Abstract
The over expression of melanogenic enzymes like tyrosinase caused many hyperpigmentaion disorders. The present work describes the synthesis of hydroxy substituted 2-[(4-acetylphenyl)amino]-2-oxoethyl derivatives 3a–e and 5a–e as antimelanogenic agents. The tyrosinase inhibitory activity of synthesized derivatives 3a–e and 5a–e was determined and it was found that derivative 5c possesses excellent activity with IC50 = 0.0089 µM compared to standard kojic acid (IC50 = 16.69 µM). The presence of hydroxyl groups at the ortho and the para position of cinnamic acid phenyl ring in compound 5c plays a vital role in tyrosinase inhibitory activity. The compound 5d also exhibited good activity (IC50 = 8.26 µM) compared to standard kojic acid. The enzyme inhibitory kinetics results showed that compound 5c is a competitive inhibitor while 5d is a mixed-type inhibitor. The mode of binding for compounds 5c and 5d with tyrosinase enzyme was also assessed and it was found that both derivatives irreversibly bind with target enzyme. The molecular docking and molecular dynamic simulation studies were also performed to find the position of attachment of synthesized compounds at tyrosinase enzyme (PDB ID 2Y9X). The results showed that all of the synthesized compounds bind well with the active binding sites and most potent derivative 5c formed stable complex with target protein. The cytotoxicity results showed that compound 5c is safe at a dose of 12 µg/mL against murine melanoma (B16F10) cells. The same dose of 5c was selected to determine antimelanogenic activity; the results showed that it produced antimelenogenic effects in murine melanoma (B16F10) cells. Based on our investigations, it was proposed that compound 5c may serve as a lead structure to design more potent antimelanogenic agents.
GRAPHICAL ABSTRACT
1. Introduction
Melanogenesis is the process of melanin synthesis; most often produced by cells called melanocytesCitation1,Citation2. Melanocytes are dendritic cells formed from melanoblasts which are unpigmented cells originating from embryonic neural crest cellsCitation3,Citation4. Melanocytes are found in the basal layer of skin epidermis and hair follicles. The primary function of melanocytes is the synthesis of melanin pigment. In skin each melanocytes is surrounded by approximately 36 keratinocytesCitation5,Citation6, to which they transfer their synthesized melaninCitation6,Citation7.
Melanin is a pigment, primarily responsible for skin colour. It retains antioxidative and photoscreening effect, therefore, provides skin photo-protection, prevents from injury and, absorbs and transforms harmful UV radiations into harmless heatCitation8. Despite its advantages, increased production and accumulation of pigmentations can cause skin problems such as freckles, age spots, post-inflammatory hyperpigmentation, lentigo and melanoma. UV which stimulates melanin synthesis is reported to cause gene mutation, DNA damage, impaired immune system and cancerCitation9. Pigmentation in metastatic melanoma patients results in short overall and disease-free survivalCitation10. Melanin content is correlated with higher disease stage and seems to protect malignant melanocytes from chemo-, radio- and photodynamic therapyCitation11,Citation12. Therefore, inhibition of melanogenesis could be a rational approach for controlling metastatic melanoma, abnormal skin pigmentation and related disordersCitation10.
Tyrosinase is key an enzyme showing the rate limiting affect in melanin biosynthesis. In cytosol, L-phenylalanine may be converted to tyrosine by phenylalanine hydroxylase (PAH) in order to serve as the substrate for tyrosinaseCitation13. Tyrosinase also catalyses the hydroxylation of L-tyrosine to 3,4-dihydroxyphenylalanine (L-DOPA) and L-DOPA to dopaquinonesCitation14 which under unregulated conditions result in abnormal accumulation of melanin pigmentsCitation15,Citation16. Therefore, inhibition of tyrosinase is the simplest approach and tyrosinase inhibitors may be an attractive target to achieve depigmentation.
Additionally, antioxidants are reported critical for melanin inhibition. Reactive oxygen species (ROS) and free radicals are shown to contribute in numerous skin disorders including hyperpigmentationCitation17. During hyperpigmentation, increased level of hydrogen peroxide (H2O2) and other ROS is reported which may increase the load of oxidative stress on melanocytes. Numerous ROS scavengers and inhibitors of ROS generation are shown to interfere with oxidation processes, therefore, frequently used to reduce oxidative damage in human body and to downregulate UV-induced melanogenesisCitation17–20.
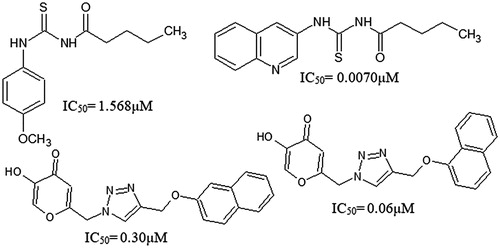
For melanin inhibition, potent tyrosinase inhibitors and antioxidant agents are always desirable. They can be obtained from a variety of sources; however, safety concerns pose a big challenge for their commercialization. There are a number of tyrosinase inhibitors; hydroquinone and kojic acid and, potent antioxidants; tert-butyl hydroxyanisole (BHA) and tert-butyl hydroxytoluene (BHT) which may show undesirable side effects; cytotoxicity, dermatitis and skin cancerCitation21. Recently, methoxy phenyl thiourea, thiazole derivatives and kojic acid derivatives have been reported as the tyrosinase inhibitor ()Citation22–24. Therefore, search for safe and effective depigmenting agents is required which may address these issues and serve as a better solution for treating pigment related dermatological disorders.
Keeping in view the importance of phenolic hydroxyl in tyrosinase inhibition,Citation25,Citation26 the present work is planned to synthesize hydroxy substituted 2-[(4-acetylphenyl)amino]-2-oxoethyl derivatives 3a–e and 5a–e to explore the role of different functionalities in tyrosinase inhibitory activity. The enzyme inhibitory kinetics of most potent compounds was determined by Line-weaver Burk Plots and Dixon Plots. The reversibility of enzyme–inhibitor complex was also determined. The computational molecular docking of the synthesized compounds was performed against target protein PDBID 2Y9X to predict the binding sites of these compounds in target protein. The molecular dynamic simulation was also performed to check the stability of enzyme-inhibitor complex computationally. The cytotoxicity and antimelanogenic activity of most potent derivative 5c was also performed against murine melanoma (B16F10) cells.
2. Results and discussion
2.1. Chemistry
The title compounds 3a–e and 5a–e were synthesized by the following already reported methodCitation27,Citation28 as shown in Schemes 1 and 2. 4-Acetyl aniline was reacted with chloroacetyl chloride in the presence of triethyl amine to synthesized intermediate 1. The final products 3a–e and 5a–e were synthesized by reacting substituted benzoic acids and cinnamic acids with intermediate 1. The formation of the final products was ascertained by their FTIR, 1HNMR and 13CNMR spectral data. The absorption for amide –NH stretching appeared at 3133–3156 cm−1 in FTIR spectra while amide carbonyl appeared at 1632–1656 cm−1. The amide –NH in 1HNMR spectra appeared as a singlet at downfield region, rest of the signals are present in the acceptable regions.
