Abstract
The arboreal rice rat of the genus Oecomys Thomas, 1906 is one of the most speciose genera of the subfamily Sigmodontinae, with 19 species currently recognized and occurring from eastern Panama to northern Argentina, Paraguay, and in northern, central and eastern Brazil. Herein we describe a new species using an integrative approach based on molecular, morphological, and morphometric data. We used in our assessment recently collected specimens from the states of Pará and Rondônia, one of the most deforested regions in Brazil. We examined 51 specimens of Oecomys from museum collections including name-bearing types from most of the distributional range of the genus. We also sequenced 32 specimens of Oecomys, and for the molecular analyses, we used the mitochondrial marker Cytochrome b and the nuclear marker intron 7 of β-fibrinogen. Our mitochondrial marker results recovered a strongly supported clade composed of two divergent clades (3.78%), one including lineages of O. bicolor and O. cleberi, and the other clade representing the new species. The topology of concatenated mitochondrial and nuclear data also recovered Oecomys sp. nov. as a sister lineage of the O. bicolor and O. cleberi clade. Also, both markers recovered new lineages from the O. bicolor and O. cleberi species group. The new species can be discriminated from other Oecomys species by pelage colour and craniodental characters, such as absent or small mastoid fenestra, and the presence of alisphenoid strut, small subsquamosal fenestra, presence of sphenopalatine vacuities, and presence of accessory loph of M1 and M2 paracones. The new species occurs exclusively in the Rondônia centre of endemism, delimited by the rivers Amazon to the north, Tapajós to the east, and Madeira to the west. The description of this new Oecomys increases the diversity, and also contributes to elevate Amazonian Sigmodontinae species richness and endemism in this still poorly known biome.
http://zoobank.org/urn:lsid:zoobank.org:7477D37A-4D02-443F-B0FB-149379BEDE92
Introduction
The genus Oecomys Thomas, 1906 currently comprises 19 valid species (Pardiñas et al. Citation2016, Citation2017; Rocha et al., Citation2018; Saldanha & Rossi, Citation2021) of small to medium-sized arboreal rodents (head and body: 90–150 mm), that occur from eastern Panama in Central America to northern Argentina, Paraguay, and northern, central and eastern Brazil in South America, with the highest diversity of species found in the Amazon basin (Carleton & Musser, Citation2015; Pardiñas et al., Citation2017). Recent studies point to the existence of species complexes in the genus (Rosa et al., Citation2012; Pardiñas et al., Citation2016; Suárez-Villota et al., Citation2018; Saldanha & Rossi, Citation2021), as attested by several species which have been recently described or revalidated (Carleton et al., Citation2009; Pardiñas et al., Citation2016; Rocha et al., Citation2018; Saldanha & Rossi, Citation2021). The most taxonomically complete study on the systematics of Oecomys included 14 of the 17 species recognized at the time, and suggested the existence of 10 species and five species complexes, among which the complex O. bicolor/O. cleberi comprises eight distinct lineages (Suárez-Villota et al., Citation2018).
Until recently, O. cleberi Locks, 1981 was recognized as restricted to the Brazilian Cerrado, but the species has been recorded in the southern portion of the Amazonia (Suárez-Villota et al., Citation2018; Saldanha et al., Citation2019) and in seasonal forests marginal to the Atlantic Forest (Brandão et al., Citation2022). Differently, O. bicolor Tomes, 1860 has a wider distribution that extends from the trans-Andean tropical forest plains in eastern Panama and western Ecuador to the east of Bolivia and central Brazil, reaching as far as eastern Amazonia (Carleton & Musser, Citation2015).
Due to great morphological similarity, O. cleberi has long been associated with O. bicolor (Carleton & Musser, Citation2015). Musser & Carleton (Citation2005) associated O. cleberi with O. bicolor and O. paricola, though the latter stands apart from the first two by exhibiting a distinct reddish or tawny brown dorsal colour and creamier grey-based ventral colour, whereas O. bicolor and O. cleberi have self-coloured white ventral colour and orangish brown dorsum (Rocha et al., Citation2012; Suárez-Villota et al., Citation2018). Rocha et al. (Citation2012) recognized the validity of O. cleberi and its sister relationship with O. bicolor based on sequences of the mitochondrial gene Cytochrome b (Cytb). Similarly, Suárez-Villota et al. (Citation2018) and Saldanha et al. (Citation2019) recognized the close relationship between bicolor and cleberi based on mitochondrial and nuclear markers. The former authors recovered O. cleberi as a monophyletic group composed of two distinct lineages (central and northwestern clades), and O. bicolor as a polyphyletic group that includes six distinct lineages (central, eastern, northern, southern, western, and westernmost clades). Both species were grouped in the O. bicolor/O. cleberi species group by Suárez-Villota et al. (Citation2018). Saldanha et al. (Citation2019) also recovered O. cleberi as monophyletic and O. bicolor as polyphyletic based on Cytb sequences, but the species were recovered as reciprocally monophyletic clades in their concatenated data (Cytb + intron 7 of β-fibrinogen) topology, although specimens of O. bicolor central and western lineages (sensu Suárez-Villota et al., Citation2018) nested among specimens of O. cleberi. Recently, Brandão et al. (Citation2022) provided compelling evidence of morphological distinction between O. bicolor and O. cleberi, but specimens examined by the authors were restricted to northern clade of O. bicolor and central clade of O. cleberi (sensu Suárez-Villota et al., Citation2018), geographically corresponding to samples from the north of the state of Mato Grosso and from central Brazil, respectively.
Specimens of Oecomys apparently associated with the O. bicolor and O. cleberi species group were collected by some of the authors during field expeditions in the Jamari National Forest, municipality of Itapuã do Oeste, state of Rondônia, Brazil. Direct comparisons of these recently collected specimens with congeners, including morphological and molecular analyses, confirmed that they belong to a hitherto undescribed species occurring in the Brazilian central Amazonia, which we formally describe herein.
Materials and methods
Specimens and collections
We examined a total of 51 specimens of Oecomys (see Appendix 1), including skins, skulls and alcohol-preserved specimens deposited in nine institutions, as follows: Brazil: Coleção Zoológica da Universidade Federal de Mato Grosso (UFMT), Cuiabá; Coleção de Mamíferos da Universidade do Estado de Mato Grosso (Unemat), Cáceres; Coleção de Mamíferos da Universidade Federal de Rondônia (UFRO), Porto Velho; Coleção de Mamíferos da Universidade Federal do Pará (UFPA), Belém; Museu de Zoologia da Universidade de São Paulo (MZUSP), São Paulo; Museu Nacional (MNRJ), Rio de Janeiro; and Museu Paraense Emílio Goeldi (MPEG), Belém; USA: American Museum of Natural History (AMNH), New York; UK: British Museum of Natural History (BMNH), London. We examined the holotypes of Oecomys bicolor (Tomes, 1860; BMNH 7.1.1.96) and its junior synonyms O. benevolens (Thomas, 1901; BMNH 1.2.1.14), O. dryas (Thomas, 1900; BMNH 99.12.5.4), O. milleri (Allen, Citation1916; AMNH 37117), O. nitedulus Thomas, 1910 (BMNH 6.4.8.1), O. rosilla (Thomas, 1904; BMNH 4.5.7.37), and holotype of O. cleberi Locks, 1981 (MNRJ24131). Specimens with field numbers prefixed by “MSF” belong to the UNEMAT and eventually will be catalogued. Specimens collected in the field were captured with either Sherman or pitfall traps between 2011 and 2013. Permits to collect the specimens and to conduct fieldwork in the protected areas were duly provided by the Brazilian Federal authorities of ICMBio (SISBIO number 30902-1).
Morphological and morphometric analyses
In the specimens examined morphologically, we observed the patterns of the pelage of the head, dorsum, venter, manus, and feet, as well as the colouration and arrangement of hairs on the tail. Cranially, we analysed the morphology of the orbital and supraorbital region, palate, basicranium, and auditory region, including the presence and position of foramina and sutures. The morphology of the molars were also examined. The nomenclature used in character descriptions follows Reig (Citation1977), Voss (Citation1991), Weksler (Citation2006), and Saldanha & Rossi (Citation2021).
