biogenic polyamines by affecting expression of key enzymes of their metabolism. Biochem Biophys Res Commun 2017;483:904–9.
- Chaturvedi R, de Sablet T, Peek RM, Wilson KT. Spermine oxidase, a polyamine catabolic enzyme that links Helicobacter pylori CagA and gastric cancer risk. Gut Microbes 2012;3:48–56.
- Pledgie A, Huang Y, Hacker A, et al. Spermine oxidase SMO(PAOh1), Not N1-acetylpolyamine oxidase PAO, is the primary source of cytotoxic H2O2 in polyamine analogue-treated human breast cancer cell lines. J Biol Chem 2005;280:39843–51.
- Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut 1997;40:297–301.
- Mastrantonio R, Cervelli M, Pietropaoli S, et al. HIV-Tat induces the Nrf2/ARE pathway through NMDA receptor-elicited spermine oxidase activation in human neuroblastoma cells. PLoS One 2016;11:e0149802.
- Uemura T, Takasaka T, Igarashi K, Ikegaya H. Spermine oxidase promotes bile canalicular lumen formation through acrolein production. Sci Rep 2017;7:14841.
- Hu T, Sun D, Zhang J, et al. Spermine oxidase is upregulated and promotes tumor growth in hepatocellular carcinoma. Hepatol Res 2018;48:967–77.
- Vujcic S, Diegelman P, Bacchi CJ, et al. Identification and characterization of a novel flavin-containing spermine oxidase of mammalian cell origin. Biochem J 2002;367:665–75.
- Wang Y, Murray-Stewart T, Devereux W, et al. Properties of purified recombinant human polyamine oxidase, PAOh1/SMO. Biochem Biophys Res Commun 2003;304:605–11.
- Moriya S, Iwasaki K, Samejima K, et al. A mass spectrometric method to determine activities of enzymes involved in polyamine catabolism. Anal Chim Acta 2012;748:45–52.
- Seiler N, Renault J, Gosse F, et al. Cytotoxicity of the polyamine oxidase inactivator MDL 72527 to cancer cells: comparison with a saturated structural analogue. Int J Oncol 2005;27:1669–76.
- Grigorenko NA, Khomutov MA, Simonian AR, et al. Synthesis of 2,11-bis(methylidene)spermine, a new inhibitor of spermine oxidase. Russ J Bioorgan Chem 2016;42:423–7.
- Cervelli M, Angelucci E, Stano P, et al. The Glu(2)(1)(6)/Ser(2)(1)(8) pocket is a major determinant of spermine oxidase substrate specificity. Biochem J 2014;461:453–9.
- Bianchi M, Polticelli F, Ascenzi P, et al. Inhibition of polyamine and spermine oxidases by polyamine analogues. FEBS J 2006;273:1115–23.
- Tavladoraki P, Cervelli M, Antonangeli F, et al. Probing mammalian spermine oxidase enzyme-substrate complex through molecular modeling, site-directed mutagenesis and biochemical characterization. Amino Acids 2011;40:1115–26.
- Cervelli M, Bellavia G, Fratini E, et al. Spermine oxidase (SMO) activity in breast tumor tissues and biochemical analysis of the anticancer spermine analogues BENSpm and CPENSpm. BMC Cancer 2010;10:555.
- Murray-Stewart TR, Woster PM, Casero RA, Jr. Targeting polyamine metabolism for cancer therapy and prevention. Biochem J 2016;473:2937–53.
- Kaserer T, Beck KR, Akram M, et al. Pharmacophore models and pharmacophore-based virtual screening: concepts and applications exemplified on hydroxysteroid dehydrogenases. Molecules 2015;20:22799–832.
- Yunlan L, Juan Z, Qingshan L. Antitumor activity of di-n-butyl-(2,6-difluorobenzohydroxamato)tin(IV) against human gastric carcinoma SGC-7901 cells via G2/M cell cycle arrest and cell apoptosis. PLoS One 2014;9:e90793.
- Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science 1998;281:1322–6.
- Li Y, Liu J, Li Q. Mechanisms by which the antitumor compound di-n-butyl-di-(4-chlorobenzohydroxamato)tin(IV) induces apoptosis and the mitochondrial-mediated signaling pathway in human cancer SGC-7901 cells. Mol Carcinog 2010;49:566–81.
- Erdélyi P, Borsos E, Takács-Vellai K, et al. Shared developmental roles and transcriptional control of autophagy and apoptosis in Caenorhabditis elegans. J Cell Sci 2011;124:1510–8.
- Nakagawa S, Ueno T, Manabe T, Kawasaki K. Imidazolines increase the levels of the autophagosomal marker LC3-II in macrophage-like RAW264.7 cells. Can J Physiol Pharmacol 2018;96:845–9.
- Duran A, Amanchy R, Linares JF, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell 2011;44:134–46.