2.2. Enzyme inhibitory kinetics
Hydroxy substituted 2-[(4-acetylphenyl)amino]-2-oxoethyl derivatives 3a–e and 5a–e have been designed and synthesized to explore their inhibitory effects on tyrosinase activity. Kojic acid a clinically applied agent was used as a standard for comparison purpose. The presence of phenolic hydroxyls is of special interest because of their high tyrosinase inhibitory activity. It has been exposed from our bioassay results that the major determining factor of inhibitory activity is the presence of phenolic hydroxyls at cinnamic acid moiety (). Interestingly, compound 5c bearing 2,4-dihydroxy substituted cinnamic acid moiety exhibited excellent activity (IC50 = 0.0089 µM) and is also more active than standard kojic acid (IC50 = 16.69 µM). The compound 5d also exhibited good activity (IC50 = 8.26 µM) compared to standard kojic acid. The bioassay results proved that derivatives with hydroxy substituted cinnamic acid moiety (5a–d) displayed greater tyrosinase inhibitory activity compared to hydroxy substituted benzoic acid derivatives (3a–e).
Table 1. Tyrosinase inhibitory activity of the synthesized compounds 3a–e and 5a–e.
To understand the mechanism of mushroom tyrosinase inhibition by compounds 5c and 5d, kinetic studies were conducted. Kinetic studies showed a concentration-dependent mushroom tyrosinase inhibition by tested inhibitors. Constant and continual monitoring of the reaction showed a marked decrease of the reaction rate in the presence of the inhibitors, which ultimately indicated the decrease of the final absorbance when compared with controls without inhibitor. The potency of inhibition exhibited by these compounds changed depending on the presence and position of substituents and on the class of compoundsCitation29. Enzyme inhibition kinetics was analysed by Lineweaver–Burk plot and Dixon plots to determine the type of inhibition and inhibition constant (KI).
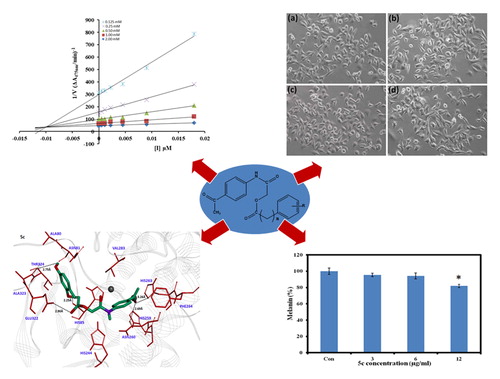
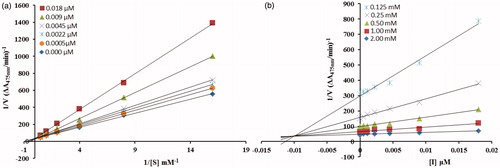
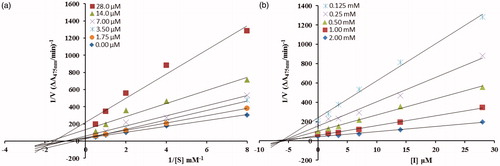
From the kinetic analyses, the Lineweaver–Burk plot of 1/Vmax versus 1/[S] in the presence of different concentrations of 5c showed that Km value change while that of 1/Vmax remained the same and in case of compound 5d both the Km and 1/Vmax reached to new values as shown in and , respectively. This graphical representation showed that the compound 5c is a competitive inhibitor and the compound 5d is a mixed-type inhibitor. On the other hand, in the Dixon plot, the dissociation constant Ki value for 5c with 0.01 µM and for 5d with 6 µM was calculated from the plots for uninhibited enzyme and with different concentrations of inhibitors as shown in and , respectively.
Figure 2. (a) Lineweaver–Burk plots for the inhibition of the diphenolase activity of mushroom tyrosinase by various concentrations 0.000, 0.0005, 0.0022, 0.0045, 0.009 and 0.018 µM of compound 5c in the presence of different concentrations 0.062, 0.125, 0.25, 0.5, 1 and 2 mM, of L-DOPA. (b) Dixon plots for the inhibition of the diphenolase activity of mushroom tyrosinase by various concentrations of compound 5c in the presence of different concentrations 0.125, 0.25, 0.5, 1 and 2 mM, of L-DOPA.
The inhibitory mechanism of mushroom tyrosinase by compounds 5c (in concentrations: 0.0, 0.0011, 0.0022, 0.0045, 0.0090 and 0.018 µM) and 5d (in concentrations: 0.0, 1.75, 3.5, 7.0, 14, 28 and 56 µM) were investigated. The plots of the remaining enzyme activity versus the concentration of enzyme (1.25, 2.5 and 5 µg/mL) at different inhibitor concentrations for the catalysis of L-DOPA gave a series of parallel straight lines with the same slopes indicating that the inhibitory effect of 5c on the tyrosinase was irreversible (. Identical research was carried out for compound 5d and the same conclusion was drawn that 5d was irreversible inhibitor (. These results suggested that both the compounds effectively inhibited the enzyme by binding to its binuclear active site irreversiblyCitation30.
Figure 3. (a) Lineweaver–Burk plots for the inhibition of the diphenolase activity of mushroom tyrosinase by various concentrations 0.00, 1.75, 3.5, 7, 14 and 28 µM of compound 5d in the presence of different concentrations 0.125, 0.25, 0.5, 1 and 2 mM, of L-DOPA. (b) Dixon plots for the inhibition of the diphenolase activity of mushroom tyrosinase by various concentrations of compound 5d in the presence of different concentrations 0.125, 0.25, 0.5, 1 and 2 mM, of L-DOPA.
2.3. Cytotoxicity and antimelanogenic activity
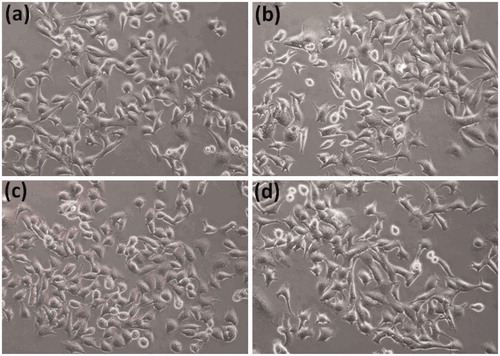
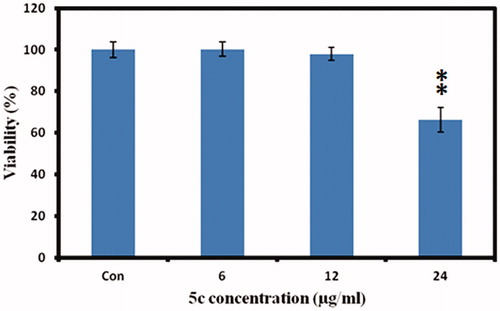
To investigate the effect of compound 5c on cellular melanin synthesis, we first assessed the safe treatment dose of tested derivative 5c using MTT cell viability assay. For viability assay, murine melanoma (B16F10) cells were incubated with various concentrations of compound 5c for 24 h and then MTT assay was performed as described in the methods section. The result showed concentration dependent 5c effect on cell viability as shown in . The 5c compound showed significant toxic effect (33.7% compared to control normalized to 100%) at 24 µg/ml concentrations however no significant effect at 12 µg/ml concentration was observed. Therefore, we selected 12 µg/ml concentration (safe concentration) of 5c for further analysis. Later, we took cellular images which showed no effect on cell morphology after incubation with 5c (.
Figure 4. Relationship between the catalytic activity of tyrosinase and various concentrations of compound 5c shown in (a) and 5d shown in (b).