External measurements and body mass were transcribed from specimen labels and include: length of head and body (HBL), tail length (TL), length of hind foot (HF), length of ear (Ear) and weight (in grams). Craniodental measurements were recorded to the nearest 0.01 mm using digital callipers. The following 31 dimensions were recorded following Saldanha & Rossi (Citation2021): occipitonasal length (ONL), condylo-incisive length (CIL), braincase height (BH), condylo-zygomatic length (CZL), zygomatic length (ZL), length of upper diastema (LD), breadth of the zygomatic plate (BZP), length of nasals (LN), length of rostrum (LR), breadth of rostrum (BR), least interorbital breadth (LIB), orbital length (OL), greatest zygomatic breadth (ZB), length of interparietal (LIP), breadth of interparietal (BIP), breadth across the incisive foramina (BIF), length of incisive foramen (LIF), breadth of rostrum at palate (BRP), breadth of palatal bridge (BPB), length of palatal bridge (LPB), mastoid breadth (MB), breadth across the exoccipital condyles (BOC), bullar breadth (BB), breadth of upper incisor (BI), breadth of M1 (BM1), coronal length of upper molars (CLM), breadth of m1 (Bm1), coronal length of lower molars (CLLM), length of lower diastema (LLD), least condyloid Incisor breadth (LCIB), and mandible height (MH).
For the morphometric analyses, specimens were grouped into five different age classes following Voss (Citation1991). Only specimens considered as adults, i.e., with the third upper molar fully erupted (age classes 3–5), were included in the statistical analyses. Standard descriptive univariate statistics were calculated for all external and craniodental measurements. Principal Component Analyses (PCA) and Discriminant Function Analysis (DFA) were performed in order to assess whether morphometric data are congruent with our molecular results as well as with the qualitative morphological comparisons among the closely related taxa, and with previously published molecular data. In DFA, the specimens were identified a priori as O. bicolor, O. cleberi, and the new species herein described. For these analyses, craniodental measurements were log-transformed, and for PCA the principal components were extracted from the variance-covariance matrix. For DFA, the canonical variates were extracted from the variance-covariance matrix. Statistical analyses were performed using SPSS v. 22.0 for Windows with a significance level of 5%.
Phylogenetic analyses
The genomic DNA was obtained from ethanol-preserved tissues of seven Oecomys specimens recently collected from the Jamari National Forest – Rondônia, one specimen from Juruti – Pará, and 24 specimens of the O. bicolor/O. cleberi species group. The extraction was performed using the saline protocol (Aljanabi & Martinez, Citation1997), with modifications. For molecular analyses, we used the mitochondrial Cytochrome b gene (Cytb) with MVZ05 and MVZ16 primers (Smith & Patton, Citation1993), and the intron 7 of β-fibrinogen nuclear gene (i7-Fgb) with βI7-mammL and βfib-mammU primers (Matocq et al., Citation2007). The amplification and sequencing were carried out following Saldanha et al. (Citation2019). These sequences were aligned to assemble the consensus in the software Geneious v. 7.1.3 (Biomatters, available at http://www.geneious.com; Kearse et al., Citation2012), and deposited in GenBank under the accession numbers OR339662–OR339700 (Supplemental Table S1). In addition, sequences of 101 specimens of the O. bicolor/O. cleberi species group and 41 sequences of other species of Oecomys were also downloaded from GenBank. Subsequently, the sequences were edited and aligned by the ClustalW tool in BioEdit 7.0.5.3 (Hall, Citation1999).
Molecular analyses were performed using a matrix of Cytb with sequences of Euryoryzomys macconnelli, E. nitidus, Handleyomys intectus, “Handleyomys” melanotis, Hylaeamys megacephalus and Oligoryzomys utiaritensis used as outgroup (Supplemental Table S1). We also created a concatenated matrix of Cytb and i7-Fgb sequences with Euryoryzomys nitidus, “Handleyomys” melanotis, Hylaeamys megacephalus and Oligoryzomys utiaritensis as outgroup (Supplemental Table S1). The best model of nucleotide substitution was GTR + I + G for Cytb and TPM2uf + G for i7-Fgb, estimated in JModeltest 2.1.6 (Darriba et al., Citation2012), based on the Akaike Information Criterion (AIC).
Phylogenetic relationships based on Cytb only and concatenated data matrices were analysed separately, using a Bayesian approach (BI) in MrBayes 3.2.6 (Ronquist et al., Citation2012) with two runs, four chains, 30 million generations, sampled every 1000 generations with 25% of burnin. The trees were estimated with 50% majority rule consensus and generate Bayesian Posterior Probabilities (pp). We also used a Maximum likelihood (ML) approach in Garli 2.0 (Zwickl, Citation2006) with bootstrap of 1000 replicates. The analyses were conducted at online platform CIPRES (Miller et al., Citation2010). The trees of bootstrap were summarized with 50% majority rule consensus on SumTrees (Sukumaran & Holder, Citation2010). The genetic distances within and among the clades were calculated in MEGA7 (Kumar et al., Citation2016) by uncorrected p-distance method.
Results
According to Carleton & Musser (Citation2015), 10 nominal taxa are currently recognized as synonym of O. bicolor and among them is O. milleri Allen, Citation1916, which deserves special attention as the species was described based on specimens from Barão de Melgaço, state of Rondônia, Brazil, approximately 400 km from our specimens from Jamari National Forest. As the skull of the holotype of O. milleri (AMNH 37117) is damaged, we were not able to include it in morphometric analyses. However, a detailed morphological examination of the holotype’s skin (Supplemental Fig. S1) by one of the authors, along with the craniodental dimensions provided in the original description, led us to consider that O. milleri cannot be assigned to the new species analysed in the present study, as discussed below. In this sense, the specimens recently collected represent an undescribed species of Oecomys with no name available in the literature, which we formally describe herein. Relevant summaries of morphometric variation and qualitative traits are provided in and , respectively.
Oecomys jamari sp. nov.
Jamari arboreal rice rat
()
Figure 2. Dorsal and ventral views of the skin of Oecomys jamari sp. nov. (UFMT4887, female, holotype). Scale: 50 mm.
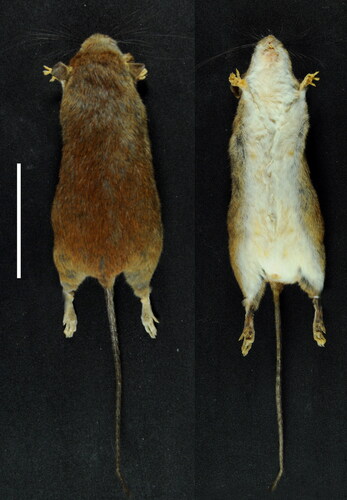
Figure 3. Dorsal, ventral, and lateral cranial views and lateral view of the mandible of Oecomys jamari sp. nov. (UFMT4887, female, holotype): Scale: 10 mm.
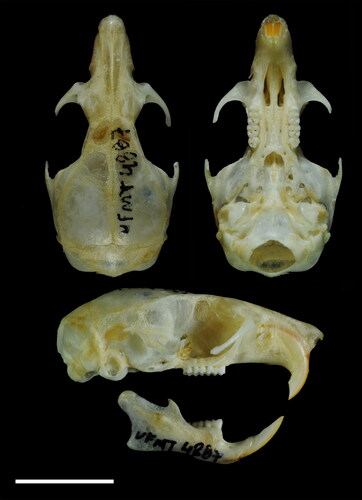
Figure 1. The paratype of Oecomys jamari sp. nov. (UFROM619), an adult female in life. Photo by R.F.B. Mendonça.

Table 1. External, cranial, and dental measurements (in mm) and weight (in grams) of adult specimens of Oecomys jamari sp. nov., O. bicolor lato sensu, and O. cleberi lato sensu.
Table 2. External and craniodental character comparisons among Oecomys jamari sp. nov., O. bicolor lato sensu, and O. cleberi lato sensu.
Holotype
UFMT4887 (ex UFROM378, field number FJP1RM013), adult female, with skin and skull, collected by Ravena F. Braga de Mendonça and Eduardo Sousa on 17 October 2011, with Sherman trap placed in the understorey (). A partial (801 bp) Cytb sequence of the hologenotype is deposited in GenBank (accession number OR339669).