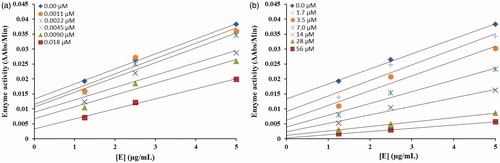
Figure 5. Cell viability measurements. The murine melanoma (B16F10) cells were incubated with indicated concentrations of 5c, and the cell viability was measured by MTT assay. The data was expressed as a percentage of the control (normalized to 100%) with mean ± standard deviation and was analysed using Student’s t-tests, **p < 0.005.
For melanin measurement, B16F10 cells were incubated in cell culture medium containing various concentrations of 5c for 24 h. Interestingly, cellular melanin analysis showed decrease of melanin content with increasing concentration of 5c (. The significant decrease of cellular melanin content was observed at 12 µg/ml where we got 82% melanin content as compared to control normalized to 100% as given in . The melanoma cells are widely used for melanin investigations and various studies have shown the effect of compound concentration on melanogenesisCitation31–35. In conclusion, our results indicate pronounced ability of 5c compound to inhibit melanin synthesis in cells in vitro which may show promising future in the field of cosmetics or medicine.
2.4. Structural assessment of target protein
Mushroom tyrosinase (Agaricusbisporus) is a class of oxidoreductase copper containing protein comprises 391 amino acids. The structure architecture of mushroom tyrosinase showed that it consists of 39% helices (154 residues) and 14% β sheets (57 residues) and 46% coils (180 residues). The X-ray diffraction study confirmed its resolution 2.78Ȧ, R-value 0.238 and unit cell crystal dimensions like length and angles of coordinates. The unit cell length values were observed for a = 103.84, b = 104.82 and c = 119.36 with angles 90°, 110.45° and 90° for all α, β and γ dimensions, respectively. The Ramachandran plots and values indicated that 95.90% of residues were in favoured regions and 100.0% residues were lies in allowed regions (Figure S1). This selected Ramachandran graph values showed the good accuracy of phi (φ) and psi (ψ) angles among the coordinates of receptor molecules and most of residues plunged in acceptable region.
2.5. Chemo-informatics properties and Lipinski’s rule
The predicted chemo-informatics properties such as molar volume, density, polarizability and surface tension were evaluated by computational approach. Literature study established a standard value for molar refractivity (40–130), molecular weight (160–480) and number of atoms (20–70) Citation36. Results showed that 5c predicted values are much better than standard values and all other synthesized compounds. Moreover, the Lipinski’s rule of five (RO5) says nothing about specific chemistry or structural features found in drugs or non-drugs. The computational results showed that 5c possesses 6 HBA, 3 HBD and 2.54 LogP values which significantly justified its drug like behaviour. Moreover, their molecular weight (355.11 g/mol) was also much better than standard value (<5000 g/mol). The RO5 justifies that molecules with poor absorption are more likely to have more than 5 HBD, MWT over 500, logP over 5 and more than 10 HBA. However there are plenty of examples available for RO5 violation amongst the existing drugsCitation37. In overall results, these predicted values justify the significance of 5c synthesized compound as a good candidate molecule ().
Table 2. Chemo-informatics evaluation of synthesized compounds.
2.6. Molecular docking
The docked complexes of all the synthesized compounds (3a–3e and 5a–5e) were analysed on the basis of lowest binding energy values and hydrogen bonding analyses. Results showed that 5c was the most active compound with best binding energy value (–7.90 kcal/mol) compared to others derivatives. The graphical depiction of 5c docking complex is mentioned in .
Figure 7. Intracellular melanin measurements. The murine melanoma (B16F10) cells were incubated with indicated concentrations of 5c, and the cellular melanin content was detected. The data were expressed as a percentage of control (normalized to 100%) with mean ± standard deviation and, and were analysed using Student’s t-tests, *p < 0.05.
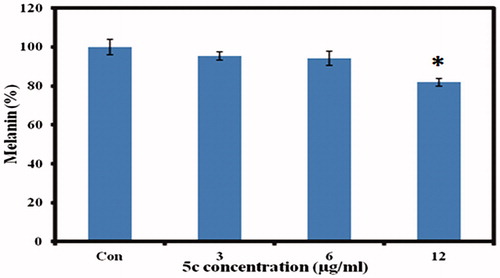
Figure 8. Docking interaction between 5c and target protein. The 5c is mentioned in green colour with embedded oxygen functional groups in red and nitrogen in purple colours. The target protein is highlighted in line ribbon format with light grey colour. The active binding site amino acids are highlighted in red colour. Two copper ions are also mentioned in black colour. Five hydrogen bonds were observed between 5c and receptor amino acids like HIS263, ASN260, HIS85, GLU322 and ALA323 with bonding distances 3.26 Ȧ, 2.69 Ȧ, 2.25 Ȧ, 2.90 Ȧ and 2.75 Ȧ, respectively. The black dotted lines show the binding distance in angstrom (Ȧ).
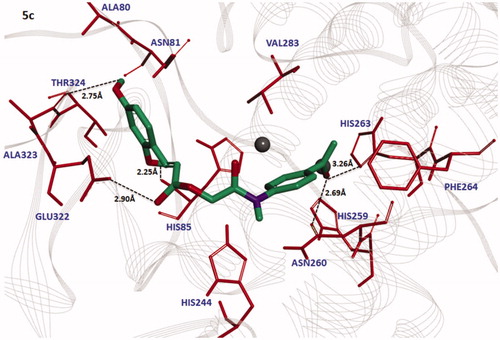
The bonding analysis of all compounds against mushroom tyrosinase showed that 5c compound is directly interacts with active residues of targeted protein. The structure activity relationship (SAR) analysis shows that 5c builds five hydrogen bonds at specific residues (HIS263, ASN260, HIS85, GLU322 and ALA323) against the target protein. The carbonyl oxygen of benzene ring of 5c interacts with HIS263and ASN260 positions having bonds lengths 3.26 Å and 2.69 Å. The ester oxygen of the same compound also interacts with GLU322 with bonding distance 2.90 Å. The hydroxyl groups of the same compounds form bonds with HIS85 and ALA323 with bonds length 2.25 Å and 2.75 Å, respectively. Literature study also justified that these interacted residues are significant in the downstream signalling pathwaysCitation38. The graphical representations of all other interacted compounds (3a–3e, and 5a, 5b, 5d and 5e) against target protein are mentioned in Figures S2–S3 (see Supplementary materials).
Moreover, comparative analysis showed that 5c has good binding energy values (−7.90 kcal/mol) as compared other derivatives. The other candidate compounds (3a–3e) also possess good binding energy values −7.60 kcal/mol, −7.70 kcal/mol, −7.60 kcal/mol, −7.40 kcal/mol and −7.60 kcal/mol, while 5a, 5b, 5d and 5e showed −7.10 kcal/mol, −6.90 kcal/mol, −6.40 kcal/mol and −7.10 kcal/mol binding energy values, respectively (). Here, it is noticeable that most of the candidate compounds bind in the active binding regions but little fluctuated with their conformational positions. The comparative binding analysis and bioassay study showed that 5c was the most active and significant compound as compared to other synthesized compounds.
Table 3. Docking results of synthesized compounds using PyRx
3. Conclusion
The hydroxy substituted 2-[(4-acetylphenyl)amino]-2-oxoethyl derivatives 3a–e and 5a–e with various hydroxyl moieties have been synthesized to explore their role in tyrosinase inhibitory activity. The analogue 5c showed excellent activity with IC50=0.0089 µM better than standard kojic acid IC50 =16.69 µM. The enzyme inhibitory kinetics results confirmed that compounds 5c, a competitive inhibitor, and 5d, a mixed type inhibitor. Both compounds 5c and 5d inhibit the enzyme by irreversible mode of binding. The computational molecular dynamic simulation results revealed that synthesized compounds bind well with the active binding sites and most potent derivative 5c formed stable complex with target protein. The cytotoxicity evaluation performed against murine melanoma (B16F10) cells showed that most potent derivative 5c is nontoxic at a dose of 12 µg/mL. The same dose of 5c was selected to determine antimelanogenic activity; the results showed that it produced antimelenogenic effects in murine melanoma (B16F10) cells. It is proposed that analogue 5c may serve as lead structure to design highly potent antimelanogenic agents.
4. Experimental
4.1. Synthesis of title compounds 3a–e and 5a–e
4-Actylaniline (0.01 mol) was reacted with chloroacetyl chloride (0.01 mol) in the presence of triethylamine (0.01 mol) in anhydrous dichloromethane (20 ml) at 0 to −5 °C. The reaction mixture was stirred at room temperature for 4 h, extracted with ethyl acetate, washed with dilute hydrochloric acid and 5% sodium hydroxide solution. The organic layer was removed under reduced pressure to afford the intermediate 1 as solid with melting point 154–155 °C; FTIR νmax cm−1: 3178 (–NH), 2922 (sp2 C–H), 2812 (sp3 C–H), 1634 (C=O amide) and 1599 (C = C aromatic). The title amides 3a–e were obtained by treating intermediate 1 with hydroxy substituted benzoic acids 2a–e (0.01 mole), triethyl amine (0.01 mol) and potassium iodide (0.01 mol) in dimethylformamide (DMF, 10 ml) at room temperature, while compounds 5a–e were synthesized by reacting intermediate 1 with acids 4a–e under the same conditions. After the completion of reaction, the mixture was poured into ice cold water with stirring and extracted with ethyl acetate (4 × 20 ml). The combined organic layer was washed with dilute hydrochloric acid (5%), sodium carbonate solution (5%) and finally with aqueous NaCl solution. The organic layer was dried over anhydrous magnesium sulphate, filtered and the solvent was removed under reduced pressure to afford the crude title amides 3a–e and 5a–e purified by silica gel column chromatography.
2-[(4-Acetylphenyl)amino]-2-oxoethyl 3-hydroxybenzoate (3a)
Solid; reaction time, 24 h; yield 76%; melting point 166–168 °C; Rf 0.52 (n-hexane:ethyl acetate 2:1), FTIR νmax cm−1: 3336 (−OH), 3132 (−NH), 2936 (sp2 C–H), 2835(sp3 C–H), 1731 (C=O ester), 1656 (C=O amide), 1596 (C=C aromatic), 1147 (C–O, ester); ESI-MS: m/z 336 [M + 23] (M + Na)+; 1H NMR (400 MHz, DMSO, ppm) δ: 10.63 (s, 1H, −NH), 9.90 (s, 1H, −OH), 7.94 (d, J = 8.2 Hz, 2H, H-17, H-19), 7.74 (d, J = 8.2 Hz, 2H, H-16, H-20), 7.48–7.43 (m, 2H, H-1, H-6), 7.35 (t, J = 2.4 Hz, 1H, H-3), 7.09–7.07 (m, 1H, H-5), 4.96 (s, 2H, H-11, −CH2), 2.54 (s, 3H, H-23, −CH3); 13 C NMR (100 MHz, DMSO, ppm) δ: 196.98 (C-21, C=O, ketone), 166.47 (C-12, C=O, amide), 165.91 (C-8, C=O, ester), 158.03 (C-4), 143.28 (C-15), 132.4 (C-18), 130.8 (C-2), 130.4 (C-6), 130 (C-17,19), 121.1 (C-1), 120.6 (C-5), 120.2 (C-4), 118.9 (C-16,20), 116.3 (C-3), 60.6 (C-11, –CH2), 26.9 (C-23, –CH3); Anal Calcd For C17H15NO5: C, 65.17; H, 4.97; Found C, 65.14; H, 4.95.
2-[(4-Acetylphenyl)amino]-2-oxoethyl 4-hydroxybenzoate (3b)
Solid; reaction time, 24 h; yield 82%; melting point 184–186 °C; Rf 0.53 (n-hexane:ethyl acetate 2:1), FTIR νmax cm−1: 3365 (–OH), 3133 (–NH), 2983 (sp2 C–H), 2885(sp3 C–H), 1715 (C=O ester), 1648 (C=O amide), 1599 (C=C aromatic), 1152 (C–O, ester); ESI-MS: m/z 336 [M + 23] (M + Na)+; 1H NMR (400 MHz, DMSO, ppm) δ: 10.55 (s, 1H, –NH), 10.42 (s, 1H, –OH), 7.96 (d, J = 7.2 Hz, 2H, H-17, H-19), 7.88 (d, J = 4.4 Hz, 2H, H-1, H-3), 7.73 (d, J = 6.8 Hz, 2H, H-16, H-20), 6.89 (d, J = 4.4 Hz, 2H, H-5, H-6), 4.92 (s, 2H, H-11, –CH2), 2.54 (s, 3H, H-22, –CH3); 13 C NMR (100 MHz, DMSO, ppm); δ 196.98 (C-21, C=O, ketone), 166.73 (C-12, C=O, amide), 165.69 (C-5), 162.76 (C-8, C=O, ester), 143.29 (C-15), 132.43 (C-18), 132.26 (C-1,3), 130.01 (C-17,19), 120.17 (C-2), 118.98 (C-16,20), 115.89 (C-4,6), 63.27 (C-11, –CH2), 26.9 (C-22, –CH3); Anal Calcd For C17H15NO5: C, 65.17; H, 4.97; Found C, 65.13; H, 4.93.
2-[(4-Acetylphenyl)amino]-2-oxoethyl 2,4-dihydroxybenzoate (3c)
Solid; reaction time, 24 h; yield 73%; melting point 227–229 °C; Rf 0.47 (n-hexane:ethyl acetate 2:1), FTIR νmax cm−1: 3354 (–OH), 3155 (–NH), 2951 (sp2 C–H), 2856(sp3 C–H), 1728 (C=O ester), 1637 (C=O amide), 1597 (C=C aromatic), 1158 (C–O, ester); ESI-MS: m/z 352 [M + 23] (M + Na)+; 1H NMR (400 MHz, DMSO, ppm) δ: 10.59 (s, 1H, H-7, –OH),10.54 (s, 1H, H-8, –OH), 10.46 (s, 1H, H-15, –NH), 7.96 (d, J = 8.4 Hz, 2H, H-18, H-20), 7.76 (d, J = 8.4 Hz, 2H, H-17, H-21), 7.73 (d, J = 8.8 Hz, 1H, H-3), 6.43 (dd, J1 = 8.8 Hz, J2=2.0 Hz, 1H, H-4), 6.34 (d, J = 2.0 Hz, 1H, H-6), 4.96 (s, 2H, H-12, –CH2), 2.54 (s, 3H, H-23, –CH3); 13 C NMR (100 MHz, DMSO, ppm) δ: 196.98 (C-22, C=O, ketone), 168.66 (C-13, C=O, amide), 166.36(C-9, C=O, ester), 164.95 (C-5), 163.12 (C-1), 143.18 (C-16), 132.58 (C-19), 132.51 (C-3), 130.03 (C-19,20), 119.01 (C-17,21), 108.97 (C-4), 104.33 (C-2), 103.07 (C-6), 63.37 (C-12, –CH2), 26.91 (C-23, –CH3); Anal Calcd For C17H15NO5: C, 62.00; H, 4.55; Found C, 61.95; H, 4.52.