Paratypes
UFROM619 (field number RMFJMDF004), adult female, preserved in alcohol as skin and dry skull, collected in Sherman trap on the ground by Ravena F. Braga de Mendonça on 5 December 2012 at Jamari National Forest, municipality of Itapuã do Oeste, state of Rondônia, Brazil (Coordinates 09°08'12”S, 63°02’07”W). Paragenotype Cytb sequences deposited in GenBank (accession number OR339670). UFROM622 (field number RMFJMDF010), adult female, skin only, collected in Sherman trap in the understorey by Ravena Fernanda Braga de Mendonça on 8 December 2012 at Jamari National Forest, municipality of Itapuã do Oeste, state of Rondônia, Brazil (coordinates 09°09’10”S, 63°00’41”W). Paragenotypes Cytb and i7-Fgb are deposited in GenBank (accession number OR339671 and OR339662, respectively). UFROM624 (field number: RMFJMDF012), adult female, skin only, collected in Sherman trap in the understorey by Ravena F. Braga de Mendonça on 9 December 2012 at Jamari National Forest, municipality of Itapuã do Oeste, state of Rondônia, Brazil (coordinates 09°08’30.4”S, 63°00’41”W). Paragenotypes Cytb and i7-Fgb deposited in GenBank (accession number OR339672 and OR339663, respectively). UFROM631 (field number RMFJMDF019), adult male, skin only, collected in pitfall trap on the ground by Uécson Suendel on 12 December 2012 at Jamari National Forest, municipality of Itapuã do Oeste, state of Rondônia, Brazil (coordinates 09°16’12”S, 62°52’51”W). Paragenotype Cytb deposited in GenBank (accession numbers OR339673 and OR339664). UFROM686 (field number PFJ007), adult female, skin only, collected by Ravena F. Braga de Mendonça on 13 October 2013 at Jamari National Forest, municipality of Itapuã do Oeste, state of Rondônia, Brazil (coordinates 09°16’12”S, 62°52’51”W). Paragenotype Cytb deposited in GenBank (accession number OR339674). UFROM785 (field number RPK-FJAM-01), adult male, skin only, collected by Rafael Kautzmann at Jamari National Forest, municipality of Itapuã do Oeste, state of Rondônia, Brazil (coordinates 09°16’12”S, 62°52’51”W). Paragenotypes Cytb and i7-Fgb deposited in GenBank (accession number OR339675 and OR339665, respectively).
Type locality
Jamari National Forest, municipality of Itapuã do Oeste, state of Rondônia, Brazil (coordinates 09°10’48”S, 62°57’11”W; ).
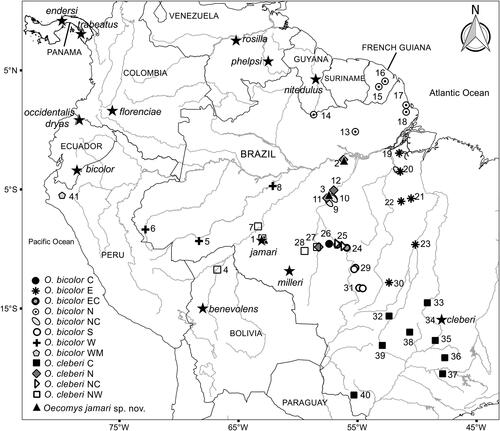
Figure 4. Map of collection localities of specimens of Oecomys jamari sp. nov., and O. bicolor and O. cleberi lineages included in this study. Stars indicate type localities of Oecomys jamari sp. nov. and the nominal taxa associated with O. bicolor according to Carleton & Musser (Citation2015). Lineages names proposed by Suarez-Villota et al. (2018), and this study (C = central; E = eastern; EC = east-central; N = northern; NC = north-central; NW = northwestern; S = southern; W = western; WM = westernmost). Locality data are provided in Supplemental Table S1.
Geographic distribution
The species is known from three localities in the Brazilian states of Pará and Rondônia. In Pará state, it occurs in the municipalities of Juruti and Jacareacanga, south of the Amazon River and west of the Tapajós River. In Rondônia, it has been recorded in the municipality of Itapuã do Oeste, east of the Madeira River. Based on these records, the species seems to be restricted to the area between the Madeira and Tapajós rivers in Brazil and to occur in sympatry with the northwestern clade of O. cleberi ().
Etymology
The specific epithet jamari is to be treated as a noun in apposition. The species name refers to the type locality in the Jamari National Forest, which is a federal protected area pioneer in activities of sustainable use in Brazil and one of the last remaining preservation areas in the state of Rondônia, which is under constant threat due to human activities (Dall’Igna & Maniesi, Citation2022).
Diagnosis
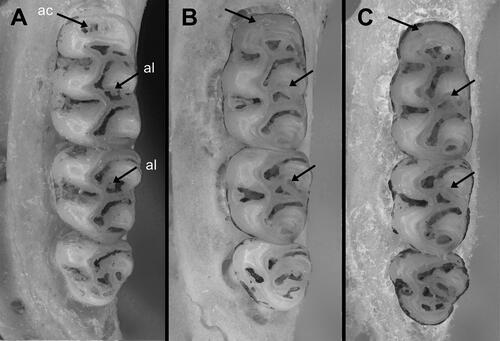
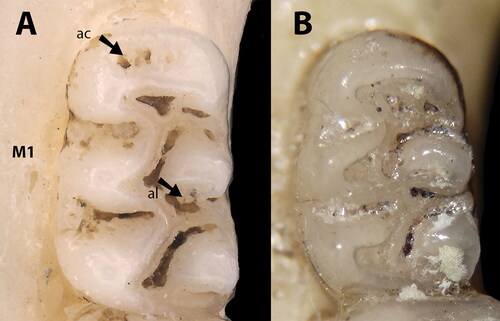
Oecomys jamari sp. nov. is a small species of Oecomys (mean HBL = 96.95 mm, ) that differs from congeneric taxa by the following combination of craniodental traits: Soft and dull orangish brown dorsal fur, composed of short grey-based hairs (5–8 mm); dirty white ventral fur composed of very light grey-based hairs, except for the throat, which is self-white; thin but conspicuous lateral band of grey-based hairs; caudal tuft present (about 5 mm, ). Small skull (ONL = 26.66–27.9 mm, n = 2); U-shaped posterior edge of the nasals, aligned with or surpassing the maxillary–frontal suture; mastoid fully ossified or with a small fenestra that does not contact the exoccipital; subsquamosal fenestra, alisphenoid strut, and sphenopalatine vacuities present; accessory loph in the paracone of the first and second upper molars present; primitive pattern of carotid circulation (pattern 1 sensu Voss Citation1988, ).
Morphological description
Dark brown rostrum with orange tones in orbital region. Abundant mystacial vibrissae extending beyond the distal edge of the ears when folded close to the body. Ears with short dark brown hairs on external face. Dorsal pelage soft and short, with dark grey-based and reddish brown-tipped hairs ranging from 5–8 mm; dark brown-tipped hairs also present on the midline of the dorsum. Lateral pelage of the body lighter than the dorsal pelage. Ventral pelage composed of very light grey-based hairs, giving a dirty white appearance on chest and abdomen, sometimes also on inguinal region; self-white hairs restricted to the throat, sometimes also present on the inguinal region; narrow but conspicuous lateral band of grey-based hairs, extending from the forelimbs to the hind limbs (). Four pairs of mammae, arranged as follows: inguinal, abdominal, postaxial, and pectoral (Voss & Carleton, Citation1993). Manus short and dorsally covered with grey hairs on metatarsals and white hairs on tarsus. Hind foot broad and usually covered with both white and black hairs, with grizzled appearance. Both manus and pes with claws moderate in length (1–2 mm) and with pads as for the genus: manus with two carpal and three plantar pads; hindfeet with a light brown patch on the metatarsals upper surface, central digits (II, III, and IV) longer than the outer digits (I and V), digit V semi-opposable, six large plantar pads (fleshy and with interdigitals 1–4 set close together) with smooth skin between. Dark brown tail, unicoloured from basis to tip, with scales in a circular arrangement; three hairs associated with each caudal scale, the central one thicker and longer, exceeding two scales in length; small dark brown caudal tuft present, about 5 mm in length.
Skull delicate, rostrum slightly longer and narrow, with a relatively long nasal (; ). Anterior margins of nasal bones rounded and posterior margin U-shaped, with posterior end aligned or extending beyond the maxillary-frontal suture. Frontal bones converging anteriorly, with supraorbital ridges developed and restricted to the frontals. Temporal ridges absent. Zygomatic notch distinctly shallow (notch not visible dorsally). Lacrimal in contact with maxillary and frontal bones. Rounded cranial vault. Interparietal wider than the posterior border of frontals, but not contacting the squamosal. Zygomatic plate narrow, with flat anterior edge aligned with the maxillary root of the zygomatic arch, and posterior margin positioned anteriorly to M1. Maxillary and squamosal processes of zygomatic arch not overlapping. Parietal expanded below the lateral edges of the braincase. Subsquamosal fenestra present, but smaller than the postglenoid foramen. Hamular process narrow. Lambdoidal crests poorly developed. Mastoid completely ossified or with a small fenestra that does not contact the exoccipital. Incisive foramina medium-sized, with convex lateral margins, wider in the median portion, acute anteriorly and rounded posteriorly; posterior margin of incisive foramina aligned with the anterior margin of M1. Palate of intermediate length (sensu Weksler Citation2006); posterolateral palatal pits small and simple. Anterior portion of mesopterygoid fossa U-shaped, reaching the maxillary margin, but not the molars. Small sphenopalatine vacuities present. Parapterygoid fossa slightly excavated. Alisphenoid strut present, separating the buccinator-masticatory foramen and the accessory oval foramen. Posterior opening of the alisphenoid canal developed. Carotid circulation pattern 1 (Voss Citation1988). Tegmen tympani not contacting the squamosal. Small ectotympanic bulla; periotic canal contributing to the wall of carotid canal; anterior bullae process largely overlapping the parapterygoid plate.
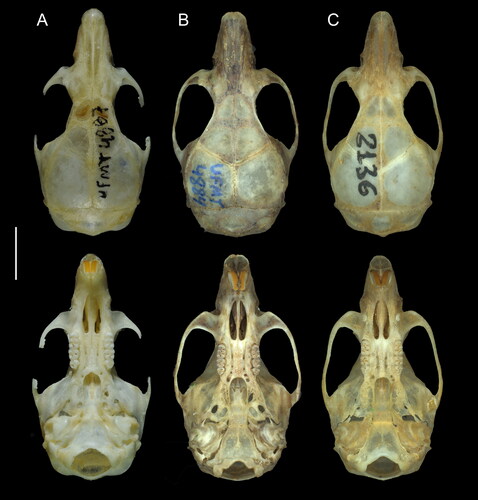
Figure 5. Dorsal and ventral views of the skull of A: Oecomys jamari sp. nov. (UFMT 4887, holotype); B: Oecomys bicolor (southern lineage; UFMT 4884) and C: Oecomys cleberi (north-central lineage; MSF2136). Scale: 5 mm.
Mandible with capsular process of lower incisor moderately developed, as a slight rounded elevation (). Masseteric ridges convergent anteriorly, and fused in the anterior portion below m1. Mental foramen laterally placed, and ahead of the confluence between the upper and lower ridges of the masseteric crests. Condyloid process robust, ending posteriorly to the angular process, which is very short; coronoid process short and posterodorsally oriented, below the level of the upper margin of the condyloid process. Sigmoid and angular notches shallow.
Upper incisors opisthodont, with yellow-orange anterior enamel surfaces. Molars pentalophodont; upper molar rows arranged in parallel. Upper first molar (M1): anterior border of the procingulum with a lower relief anteriorly to the anterocone (anterior cingulum, see Pires et al., Citation2016; ); anterocone not divided by anteromedian flexus; procingulum with a central fossete; anteroflexus present; anteroloph present with parastyle loosely linked to the paracone; long paraflexus; paracone with an accessory loph and paralophule, the latter connected to the mesoloph; mesoloph with mesostyle extending to the metacone; posteroloph and posteroflexus present; hypocone connected to the mesoloph and protocone by a median mure; protocone not directly connected to the paracone, the latter connected punctually to the median mure; hypoflexus present; enteroloph absent; enterostyle present; protoflexus absent; protostyle present; labial accessory root present in only one specimen (UFMT4887, holotype). M2: anteroloph present and connected to the protocone; protoflexus present; paracone with an accessory loph; mesoflexus present; mesoloph present and connected to the paracone and metacone by mesostyle; posteroloph present and connected to the metacone; median mure connected to the hypocone, protocone, paracone, and mesoloph; hypoflexus present; enterostyle present. M3: mesoloph and metaflexus present; metacone present; posteroloph and posteroflexus present; hypocone extremely reduced; protocone present. First lower molar (m1): anteromedian flexid absent; anteroconid with a well-developed anterofossettid; posterostylid present and connected to the anteroconid, forming a fossettid (protoflexid); anterolabial cingulum present, but not connected to the protoconid; ectolophid present, connected to the hypoconid; posterolophid present, connected to the entoconid; posteroflexid present (fossette); mesolophid present, connecting entoconid with metaconid; mesoflexid present (as a fossettid); anterolophid present, connected to the metaconid and forming a fossettid (anteroflexid); metaflexid present; anterior murid connecting anteroconid, metaconid, and protoconid; median murid connecting protoconid, ectolophid, hypoconid, entoconid, and mesolophid; accessory root absent. m2: protoflexid present; ectolophid absent; ectostylid connected to the protoconid and hypoconid; posterolophid present, connected to the entoconid; median murid connecting protoconid, hypoconid, entoconid, and mesolophid; mesolophid with a mesostylid, connecting metaconid with entoconid. m3: protoflexid present; ectolophid absent; hypoconid present; posterolophid present; posteroflexid present (as a fossettid); entoconid very reduced ().
Comparisons
On average, Oecomys jamari sp. nov. is larger than O. bicolor and O. cleberi in several external and craniodental measurements (, ). Externally, Oecomys jamari sp. nov. differs from the other two species by the dark reddish brown dorsal pelage versus fair reddish brown to orangish brown pelage; ventral fur made up of very light grey-based hairs, giving an appearance of dirty white in the region of chest and abdomen, occasionally extending until the inguinal region, versus completely white or cream; narrow but conspicuous lateral band of grey-based hairs, versus grey-based lateral band absent in O. bicolor and usually absent in O. cleberi.
Craniodentally, Oecomys jamari sp. nov. differs from O. bicolor and O. cleberi by its larger and wider cranial vault; mastoid fully ossified or with a small fenestra that does not contact the exoccipital, versus a usually large fenestra in contact with the exoccipital; small subsquamosal fenestra, versus a large subsquamosal fenestra; alisphenoid strut present, versus absent in O. bicolor and usually absent in O. cleberi (present in UFMT1372 and MSF2130); small sphenopalatine vacuities present, versus usually ossified in O. cleberi and variable in O. bicolor; accessory loph in the paracone of M1 and M2 present, versus absent in O. bicolor and usually absent in O. cleberi (present in UFMT1363, UFMT1372, MSF2341; ). Oecomys jamari sp. nov. can also be discriminated from O. cleberi by exhibiting posterior margins of the nasals aligned or surpassing the maxillary-frontal suture, versus aligned but not surpassing the maxillary-frontal suture in the latter species ().
Considering that Oecomys jamari sp. nov. is possibly restricted to the Madeira-Tapajós interfluve, a region that encompasses the type locality of O. milleri Allen, Citation1916 from Barão de Melgaço, state of Rondônia (), comparisons between these two forms deserve attention. The skin of the holotype of O. milleri (AMNH37117) is dirty in the venter, appearing to be greyish. However, a closer look allows one to see self-white fur in part of the abdomen and chin (Supplemental Fig. S1). Therefore, the holotype of O. milleri differs from Oecomys jamari sp. nov. by exhibiting self-white venter, with no conspicuous lateral bands of grey-based hairs, versus a dirty white venter composed of very light grey-based hairs on chest and abdomen, sometimes also on inguinal region (never on the throat), and conspicuous narrow lateral bands of grey-based hairs. Cranially, the holotype of O. milleri exhibits posterior edge of the nasals aligned with the maxillary-frontal suture, versus aligned with or surpassing the maxillary-frontal suture in Oecomys jamari sp. nov.; and a large fenestra on the mastoid, versus a small or absent fenestra on the mastoid in Oecomys jamari sp. nov. The M1 accessory loph in the paracone of the first and second upper molars is present in Oecomys jamari sp. nov. and absent in O. milleri (). Additionally, several craniodental dimensions of the holotype of O. milleri provided by Allen (Citation1916) in the original description do not overlap with those exhibited by Oecomys jamari sp. nov. In fact, the occipitonasal length (ONL), the length of upper diastema (LD), the greatest zygomatic breadth (ZB), and the coronal length of upper molars (CLM) are smaller in the holotype of O. milleri, while the breadth across the incisive foramina (BIF) and the length of incisive foramen (LIF) are larger when compared with Oecomys jamari sp. nov. ().