2-[(4-Acetylphenyl)amino]-2-oxoethyl 3,4-dihydroxybenzoate (3d)
Solid; reaction time, 24 h; yield 70%; melting point 205–207 °C; Rf 0.46 (n-hexane:ethyl acetate 2:1), FTIR νmax cm−1: 3314 (–OH), 3156 (–NH), 2934 (sp2 C–H), 2843 (sp3 C–H), 1730 (C=O ester), 1642 (C=O amide), 1592 (C=C aromatic), 1148 (C–O, ester); ESI-MS: m/z 352 [M + 23] (M + Na)+; 1H NMR (400 MHz, DMSO, ppm) δ: 10.56 (s, 1H, H-14, –NH), 9.88(s, 1H, H-7, –OH), 9.44 (s, 1H, H-8, –OH), 7.98 (d, J = 5.2 Hz, 2H, H-18, H-20), 7.75 (d, J = 5.2 Hz, 2H, H-17, H-21), 7.43 (d, J = 2.0 Hz, 1H, H-2), 7.40 (dd, J1 = 8.4, J2=2.0 Hz, 1H, H-6), 6.86 (d, J = 8.0 Hz, 1H, H-5), 4.86 (s, 2H, H-12, –CH2), 2.51 (s, 3H, H-23, –CH3); 13 C NMR (100 MHz, DMSO, ppm) δ: 196.98 (C-22, C=O, ketone), 166.78 (C-13, C=O, amide), 165.8(C-9, C=O, ester), 151.26(C-4), 145.6 (C-3), 143.32 (C-16), 132.42 (C-19), 130.01 (C-18,20), 122.69 (C-1), 120.39(C-6), 118.96 (C-17,21), 117.01 (C-5),115.85 (C-2), 63.26 (C-12, –CH2), 26.90 (C-23, –CH3); Anal Calcd For C17H15NO5: C, 62.00; H, 4.55; Found C, 61.96; H, 4.52.
2-([(4-Acetylphenyl)amino]-2-oxoethyl 3,5-dihydroxybenzoate (3e)
solid; reaction time, 24 h; yield 74%; melting point 249–251 °C; Rf 0.45 (n-hexane:ethyl acetate 2:1), FTIR νmax cm−1: 3365 (–OH), 3145 (–NH), 2949 (sp2 C–H), 2862 (sp3 C–H), 1744 (C=O ester), 1623 (C=O amide), 1601 (C=C aromatic), 1147 (C–O, ester); ESI-MS: m/z 352 [M + 23] (M + Na)+; 1H NMR (400 MHz, DMSO, ppm) δ: 10.57 (s, 1H, H-14, –NH), 9.69 (s, 2H, H-7, H-8 –OH), 7.97 (d, J = 1.6 Hz, 2H, H-18, H-20), 7.74 (d, J = 1.6 Hz, 2H, H-17, H-21), 6.90 (d, J = 2.0 Hz, 2H, H-2, H-6), 6.49 (s, 1H, H-4), 4.92 (s, 2H, H-12, –CH2), 2.54 (s, 3H, H-23, –CH3); 13 C NMR (100 MHz, DMSO, ppm) δ: 196.98 (C-22, C=O,ketone), 166.50 (C-13, C=O, amide), 165.96 (C-9, C=O, ester), 159.06 (C-3,5), 143.27 (C-16), 132.46 (C-19), 131.25 (C-18,20), 130.03 (C-1), 118.97 (C-17,21), 108.1 (C-4), 107.86 (C-2,6), 63.57 (C-12, –CH2), 26.90 (C-23, –CH3); Anal Calcd For C17H15NO5: C, 62.00; H, 4.55; Found C, 61.97; H, 4.51.
2-[(4-Acetylphenyl)amino]-2-oxoethyl cinnamate (5a)
Solid; reaction time, 24 h; yield 78%; melting point 164–166 °C; Rf 0.52 (n-hexane:ethyl acetate 2:1), FTIR νmax cm−1: 3144 (–NH), 2918 (sp2 C–H), 2820 (sp3 C–H), 1728 (C=O ester), 1650 (C=O amide), 1593 (C=C aromatic), 1146 (C–O, ester); ESI-MS: m/z 346 [M + 23] (M + Na)+; 1H NMR (400 MHz, DMSO, ppm) δ: 10.54 (s, 1H, H-14, –NH), 7.96 (d, J = 2.0 Hz, 2H, H-18, H-20), 7.75 (m, 5H, H-17, H-21, H-7, H-6, H-2), 7.46 (m, 3H, H-3, H-4, H-5), 6.78 (d, J = 16.0 Hz, 1H, H-8), 4.86 (s, 2H, H-12, –CH2), 2.54 (s, 3H, H-23, –CH3); 13 C NMR (100 MHz, DMSO, ppm) δ: 196.98 (C-22, C=O, ketone), 166.56 (C-13, C=O, amide), 166.27 (C-9, C=O, ester), 145.91 (C-7), 143.26 (C-16), 134.4 (C-19), 132.46 (C-1), 131.16 (C-3,5), 130.01 (C-4), 129.8 (C-2,6), 119.02 (C-17,21), 117.86 (C-8), 63.19 (C-12, –CH2), 26.90 (C-23, –CH3); Anal Calcd For C19H17NO4: C, 70.58; H, 5.01; Found C, 70.53; H, 4.98.
2-[(4-Acetylphenyl)amino]-2-oxoethyl (E)-3-(4-hydroxyphenyl)acrylate (5b)
Solid; reaction time, 24 h; yield 72%; melting point 226–228 °C; Rf 0.45 (n-hexane:ethyl acetate 2:1), FTIR νmax cm−1: 3366 (–OH), 3173 (–NH), 2981 (sp2 C–H), 2867 (sp3 C–H), 1732 (C=O ester), 1639 (C=O amide), 1593 (C=C aromatic), 1147 (C–O, ester); ESI-MS: m/z 362 [M + 23] (M + Na)+; 1H NMR (400 MHz, DMSO, ppm) δ: 10.57 (s, 1H, H-15, –NH), 10.11 (s, 1H, H-7, –OH), 7.95 (d, J = 2.0 Hz, 2H, H-19,21), 7.74 (d, J = 2.0 Hz, 2H, H-18, H-22), 7.63 (m, 3H, H-2, H-6, H-8), 6.83 (d, J = 8.4 Hz, 2H, H-3, H-5), 6.52 (d, J = 16.0 Hz, 1H, H-9), 4.82 (s, 2H, H-13, –CH2), 2.54 (s, 3H, H-25, –CH3); 13C NMR (100 MHz, DMSO, ppm) δ: 196.99 (C-23, C=O, ketone), 166.74 (C-14, C=O, amide), 166.64 (C-10, C=O, ester), 160.56 (C-4), 146.13 (C-8), 143.32 (C-17), 132.42 (C-20), 130.96 (C-2,6), 129.99 (C-19,21), 125.46 (C-1), 118.99 (C-18,22), 116.31 (C-9), 113.87 (C-3,5), 63.0 (C-13, –CH2), 26.91 (C-25, –CH3); Anal Calcd For C19H17NO5: C, 67.25; H, 5.01; Found C, 67.21; H, 4.97.