Natural history
At the type locality (Jamari National Forest), seven specimens were captured in submontane open ombrophilous forest, with palm trees and vines (Supplemental Fig. S2). Two refers to males (one juvenile and one adult), four are lactating females collected in December, and one is a female with offspring collected in October. Captures took place in different strata, either through pitfall-traps, Sherman or cage traps, both installed on the ground and the understorey (approximately 1.5 metres high) and baited with pieces of pineapple soaked in cotton with cod liver oil. With a collection effort of 27,564 trap-nights, the capture success of these species for the area was 0.02%. Other small non-volant mammal species captured in the same sites of Oecomys jamari sp. nov. are the didelphid marsupials Didelphis marsupialis, D. albiventris, Marmosa sp., M. constantiae, Marmosops sp., Marmosops noctivagus, Metachirus myosuros, Monodelphis emiliae, M. glirina, M. saci, the cricetid rodents O. cleberi, Rhipidomys sp., the echimyid rodents Proechimys sp., Mesomys cf. hispidus, and the exotic murid rodent Rattus rattus.
Phylogenetic analyses
The phylogenetic analyses of Bayesian Inference (BI) and Maximum Likelihood (ML) with 174 Cytb sequences of Oecomys (matrix size = 801 bp) recovered all lineages of the O. bicolor/O. cleberi species group (sensu Suárez-Villota et al., Citation2018) in a single clade (posterior probability [pp] = 1; bootstrap = 60%; ). Most lineages could be associated with those recognized by Suárez-Villota et al. (Citation2018), whose names are adopted here. However, three new lineages were recovered within the species group, two of which (herein named northern and north-central O. cleberi; ) are nested in a monophyletic group with high support (pp = 0.99) that also includes the two O. cleberi lineages (pp = 1; bootstrap 95%) and the northern lineage of O. bicolor. The third new lineage (herein named north-central O. bicolor; ) appeared as sister to the southern O. bicolor lineage with high branch support (pp = 0.99; bootstrap = 73%). Among the clades recovered, the north-central, northwestern and central O. cleberi had very low branch support. Finally, we were not able to associate several specimens to any of the lineages in the species group. Two of these specimens (MPEG 40734-40735) are closely related to the north-central, southern and westernmost O. bicolor, and the remaining appeared in basal positions within the species group, with very low branch supports ().
Figure 8. Bayesian Inference topology (BI) based on the mitochondrial Cytochrome b gene of the genus Oecomys. Numbers above the branches indicate Bayesian posterior probabilities number and below the branches indicate maximum likelihood bootstrap values above 50%. Bold numbers indicate the samples of Oecomys jamari sp. nov. Asterisks represent samples analysed molecularly and morphologically. We follow the lineage names proposed by Suárez-Villota et al. (Citation2018), and new lineages named in this study are indicated in bold (C = central; E = eastern; EC = east-central; N = northern; NC = north-central; NW = northwestern; S = southern; W = western; WM = westernmost). Sample data are provided in Supplemental Table S1.
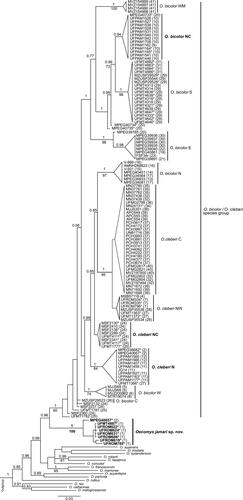
Specimens of Oecomys jamari sp. nov. were recovered in a highly supported clade (pp = 1; bootstrap = 100%) as sister to the O. bicolor and O. cleberi species group, with high support in the BI (pp = 0.96; bootstrap = 49%, ). The Oecomys jamari sp. nov. had 3.78% of genetic divergence in relation to the O. bicolor and O. cleberi species group, and from 5.58% up to 9.62% in relation to other congeners. Genetic distances were 0.91% within Oecomys jamari sp. nov. and 2.15% within the O. bicolor and O. cleberi species group ().
Table 3. Uncorrected pairwise distances of mitochondrial gene Cytochrome b within and among Oecomys species.
The topology of concatenated data (Cytb + i7-Fgb; ) with 70 Oecomys sequences (samples were included only when data were available for the two markers) and a matrix size of 1538 pb (Cytb = 801 pb + i7-Fgb = 737 pb) also showed the specimens of Oecomys jamari sp. nov. as a clade with high support (pp = 1; bootstrap 100%), in a sister relation to the O. bicolor and O. cleberi species group with low support (pp = 0.76). Lineages northern and north-central O. cleberi, and north-central O. bicolor were also recovered with high branch supports (pp = 1, 0.99, and 1, respectively; bootstrap = 85% for northern O. cleberi), and specimens basally positioned in the Cytb topology appeared as a new lineage (east-central O. bicolor; ) in a sister relationship (p = 0.99) to the lineages central and western O. bicolor, northern, north-central, north-western, and central O. cleberi (). Among the clades recovered, the lineages east-central O. bicolor and north-western O. cleberi had very low branch support.
Figure 9. Bayesian Inference topology (BI) based on the mitochondrial Cytochrome b gene and nuclear intron 7 β-fibrinogen concatenated of species of the genus Oecomys. Numbers above the branches indicate Bayesian posterior probabilities numbers and below the branches indicate maximum likelihood bootstrap values, above 50%. Bold numbers indicate the samples of Oecomys jamari sp. nov. Asterisks represent samples analysed molecularly and morphologically. We follow the lineage names proposed by Suárez-Villota et al. (Citation2018), and new lineages named in this study are indicated in bold (C = central; E = eastern; EC = east-central; N = northern; NC = north-central; NW = northwestern; S = southern; W = western; WM = westernmost). Sample data are provided in Supplemental Table S1.
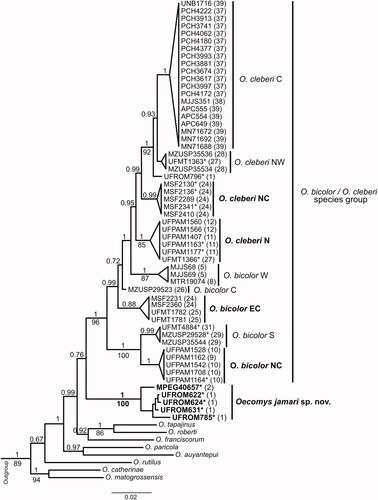
Similarly to the Cytb topology, clades northern, north-central O. cleberi were closely related to the clades western O. bicolor, northwestern and central O. cleberi, forming a highly supported (pp = 1) monophyletic group, as well as the clades north-central and southern O. bicolor (pp = 1; bootstrap = 100%; ). Sequences of i7-Fgb were not available for the northern, eastern, and westernmost O. bicolor clades.
Morphometric analysis
Based on our concatenated topology, we considered specimens of the lineages northern, north-central, northwestern, and central O. cleberi as O. cleberi lato sensu in the morphometric analyses. And for analytical purposes, we considered as O. bicolor lato sensu the lineages southern and north-central O. bicolor, plus the specimens MPEG 40734-40735, which forms a monophyletic clade in our Cytb topology. In the PCA for Oecomys jamari sp. nov. (n = 4), O. bicolor lato sensu (n = 24), and O. cleberi lato sensu (n = 17), the first two principal components (PC1 and PC2) together accounted for 64.56% of total variation. Based on this analysis, the Oecomys jamari sp. nov. partially overlaps with O. bicolor lato sensu and O. cleberi lato sensu (; ). Along the PC1 axis, Oecomys jamari sp. nov. overlaps only with the larger specimens of O. bicolor lato sensu and O. cleberi lato sensu, revealing a slightly larger size of the new species. With regard to the PC2, Oecomys jamari sp. nov. almost entirely separates from O. bicolor lato sensu, but not from O. cleberi lato sensu, revealing a more similar shape of the skull with the latter. Among the most important variables are the condyle-incisive length (CIL), the condylo-zygomatic length (CZL), and the greatest zygomatic breadth (ZB) for PC1, all related to skull size, and the breadth of palatal bridge (BPB), the least condyloid-incisor breadth (LCIB), and the breadth of incisor (BI) for PC2, all associated with skull shape ().
Figure 10. Scatterplot of the first (PC1) and second (PC2) principal component analysis (PCA) of 31 log-transformed craniodental measurements of Oecomys jamari sp. nov., O. bicolor lato sensu, and O. cleberi lato sensu analysed in this study.