2-[(4-Acetylphenyl)amino]-2-oxoethyl (E)-3-(2,4-dihydroxyphenyl)acrylate (5c)
Solid; reaction time, 24 h; yield 70%; melting point 236–238 °C; Rf 0.43 (n-hexane:ethyl acetate 2:1), FTIR νmax cm−1: 3366 (–OH), 3154 (–NH), 2959 (sp2 C–H), 2844(sp3 C–H), 1726 (C=O ester), 1654 (C=O amide), 1592 (C=C aromatic), 1157 (C–O, ester); ESI-MS: m/z 378 [M + 23] (M + Na)+; 1H NMR (400 MHz, DMSO, ppm) δ: 10.48 (s, 1H, H-8, –OH), 10.22 (s, 1H, H-16, –NH), 9.93 (s, 1H, H-7, –OH), 7.96 (d, J = 2.0 Hz, 2H, H-20, H-22), 7.85 (d, J = 16.0 Hz, 1H, H-9), 7.73 (d, J = 2.0 Hz, 2H, H-19, H-23), 7.46 (d, J = 8.4 Hz, 1H, H-6), 6.5 (d, J = 16.0 Hz, 1H, H-10), 6.38 (s, 1H, H-3), 6.3 (d, J = 8.4 Hz, 1H, H-5), 4.78 (s, 2H, H-14, –CH2), 2.54 (s, 3H, H-25, –CH3); 13 C NMR (100 MHz, DMSO, ppm) δ: 196.98 (C-24, C=O, ketone), 167.22 (C-15, C=O, amide), 166.91(C-11, C=O, ester), 161.69 (C-2), 159.21 (C-4), 143.32 (C-18), 141.94 (C-9), 132.41 (C-21), 131.07 (C-6), 130 (C-20,22), 118.99 (C-19,23), 116.31 (C-9), 113.01 (C-10), 112.59 (C-1), 108.38 (C-5), 102.96 (C-3), 62.86 (C-14, –CH2), 25.6 (C-25, –CH3); Anal Calcd For C19H17NO6: C, 64.22; H, 4.78; Found C, 64.18; H, 4.73.
2-[(4-Acetylphenyl)amino]-2-oxoethyl-(E)-3-(4-chlorophenyl)acrylate (5d)
solid; reaction time, 24 h; yield 78%; melting point 161–163 °C; Rf 0.50 (n-hexane:ethyl acetate 2:1), FTIR νmax cm−1: 3136 (–NH), 2923 (sp2 C–H), 2855 (sp3 C–H), 1719 (C=O ester), 1648 (C=O amide), 1591 (C=C aromatic), 1150 (C–O, ester); ESI-MS: m/z 382.5 [M + 23] (M + Na)+; 1H NMR (400 MHz, DMSO, ppm) δ: 10.52 (s, 1H, H-15, –NH), 7.96 (d, J = 1.6 Hz, 2H, H-19, H-21), 7.82 (d, J = 1.6 Hz, 2H, H-18, H-22), 7.75 (m, 3H, H-2, H-6, H-8), 7.51 (d, J = 6.4 Hz, 2H, H-3, H-5), 6.84 (d, J = 16.0 Hz, 1H, H-9), 4.85 (s, 2H, H-13, –CH2), 2.54 (s, 3H, H-25, –CH3); 13 C NMR (100 MHz, DMSO, ppm) δ: 196.97 (C-23, C=O, ketone), 166.5 (C-14, C=O, amide), 166.14 (C-10, C=O, ester), 144.48 (C-8), 143.24 (C-17), 135.69 (C-20), 133.37 (C-4), 132.47 (C-1), 130.69 (C-19,21), 130 (C-2,6), 129.48 (C-3,5), 119.01 (C-18,22), 118.69 (C-9), 60.21 (C-13, –CH2), 26.89 (C-25, –CH3); Anal Calcd For C19H18NO4: C, 63.42; H, 5.00; Found C, 63.38; H, 4.95.
2-[(4-Acetylphenyl)amino]-2-oxoethyl 3-(4-hydroxyphenyl)propanoate (5e)
Solid; reaction time, 24 h; yield 74%; melting point 234–236 °C; Rf 0.45 (n-hexane:ethyl acetate 2:1), FTIR νmax cm−1: 3356 (–OH), 3149 (–NH), 2944 (sp2 C–H), 2856(sp3 C–H), 1721 (C=O ester), 1643 (C=O amide), 1590 (C=C aromatic), 1147 (C–O, ester); ESI-MS: m/z 364 [M + 23] (M + Na)+; 1H NMR (400 MHz, DMSO, ppm) δ: 10.43 (s, 1H, H-15, –NH), 9.18 (s, 1H, H-7, –OH), 7.95 (d, J = 2.0 Hz, 2H, H-19,21), 7.71 (d, J = 2.0 Hz, 2H, H-18, H-22), 7.05 (d, J = 2.0 Hz, 2H, H-2, H-6), 6.68 (d, J = 2.0 Hz, 2H, H-3, H-5), 4.70 (s, 2H, H-13, –CH2), 2.78 (t, J = 7.6 Hz, 2H, H-8), 2.66 (t, J = 1.2 Hz, 2H, H-9), 2.54 (s, 3H, H-25, –CH3); 13 C NMR (100 MHz, DMSO, ppm) δ: 196.99 (C-23, C=O, ketone), 172.49 (C-10, C=O, ester), 166.52 (C-14, C=O, amide), 156.10 (C-4), 143.20 (C-17), 132.47 (C-20), 130.90 (C-2,6), 129.59 (C-19,21), 119.10 (C-18,22), 115.58 (C-3,5), 63.00 (C-13, –CH2), 35.66 (C-9), 29.83 (C-8), 26.90 (C-25, –CH3); Anal Calcd For C19H19NO5: C, 66.86; H, 5.57; Found C, 66.80; H, 5.52.
4.2. Anti-tyrosinase activity
The mushroom tyrosinase (EC 1.14.18.1) (Sigma Chemical Co., ST. Louis, MO) was used for in vitro bioassays as described previously with some modificationsCitation39,Citation40. Briefly, 140 µL of phosphate buffer (20 mM, pH 6.8), 20 µL of mushroom tyrosinase (30 U/mL) and 20 µL of the inhibitor solution were placed in the wells of a 96-well micro plate. After pre-incubation for 10 min at room temperature, 20 µL of L-DOPA (3,4-dihydroxyphenylalanine) (0.85 mM) was added and the plate was further incubated at 25 °C for 20 min. Subsequently the absorbance of dopachrome was measured at 492 nm using a micro plate reader (OPTI Max, Tunable). Kojic acid was used as a reference inhibitor and for negative tyrosinase inhibitor phosphate buffer was used instead of the inhibitor solution. The extent of inhibition by the test compounds was expressed as the percentage of concentration necessary to achieve 50% inhibition (IC50). Each concentration was analysed in three independent experiments run in triplicate. The IC50 values were determined by the data analysis and graphing software Origin 8.6, 64-bit.