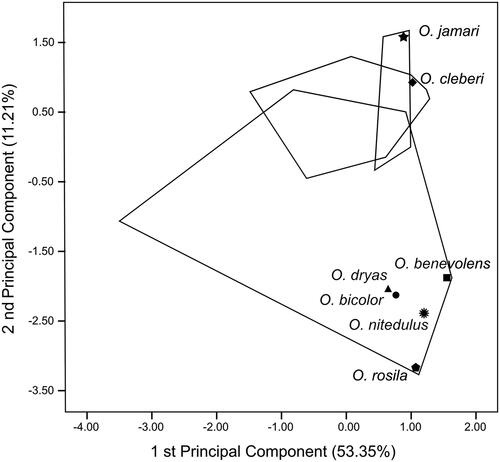
Table 4. Coefficient results of the first three components of Principal Component (PC) Analysis of adult specimens of Oecomys jamari sp. nov. (n = 4), O. bicolor lato sensu (n = 24), and O. cleberi lato sensu (n = 17) based on 31 craniodental variables.
In relation to the DFA for Oecomys jamari sp. nov. (n = 4), O. bicolor lato sensu (n = 24), and O. cleberi lato sensu (n = 17), the first discriminant function (DF1) accounted for 79.2% of the total variation, while DF2 accounted for 20.8% of the variation (; ). Based on this analysis, Oecomys jamari sp. nov. completely separates from O. bicolor lato sensu and O. cleberi lato sensu, and O. bicolor lato sensu separates from O. cleberi lato sensu in both axes (). Among the most important measurements to discriminate the species are the condylo-zygomatic length (CZL), the orbital length (OL), and the condylo-incisive length (CIL) in DF1, plus the condylo-incisive length (CIL), the condylo-zygomatic length (CZL), and the least condyloid-incisor breadth (LCIB) in DF2. Furthermore, the DFA indicated 100% correct classification of specimens of Oecomys jamari sp. nov., O. bicolor lato sensu, and O. cleberi lato sensu (Supplemental Table S2).
Figure 11. Scatterplot of the first (DF1) and second (DF2) canonical variates of the discriminant function analysis (DFA) of 31 log-transformed craniodental measurements of Oecomys jamari sp. nov., O. bicolor lato sensu, and O. cleberi lato sensu analysed in this study.
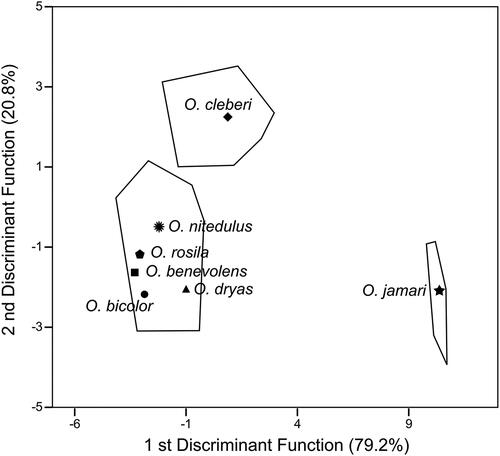
Table 5. Coefficient of the first two functions of the Discriminant Function Analysis of adult specimens of Oecomys jamari sp. nov. (n = 4), O. bicolor lato sensu (n = 24), and O. cleberi lato sensu (n = 17) based on 31 craniodental variables.
For both morphometric analyses, all the holotypes of nominal taxa under synonym of O. bicolor are included in the morphospace delimited by this species, not overlapping with O. cleberi and Oecomys jamari sp. nov. morphospaces.
The results obtained in our molecular-based tree and in the DFA analyses indicate that Oecomys jamari sp. nov. represents a separate species from O. bicolor lato sensu and O. cleberi lato sensu, sister to the clade formed by the latter two species. Moreover, external morphology, craniodental analyses and our morphometric results presented above show the existence of an exclusive set of traits that support the description of this new species.
Discussion
Our topologies of Cytb + i7-Fgb recovered Oecomys jamari sp. nov. as a sister species to the clade comprising all the lineages of O. bicolor and O. cleberi. The genetic distance between Oecomys jamari sp. nov. and the O. bicolor and O. cleberi group (3.78%) is lower than values reported for sister species within the genus, such as 4.5% to 6.6% between representative lineages of O. franciscorum and O. mamorae (Pardiñas et al., Citation2016) and 5.8% between O. tapajinus and O. roberti (Rocha et al., Citation2018).
Similarly to previous studies (Saldanha et al., Citation2019; Suárez-Villota et al., Citation2018), the O. bicolor and O. cleberi group was recovered as monophyletic, the central and northwestern O. cleberi lineages appeared as sister clades, and lineages associated to O. bicolor by Suárez-Villota et al. (Citation2018) did not form a monophyletic group in both Cytb and concatenated data topologies. A close relation between the northern O. bicolor clade and the two O. cleberi clades based on Cytb sequences was also reported by Suárez-Villota et al. (Citation2018) and Saldanha et al. (Citation2019), as well as a close relation among the central and western O. bicolor clades and the central and northwestern O. cleberi clades based on concatenated data. This led Saldanha et al. (Citation2019: ) to include the central and western O. bicolor clades in their concept of O. cleberi.
By adding new samples of specimens of the O. bicolor and O. cleberi species group, our phylogenetic analyses recovered four new lineages, three of which (northern, north-central O. cleberi, and east-central O. bicolor; and ) are nested in a monophyletic group that includes the two O. cleberi lineages plus either the northern lineage of O. bicolor in the Cytb topology or the central and western lineages of O. bicolor in the concatenated topology. Another new lineage (north-central O. bicolor; and ) appeared as sister to the southern O. bicolor clade in both topologies.
Among the clades recovered in this study, some had very low branch support, as follows: north-central and central O. cleberi in the Cytb topology; east-central O. bicolor in the concatenated topology; and northwestern O. cleberi in both topologies. Similar results were recorded for the northwestern O. cleberi clade by Suárez-Villota et al. (Citation2018). By contrast, the western O. bicolor lineage had high branch support in our concatenated topology, while presented very low support in the corresponding topology of Suárez-Villota et al. (Citation2018).
Altogether, the results of our phylogenetic analyses corroborate that both O. bicolor and O. cleberi represent species complexes, whose species richness and delimitations can only be appropriately defined based on additional molecular and morphological studies. As previously mentioned by Suárez-Villota et al. (Citation2018), and considering the Cytb and concatenated topologies herein provided, the name O. cleberi Locks, 1981 applies to the central O. cleberi lineage. Considering the Cytb topology and the geographic proximity to the type locality of O. bicolor (Tomes, 1860; see ), this may be the name to be associated with specimens of the westernmost O. bicolor clade.
The specimens of Oecomys jamari sp. nov. can be distinguished from O. bicolor lato sensu (southern and north-central O. bicolor lineages, plus the specimens MPEG 40734-40735, which forms a monophyletic clade in the Cytb topology) and O. cleberi lato sensu (northern, north-central, northwestern, and central O. cleberi lineages) by craniodental dimensions and external and craniodental morphology. The DFA showed that Oecomys jamari sp. nov. separates completely apart from those taxa in the morphospace and indicated 100% correct classification of Oecomys jamari sp. nov., O. bicolor sensu lato, and O. cleberi sensu lato. Morphologically, Oecomys jamari sp. nov. can be differentiated from its sister taxa by a unique combination of characters ().
The species herein described contributes to the knowledge of the taxonomy of Oecomys, increasing to 20 species known for the genus. This new species occurs in one of the least known area of endemism located in the Amazon basin, the Rondônia center of endemism, delimited by the Amazon rivers to the north, Tapajós to the east, and Madeira to the west (Cracraft, Citation1985).
Regarding sympatry, Oecomys jamari sp. nov. and the northwestern lineage of O. cleberi were both recorded in the municipality of Juruti, state of Pará, and in the Jamari National Forest, Rondônia, reinforcing our hypothesis that these are distinct species. Suárez-Villota et al. (Citation2018) pointed out that sympatry is more common between Oecomys species of distinct body sizes. Nonetheless, the occurrence of sympatry between two species of medium body size was recently reported by Rocha et al. (Citation2014, Citation2018) and Saldanha et al. (Citation2019), and between two small-sized species by Patton et al. (2000), besides the present study. Based on the present study and Suárez-Villota et al. (Citation2018), Rocha et al. (Citation2018), Saldanha et al. (Citation2019), and Saldanha & Rossi (Citation2021), we estimate that the new species here described may occur in sympatry also with seven other congeners, such as: the small O. cleberi northern lineage, the medium-sized O. paricola, O. roberti, O. tapajinus, and O. trinitatis, and the large-sized O. catherinae westernmost lineage and O. matogrossensis, which have their distribution overlapping in some degree.
The description of a new species of Oecomys clearly demonstrates the need for continuous inventories and taxonomic studies in the Brazilian Amazonia, even in areas that have been widely deforested, as in the state of Rondônia. It is important to note that the species possibly might be endangered in some degree mainly by the loss of their habitat caused by the expansion of the agricultural frontier in the so-called ‘arc of deforestation’ of Amazonia, which includes the entire state of Rondônia (Dall’Igna & Maniesi, Citation2022) and by the existence of two federal roads that cross the Rondônia centre of endemism, which may soon be paved: BR-230 (Transamazonian Highway) and BR-319 (Manaus-Porto Velho Highway). According to Barber et al. (Citation2014), around 95% of all deforestation in the Brazilian Amazonia is located in a strip of 5.5 km along roads and 1 km along rivers, and according to Ferrante et al. (Citation2021), paving roads is the main source of access to unprotected land in the region. Another threat to the species is the weakening of the Brazilian environmental agencies and forest code, which generates expectations of impunity that encourages deforestation acts (Ferrante & Fearnside, Citation2019), even in well-established protected areas, as observed in the state of Rondônia since 2020 (Messias, M.R., pers. obs.).
Finally, the creation and maintenance of additional protected areas in the Tapajós-Madeira interfluve is one of the measures needed for Oecomys jamari sp. nov. to be safeguarded, along with many other vertebrates that are endemic to this portion of Amazonia, such as Rondon’s marmoset (Mico rondoni) and Prince Bernhard’s Titi monkey (Plecturocebus bernhardi).
Associate Editor: Dr Susan Tsang
Supplemental Material - Fig. S1
Download JPEG Image (62.6 KB)Supplemental Material - Fig. S2
Download TIFF Image (7.9 MB)Supplemental Material - Tables
Download MS Word (36.1 KB)Acknowledgements
We thank Tamara Flores for providing extraction and sequences; Manoel Santos-Filho (Unemat), João Alves de Oliveira (MNRJ), and Alexandra Maria Ramos Bezerra and José Silva Junior (MPEG) and Robert S. Voss (AMNH) for allowing us to access the mammal collections; Alexandra Maria Ramos Bezerra and Cibele Rodrigues Bonvicino for kindly providing photographs of the holotype of O. milleri, which were obtained by the projects # 402176/2012-0 and 372459/2013-7 of the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). We thank the Laboratório de Scarabaeoidologia at Universidade Federal de Mato Grosso (Cuiabá, Mato Grosso, Brazil) which provided access to the stereomicroscope that we used for and , obtained through sub-project EECBio of UFMT/FINEP (agreement FINEP # 01.12.0359.00). We thank Guilherme Garbino for reading a previous version of the manuscript; JS had a fellowship provided by Programa de Capacitação em Taxonomia (PROTAX II – Edital n° 001/2015) and by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplemental material
Supplemental material for this article can be accessed here: https://doi.org/10.1080/14772000.2023.2259037.
Additional information
Funding
References
- Aljanabi, S. M., & Martinez, I. (1997). Universal and rapid salt–extraction of high quality genomic DNA for PCR–based techniques. Nucleic Acids Research, 25, 4692–4693. https://doi.org/10.1093/nar/25.22.4692
- Allen, J. A. (1916). New mammals collected on the Roosevelt Brazilian expedition. Bulletin of American Museum of Natural History, 35, 523–530.
- Barber, C. P., Cochrane, M. A., Souza, C. M., & Laurance, W. F. (2014). Roads, deforestation, and the mitigating effect of protected areas in the Amazon. Biological Conservation, 177, 203–209. https://doi.org/10.1016/j.biocon.2014.07.004
- Brandão, M. V., Garbino, G. S. T., Rezende, G. C., Tenório, S. F., & Reis, S. F. (2022). Taxonomic and natural history notes on Oecomys cleberi (Rodentia: Cricetidae) and first records in the Atlantic Forest, Brazil. Studies on Neotropical Fauna and Environment, 1–15. https://doi.org/10.1080/01650521.2022.2046962
- Carleton, M. D., & Musser, G. G. (2015). Genus Oecomys Thomas, 1906. In J. Patton, U. F. J. Pardiñas & G. D’Elía (Eds.), Mammals of South America, vol.2, rodents (pp. 393–417). University of Chicago Press.
- Carleton, M. D., Emmons, L. H., & Musser, G. G. (2009). A new species of the rodent genus Oecomys (Cricetidae: Sigmodontinae: Oryzomyini) from eastern Bolivia, with emended definitions of O. concolor and O. mamorae (Thomas). American Museum Novitates, 3661, 1–32. https://doi.org/10.1206/612.1
- Cracraft, J. (1985). Historical biogeography and patterns of differentiation within the South American avifauna: Areas of endemism. Ornithological Monographs, 36, 49–84. https://doi.org/10.2307/40168278
- Dall’Igna, F., & Maniesi, V. (2022). Spatial and temporary dynamics of heat spots in the amazon ecological corridor conservation unit: The case of intense anthropic pressure in the National Forest of Jamari/RO. Research, Society and Development, 11, e42011629271. https://doi.org/10.33448/rsd-v11i6.29271
- Darriba, D., Taboada, G. L., Doallo, R., & Posada, D. (2012). jModelTest 2: More models, new heuristics and parallel computing. Nature Methods, 9, 772–772. https://doi.org/10.1038/nmeth.2109
- Ferrante, L., & Fearnside, P. M. (2019). Brazil’s new president and ‘ruralists’ threaten Amazonia’s environment, traditional peoples and the global climate. Environmental Conservation, 46, 261–263. https://doi.org/10.1017/S0376892919000213
- Ferrante, L., Andrade, M. B. T., Leite, L., Junior, C. A. S., Lima, M., Junior, G. C., Neto, E. C. S., Campolina, D., Carolino, K., Diele-Viegas, L. M., Pereira, E. J. A. L., & Fearnside, P. M. (2021). Brazil highway BR-319: The road to the collapse of the Amazon and the violation of indigenous rights. Journal of the Geographical Society of Berlin, 152, 1–6. https://doi.org/10.12854/erde-2021-552
- Hall, T. A. (1999). BioEdit: A user–friendly biological sequence alignment editor and analisys program for Windows 85/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
- Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Meintjes, P., & Drummond, A. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England), 28, 1647–1649. https://doi.org/10.1093/bioinformatics/bts199
- Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. https://doi.org/10.1093/molbev/msw054
- Matocq, M. D., Shurtliff, Q. R., & Feldman, C. R. (2007). Phylogenetics of the woodrat genus Neotoma (Rodentia: Muridae): Implications for the evolution of phenotypic variation in male external genitalia. Molecular Phylogenetics and Evolution, 42, 637–652. https://doi.org/10.1016/j.ympev.2006.08.011
- Miller, M. A., Pfeiffer, W., Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), IEEE, New Orleans, LA, 1–8. https://doi.org/10.1109/GCE.2010.5676129
- Musser, G. G., & Carleton, M. D. (2005). Superfamily Muroidea. In D. E. Wilson & D. M. Reeder (Eds.), Mammals species of the world: A taxonomic geographic reference (3rd ed. Vol. 2, pp. 894–1531). The Johns Hopkins University Press.
- Pardiñas, U. F. J., Teta, P., Salazar-Bravo, J., Myers, P., & Galliari, C. A. (2016). A new species of arboreal rat, genus Oecomys (Rodentia, Cricetidae) from Chaco. Journal of Mammalogy, 97, 1177–1196. https://doi.org/10.1093/jmammal/gyw070
- Pardiñas, U., Ruelas, D., Brito, J., Bradley, L., Bradley, R., Garza, N. O., Kryštufek, B., Cook, J., Soto, E. C., Salazar–Bravo, J., Shenbrot, G., Chiquito, E., Percequillo, A., Prado, J., Haslauer, R., Patton, J., & León–Paniagua, L. (2017). Species Accounts of Cricetidae. In D. E. Wilson, T. E. Lacher & R. A. Mittermeier (Eds.), Handbook of mammals of the world, Vol.7, Rodents II (pp. 281–535). Lynx Edicions.
- Pires, C., Gudinho, F., & Weksler, M. (2016). Morfologia dentária de gêneros de Sigmodontinae (Rodentia: Cricetidae) com ocorrência no Cerrado brasileiro. Boletim da Sociedade Brasileira de Mastozoologia, 75, 1–32.