4.3. Kinetic analysis of the inhibition of mushroom tyrosinase
A series of experiments were performed to determine the inhibition kinetics by following methodCitation41,Citation42. Inhibitor 5c with concentrations 0.0, 0.0005, 0.0022, 0.0045, 0.009, 0.018 µM and 5d with concentrations 0.0, 1.75, 3.5, 7, 14 and 28 µM, respectively, were used. Substrate L-DOPA concentration was among 0.0625–2 mM in all kinetic study. Pre-incubation and measurement time was the same as discussed in mushroom tyrosinase inhibition assay protocol. The formation of DOPAchrome was continuously monitored at 475 nm for 5 min at a 30 s interval in the microplate reader after addition of enzyme. The inhibition type on the enzyme was assayed by Lineweaver–Burk plots of inverse of velocities (1/V) versus inverse of substrate concentration 1/[S] mM−1, and the inhibition constant Ki was determined by the Dixon plot of 1/V versus inhibitor concentrations.
4.4. Cytotoxicity and cell morphology
To test the safety level of 5c compound, MTT assay (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide) was performedCitation43. The murine melanoma (B16F10) cells (1.5 × 105 cells/ml) were seeded in 24 well plates (SPL Korea) with DMEM medium at 37 °C and 5% CO2 incubator. Test compound 5c was dissolved in DMSO and diluted with culture medium to get the final concentrations of 0 (control), 6, 12 and 24 µg/ml, and then added to the cells for 24 h further incubation. The MTT assay was then conducted, and results were calculated in percentage of control. In addition, cellular images were taken by inverted fluorescent microscope (Nikon, ECLIPSE, Tokyo, Japan) to access the 5c effect on cell morphology.
4.5. Melanin content measurement
For melanin measurement, melanoma (1.5 × 105) cells were seeded in cell culture plates for 24 h at 37 °C and 5% CO2incubator. Then cells were incubated with various concentrations of 5c in cell culture medium for further 24 h. Later, assay for melanin measurement was performed following already reported methodCitation44. Briefly, the cells were trypsinised and centrifuged for 5 min at 1000 rpm. The cell pellet was dissolved in 1 N NaOH at 60 °C for 1 h and melanin content was measured spectrophotometrically at 405 nm wavelengths using a microplate reader (BioTek, synergy HT, Winooski, VT).
4.6. Computational studies
4.6.1. Retrieval of protein structure from PDB
The crystal structure of mushroom tyrosinase (PDBID: 2Y9X) was retrieved from the Protein Data Bank (PDB) (http://www.rcsb.org). The energy minimization of target protein was carried out by employing conjugate gradient algorithm and Amber force field in UCSF Chimera 1.10.1Citation45. The stereo-chemical properties, Ramachandran graph and valuesCitation46 of mushroom tyrosinase were assessed by Molprobity serverCitation47, while the hydrophobicity graph was generated by Discovery Studio 4.1 ClientCitation48. The protein architecture and statistical percentage values of helices, beta-sheets, coils and turns were accessed by VADAR 1.8Citation49.
4.6.2. Candidate structures
The synthesized compounds 3a–e and 5a–e were sketched in drawing ACD/ChemSketch tool. The designed ligand molecules were further visualized and minimized by UCSF Chimera 1.10.1 in PDB format. The drug assessment properties of these compounds were access by various computational tools like Molinspiration (http://www.molinspiration.com/) and Molsoft (http://www.molsoft.com/). The Osiris Property Explorer (http://www.organic-chemistry.org/prog/peo/) an online tool was used to calculate their possible tumorigenic or mutagenic risks and drug-likeness values. Lipinski’s rule of five was justified by using Molsoft and Molinspiraion tools. Furthermore, different molecular properties such as molar refractivity, density, surface tension and polarizability were also accessed by chemsketch online.
4.6.3. Molecular docking
Molecular docking experiment was employed on all synthesized ligand molecules 3a–e and 5a–e against mushroom tyrosinase using diverse PyRx toolCitation50. The grid box parametric dimension values (X = 61.0781, Y = 56.3001 and Z = 63.1015) with spacing 1.0Ȧ were adjusted to attain the finest binding conformational pose of protein-ligand molecules. The maximum docking poses (100 numbers of run) for each docking were adjusted to obtain the best docking complex with good conformational pose. All compounds were docked separately against crystal structure of mushroom tyrosinase and the obtained docked complexes were further evaluated on lowest binding energy (kcal/mol) values using Discovery Studio (4.1) and UCSF Chimera 1.10.1.
Supplemental Material
Download PDF (555.8 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bonaventure J, Domingues MJ, Larue L. Cellular and molecular mechanisms controlling the migration of melanocytes and melanoma cells. Pigment Cell Melanoma Res 2013;26:316–25.
- Borovansky J, Riley PA, editors. Melanins and melanosomes: biosynthesis, structure. Physiological and pathological functions. New York: John Wiley & Sons; 2011.
- Lei TC, Virador V, Yasumoto KI, et al. Stimulation of melanoblast pigmentation by 8-methoxypsoralen: the involvement of microphthalmia-associated transcription factor, the protein kinase a signal pathway, and proteasome-mediated degradation. J Invest Dermatol 2002;119:1341–9.
- Sviderskaya EV, Hill SP, Balachandar D, et al. Agouti signaling protein and other factors modulating differentiation and proliferation of immortal melanoblasts. Dev Dynamics 2001;221:373–9.
- Seiberg M. Keratinocyte–melanocyte interactions during melanosome transfer. Pigment Cell Res 2001;14:236–42.
- Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature 2007;445:843.
- Delevoye C. Melanin transfer: the keratinocytes are more than gluttons. J Invest Dermatol 2014;134:877–9.
- Meredith P, Riesz J. Radiative relaxation quantum yields for synthetic eumelanin. Photochem Photobiol 2004;79:211–6.
- Vandamme M, Robert E, Pesnel S, et al. Antitumor effect of plasma treatment on U87 gliomaxenografts: preliminary results. Plasma Process Polym 2010;7:264–73.
- Brożyna AA, Jóźwicki W, Carlson JA, Slominski AT. Melanogenesis affects overall and disease-free survival in patients with stage III and IV melanoma. Hum Pathol 2013;44:2071–4.
- Slominski A, Paus RA, Mihm MC. Inhibition of melanogenesis as an adjuvant strategy in the treatment of melanotic melanomas: selective review and hypothesis. Anticancer Res 1998;18:3709–15.
- Brożyna AA, Jóźwicki W, Roszkowski K, et al. Melanin content in melanoma metastases affects the outcome of radiotherapy. Oncotarget 2016;7:17844.
- Videira IF, Moura DF, Magina S. Mechanisms regulating melanogenesis. An Bras Dermatol 2013;88:76–83.
- Cooksey CJ, Garratt PJ, Land EJ, et al. Evidence of the indirect formation of the catecholic intermediate substrate responsible for the autoactivation kinetics of tyrosinase. J Biol Chem 1997;272:26226–35.