- Reig, O. A. (1977). A proposed unified nomenclature for the enamelled components of the molar teeth of the Cricetidae (Rodentia). Journal of Zoology, 181, 227–241. https://doi.org/10.1111/j.1469-7998.1977.tb03238.x
- Rocha, R. G., Duda, R., Flores, T., Rossi, R., Sampaio, I., Mendes-Oliveira, A. C., Leite, Y. L. R., & Costa, L. P. (2018). Cryptic diversity in the Oecomys roberti complex: Revalidation of Oecomys tapajinus (Rodentia, Cricetidae). Journal of Mammalogy, 99, 174–186. https://doi.org/10.1093/jmammal/gyx149
- Rocha, R. G., Ferreira, E., Fonseca, C., Justino, J., Leite, Y. L. R., & Costa, L. P. (2014). Seasonal flooding regime and ecological traits influence genetic structure of two small rodents. Ecology and Evolution, 4, 4598–4608. https://doi.org/10.1002/ece3.1336
- Rocha, R. G., Fonseca, C., Zhou, Z., Leite, Y. L. R., & Costa, L. P. (2012). Taxonomic and conservation status of the elusive Oecomys cleberi (Rodentia, Sigmodontinae) from central Brazil. Mammalian Biology, 77, 414–419. https://doi.org/10.1016/j.mambio.2012.02.004
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M. A., & Huelsenbeck, J. P. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542. https://doi.org/10.1093/sysbio/sys029
- Rosa, C. C., Flores, T., Pieczarka, J. C., Rossi, R. V., Sampaio, M. I. C., Rissino, J. D., Amaral, P. J. S., & Nagamachi, C. Y. (2012). Genetic and morphological variability in South American rodent Oecomys (Sigmodontinae, Rodentia): Evidence for a complex of species. Journal of Genetics, 91, 265–277. https://doi.org/10.1007/s12041-012-0182-2
- Saldanha, J., & Rossi, R. V. (2021). Integrative analysis supports a new species of the Oecomys catherinae complex (Rodentia, Cricetidae) from Amazonia. Journal of Mammalogy, 102, 69–89. https://doi.org/10.1093/jmammal/gyaa145
- Saldanha, J., Ferreira, D. C., da Silva, V. F., Santos–Filho, M., Mendes–Oliveira, A. C., & Rossi, R. V. (2019). Genetic diversity of Oecomys (Rodentia, Sigmodontinae) from the Tapajós River basin and the role of rivers as barriers for the genus in the region. Mammalian Biology, 97, 41–49. https://doi.org/10.1016/j.mambio.2019.04.009
- Smith, M. F., & Patton, J. L. (1993). The diversification of South American murid rodents: Evidence from mitochondrial DNA sequence data for Akodontine tribe. Biological Journal of the Linnean Society, 50, 149–177. https://doi.org/10.1111/j.1095-8312.1993.tb00924.x
- Suárez-Villota, E. Y., Carmignotto, A. P., Brandão, M. V., Percequillo, A. R., & Silva, M. J. J. (2018). Systematics of the genus Oecomys (Sigmodontinae: Oryzomyini): Molecular phylogenetic, cytogenetic and morphological approaches reveal cryptic species. Zoological Journal of the Linnean Society, 184, 182–210. https://doi.org/10.1093/zoolinnean/zlx095
- Sukumaran, J., & Holder, M. T. (2010). DendroPy: A Python library for phylogenetic computing. Bioinformatics (Oxford, England), 26, 1569–1571. https://doi.org/10.1093/bioinformatics/btq228
- Voss, R. S., & Carleton, M. D. (1993). A new genus for Hesperomys molitor Winge and Holochilus magnus Hershkovitz (Mammalia, Muridae) with an analysis of its phylogenetic relationships. American Museum of Natural History, 3085, 1–39. http://hdl.handle.net/2246/4982 (Accessed 16 Jul. 2021)
- Voss, R. S. (1988). Systematics and ecology of ichthyomyine rodents (Muroidea): Patterns of morphological evolution in a small adaptative radiation. Bulletin of the American Museum of Natural History, 188, 259–493. http://hdl.handle.net/2246/927 (Accessed 16 Jul. 2021)
- Voss, R. S. (1991). An introduction to the neotropical muroid rodent genus Zygodontomys. Bulletin of the American Museum of Natural History, 210, 1–113.
- Weksler, M. (2006). Phylogenetic relationships of Oryzomine rodents (Muroidea: Sigmodontinae): Separate and combined analyses of morphological and molecular data. Bulletin of the American Museum of Natural History, 296, 1–149. https://doi.org/10.1206/0003-0090(2006)296[0001:PROORM]2.0.CO;22.0.CO;2]
- Zwickl, D. J. (2006). Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion [Ph.D. dissertation]. The University of Texas at Austin. Retrieved July 16, 2021, from http://hdl.handle.net/2152/2666
Appendix 1
SPECIMENS EXAMINED (n = 51)
Asterisks represent specimens analysed molecularly and morphologically.
Oecomys bicolor lato sensu (n = 25)
BRAZIL – Mato Grosso: Sinop (11°43’57”S, 55°21’36”W [29]): UFMT4316*, 4321*, 4333*, 4636*, 4638*, 4642*, 4646*, 4647*; Nova Ubiratã (13°16’20”S, 54°51’34”W [31]): UFMT4882*, 4883*, 4884*, 4886*; Cláudia (11°58’33”S, 55°16’67”W [29]): MZUSP29528*, 29529*. Pará: Vitória do Xingú (03°24’20”S, 51°40’00”W [20]): MPEG40733*, 40734*, 40735*; Itaituba – Rio Tapajós (05°46’00”S, 57°16’28”W [10]): UFPAM1164*, 1165*. BOLIVIA – Chimat, Upper Beni River (15°S, 68°W): BMNH 1.2.1.14 Oecomys benevolens holotype. BRAZIL – Rondônia: Barão de Melgaço, (11°51'36.0"S, 60°43'15.6"W): AMNH 37117 Oecomys milleri holotype. ECUADOR – Gualaquiza (3°24'36.0"S, 78°34'12.0"W) BMNH 7.1.1.96 Oecomys bicolor holotype; Paramba, N. Ecuador (0°49'12.0"N, 78°27'00.0"W): BMNH 99.12.5.4 Oecomys dryas holotype. GUYANA – Demerara, Lower Essequibo (04°14'24.0"N, 58°30'36.0"W): BMNH 6.4.8.31 Oecomys nitedulus holotype. VENEZUELA – La Union, Lower Orinoco (07°30'00.0"N, 65°12'00.0"W): BMNH 4.5.7.37 Oecomys rosilla holotype.
Oecomys cleberi lato sensu (n = 17)
BRAZIL – Brasília: Distrito Federal, Fazenda Água Limpa (15°57’00”S, 47°55’58”W [34]) MN24131* Oecomys cleberi, holotype. Mato Grosso: Alta Floresta (09°45’25”S, 55°58’19W [24]): [UNEMAT] MSF2130*, 2136*; (09°52’10”S, 55°53’59W [24]): [UNEMAT] MSF2341*; (09°58’40”S, 56°04’58W [24)): UFMT4111*, 4117*; Cotriguaçu – Fazenda São Nicolau (09°50’S, 58°15’W [27]): UFMT1363*, 1366*, 1372*; Paranaíta (09°29’06”S, 56°28’18”W [25]): UFMT1777*. Pará: Itaituba (05°36’36”S, 57°17’45”W [11]): UFPAM1163*; Jacareacanga (05°43’19”S, 57°21’56”W [11]): UFPAM1177*; Juruti (02°29’60”S, 56°10’58”W [2]): MPEG38682*, 40667*. Rondônia: Porto Velho – Estação Ecológica do Cuniã (08°05’54”S, 63°21’23”W [7]): UFROM334*, 335*; Itapuã do Oeste – Floresta Nacional do Jamari (09°09’54”S, 62°54’29[1]): UFROM796*.
Oecomys jamari sp. nov. (n = 9)
BRAZIL – Pará: Jacareacanga–Flexal (05°34’S, 57°13’W [3]): MPEG13167; Juruti – Ramal Pacoval (02°34’S; 56°08’W [2]): MPEG40657*. Rondônia: Itapuã do Oeste – Floresta Nacional do Jamari (09°10’48”S, 62°57’11”W [1]): UFMT4887* holotype; (09°08’12”S, 63°02’07”W [1]): UFROM619*, 624*, (09°09’10”S, 63°00’41”W [1]): UFROM622*, (09°16’12”S, 62°52’51”W [1]): UFROM631*, 686*, 785*.