- Hearing VJ, Jiménez M. Analysis of mammalian pigmentation at the molecular level. Pigment Cell Res 1989;2:75–85.
- Fairhead M, Thöny-Meyer L. Bacterial tyrosinases: old enzymes with new relevance to biotechnology. New Biotechnol 2012;29:183–91.
- Büyükokuroğlu ME, Gülçin I, Oktay M, Küfrevioğlu OI. In vitro antioxidant properties of dantrolene sodium. Pharmacol Res 2001;44:491–4.
- Shahidi F, Janitha PK, Wanasundara PD. Phenolic antioxidants. Crit Rev Food Sci Nutr 1992;32:67–103.
- Gülçin İ, Oktay M, Küfrevioğlu Öİ, Aslan A. Determination of antioxidant activity of lichen Cetraria islandica (L) Ach. J Ethnopharmacol 2002;79:325–9.
- Yasui H, Sakurai H. Age-dependent generation of reactive oxygen species in the skin of live hairless rats exposed to UVA light . Exp Dermatol 2003;12:655–61.
- Ito N, Hirose M, Fukushima S, et al. Studies on antioxidants: their carcinogenic and modifying effects on chemical carcinogenesis. Food Chem Toxicol 1986;24:1071–82.
- Ashraf Z, Rasool R, Hassan M, et al. Synthesis, bioevaluation and molecular dynamic simulation studies of dexibuprofen–antioxidant mutual prodrugs. Int J Mol Sci 2016;17:2151.
- (a) Larik FA, Saeed A, Channar PA, et al. Design, synthesis, kinetic mechanism and molecular docking studies of novel 1-pentanoyl-3-arylthioureas as inhibitors of mushroom tyrosinase and free radical scavengers. Eur J Med Chem 2017;141:273.; (b) Saeed A, Ali Mahesar P, Ali Channar P, et al. Synthesis, Molecular docking studies of Coumarinyl-pyrazolinyl substituted thiazoles as non-competitiveinhibitors of mushroom tyrosinase. Bioorg Chem 2017;74:187.
- Ashooriha M, Khoshneviszadeh M, Khoshneviszadeh M, et al. 1,2,3-Triazole-based kojic acid analogs as potent tyrosinase inhibitors: design, synthesis and biological evaluation. Bioorg Chem 2019;82:414–22.
- Menezes JCJMDS, Kamat SP, Cavaleiro JAS, et al. Synthesis and antioxidant activity of long chain alkyl hydroxycinnamates. Eur J Med Chem 2011;46:773–7.
- Miliovsky M, Svinyarov I, Mitrev Y, et al. A novel one-pot synthesis and preliminary biological activity evaluation of cis-restricted polyhydroxy stilbenes incorporating protocatechuic acid and cinnamic acid fragments. Eur J Med Chem 2013;66:185–92.
- Ashraf Z, Rafiq M, Seo S-Y, et al. Synthesis, kinetic mechanism and docking studies of vanillin derivatives as inhibitors of mushroom tyrosinase. Bioorg Med Chem 2015;23:5870–80.
- Ashraf Z, Rafiq M, Seo S-Y, et al. Kinetic and in silico studies of novel hydroxy-based thymol analogues as inhibitors of mushroom tyrosinase. Eur J Med Chem 2015;98:203–11.
- Chen Z, Cai D, Mou D, et al. Design, synthesis and biological evaluation of hydroxy-or methoxy-substituted 5-benzylidene (thio) barbiturates as novel tyrosinase inhibitors. Bioorg Med Chem 2014;22:3279–84.
- Huang HC, Chang TY, Chang LZ, et al. Inhibition of melanogenesis versus antioxidant properties of essential oil extracted from leaves of Vitex negundo Linn and chemical composition analysis by GC-MS. Molecules 2012;17:3902–16.
- Lee J, Jung E, Park J, et al. Glycyrrhizin induces melanogenesis by elevating a cAMP level in B16 melanoma cells. J Invest Dermatol 2005;124:405–11.
- Kim A, Yang Y, Lee MS, et al. NDRG2 gene expression in B16F10 melanoma cells restrains melanogenesis via inhibition of Mitf expression. Pigment Cell Melanoma Res 2008;21:653–64.
- Hamid MA, Sarmidi MR, Park CS. Mangosteen leaf extract increases melanogenesis in B16F1 melanoma cells by stimulating tyrosinase activity in vitro and by up-regulating tyrosinase gene sinase gene expression. Int J Mol Med 2012;29:209–17.
- Jung GD, Yang JY, Song ES, Park JW. Stimulation of melanogenesis by glycyrrhizin in B16 melanoma cells. Exp Mol Med 2001;33:131.
- Kadam RU, Roy N. Recent trends in drug-likeness prediction: a comprehensive review of in silico methods. Indian J Pharm Sci 2007;69:609.
- Tian S, Wang J, Li Y, et al. The application of in silico drug-likeness predictions in pharmaceutical research. Adv Drug Deliv Rev 2015;86:2–10.
- Chen WC, Tseng TS, Hsiao NW, et al. Discovery of highly potent tyrosinase inhibitor, T1, with significant anti-melanogenesis ability by zebrafish in vivo assay and computational molecular modeling. Sci Rep 2015;5:7995.
- Ashraf Z, Rafiq M, Nadeem H, et al. Carvacrol derivatives as mushroom tyrosinase inhibitors; synthesis, kinetics mechanism and molecular docking studies. PLoS One 2017;12:e0178069.
- Ashraf Z, Rafiq M, Seo SY, et al. Design, synthesis and bioevaluation of novel umbelliferone analogues as potential mushroom tyrosinase inhibitors. J Enzyme Inhib Med Chem 2015;30:874–83.
- Wang Q, Qiu L, Chen XR, et al. Inhibitory effects of phloridzin dihydrate on the activity of mushroom (Agaricus bisporus) tyrosinase. Bioorg Med Chem 2007;15:1568–71.
- Chiari ME, Vera DM, Palacios SM, Carpinella MC. Tyrosinase inhibitory activity of a 6-isoprenoid-substituted flavanone isolated from Daleaelegans. Bioorg Med Chem 2011;19:3474–82.
- Rafiq M, Saleem M, Hanif M, et al. Synthesis, structural elucidation and bioevaluation of 4-amino-1, 2, 4-triazole-3-thione’s Schiff base derivatives. Arch Pharmacal Res 2016;39:161–71.
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983;65:55–63.
- Chung KW, Park YJ, Choi YJ, et al. Evaluation of in vitro and in vivo anti-melanogenic activity of a newly synthesized strong tyrosinase inhibitor (E)-3-(2, 4 dihydroxybenzylidene) pyrrolidine-2, 5-dione (3-DBP). Biochim Biophys Acta (BBA)–General Subjects 2012;1820:962–9.
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605–12.
- Lovell SC, Davis IW, Arendall WB, et al. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 2003;50:437–50.
- Chen VB, Arendall WB, Headd JJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallograph Sect D: Biol Crystallogr 2010;66:12–21.
- Pramanik S, Roy K. Exploring QSTR modeling and toxicophore mapping for identification of important molecular features contributing to the chemical toxicity in Escherichia coli. Toxicol In Vitro 2014;28:265–72.
- Willard L, Ranjan A, Zhang H, et al. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res 2003;31:3316–9.
- Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Meth mol biol. 2015;1263:243–50.