1. Introduction
The first patient with coronavirus disease (COVID-19) was reported in December 2019 in Wuhan, Hubei province, China. Since then, more than 149.471 million COVID-19 cases have been reported worldwide, with over 3.15 million deaths. The rapid spread of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection or COVID-19 has made researchers focus on developing an effective vaccine to prevent the disease. One hundred and eighty-four vaccine candidates in pre-clinical development have been tested on different animal models of SARS-CoV-2 infection (Source WHO; Draft landscape and tracker of COVID-19 candidate vaccines (who.int)). An additional 63 vaccines are currently in clinical development. Thus, more than 200 vaccine candidates have been developed within almost a year of the first reported case of COVID-19, and of these, 10 vaccines have been approved for human use (). The first human trial of a COVID-19 vaccine started on 6 March 2020, and the rapid pace of vaccine development would not have been feasible without a combined proteomics and genomics approach against SARS-CoV-2.
Table 1. Different vaccines approved for human use to prevent COVID-19
The recent emergence of SARS-CoV-2 variants in the United Kingdom (known as 20I/501Y.V1, VOC 202,012/01, or B.1.1.7), South Africa (20 H/501Y.V2 or B.1.351), and Brazil (P.1 B.1.1.28.P1 or 20 J/501Y.V3; this variant has unique mutations, including three in the receptor-binding domains [RBDs] of the spike [S] protein) is of marked concern for vaccine efficacy against COVID-19 (). The B.1.1.7 variant, originating in the United Kingdom, has now spread globally. Infected patients induce a defective antibody (Ab) response that is unable to protect against the parent SARS-CoV-2 or its B.1.351 variant, indicating the induction of asymmetric heterotypic immunity [Citation1]. Mutant strains have also emerged in the USA. For example, the SARS-CoV-2 variant, CAL.20 C, discovered in Southern California has three mutations in its S-protein, characterizing it as a subclade of 20 C, and the S protein L452R mutation in the RBD makes it resistant to several S protein monoclonal Abs [Citation2,Citation3].
Figure 1. Schematic representation of the emergence of COVID-19 in humans and its control using proteomics-based approaches. COVID-19 is thought to be a zoonotic viral infection that has been transferred to humans from horseshoe bats. However, it is unknown whether the virus evolved the ability to infect humans directly in these bats or if the virus first infected an unknown intermediate host and subsequently infected humans causing the ongoing COVID-19 pandemic. Currently available vaccines have shown protection against the parent SARS-CoV-2 strain causing COVID-19. Proteomics-based approaches can be used to study the immunopathogenesis of COVID-19 caused by both parent and mutant strains of SARS-CoV-2 and identify novel disease-specific biomarkers, drug targets, antivirals, vaccine candidates, and immunomodulators in the future
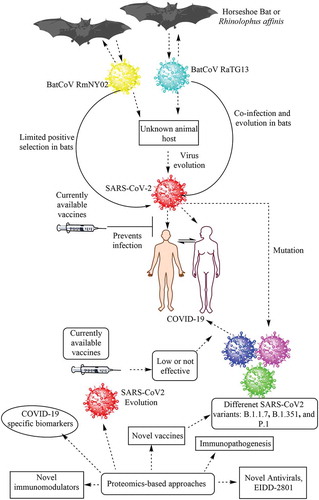
Mutant strains of SARS-CoV-2 are of great concern in terms of the efficacy of currently available vaccines. For example, SARS-CoV-2 S protein variants can escape the action of neutralizing Abs or may be less efficiently cleared [Citation4,Citation5]. The vaccine developed by Pfizer for COVID-19 has a low efficacy against the South African variant P.1. The vaccine developed by Novavax (NVX-CoV2373) is 50–60% less effective against the South African variant but has a high efficacy against the UK variant [Citation6]. The 501Y.V2 or B.1.351 variants also escape neutralization by convalescent plasma, indicating that the mutant virus can evade the immunity generated by prior SARS-CoV-2 infection [Citation7]. Thus, emerging SARS-CoV-2 variants that can escape approved vaccine-based immune responses and convalescent plasma-based therapies are of concern.
Depending on the severity of COVID-19, patients receive a combination of different drugs, including antivirals (favipiravir), antimalarials (hydroxychloroquine), antibacterials (azithromycin and doxycycline), antiplatelets, and immunomodulators [Citation8–10]. In November 2020, the World Health Organization approved the antiviral, Remdesivir (previously used for treating hepatitis C virus and Ebola virus infections), as a conditional drug. It is not possible to mention all the clinical trials associated with COVID-19 therapy. However, AstraZeneca’s Calquence (a blood cancer drug) failed in phase 2 trial, Novartis’ Ilaris (an anti-arthritis drug) showed no benefit to COVID-19 patients, and Sanofi and Regeneron’s Kevzara (a drug used for treating rheumatoid arthritis) did not perform favorably in COVID-19 patients.
2. Proteomics as a biological tool in infection and immunity research
Proteomic analysis techniques are highly sophisticated tools for identifying and quantifying cell and organism proteomes. These techniques can be used to identify immune-related proteins generated or suppressed during infection and elucidate the mechanisms through which pathogens escape the immune response of the host. Comparative proteomic analysis identifies the host immune response, including mucosal (pulmonary and gut) immunity, which plays a crucial role in COVID-19 pathogenesis. Proteomics has the potential to identify single immune cell (macrophages, dendritic cells, natural killer cells, neutrophils, mast cells, and other adaptive immune cells [T and B cells])-based host immune responses against pathogens, including SARS-CoV-2. Proteomics-based approaches are a crucial component of functional genomics and have considerable potential to establish COVID-19 severity-based biomarkers. This approach can also provide data on the effect of treatments on host metabolism and single immune cell-based future designer therapies. In addition, post-translationally modified proteins, such as phosphoproteins, glycoproteins, and lipoproteins, can be studied in detail using phosphoproteomics, glycoproteomics, and lipoproteomics. The evolution of proteogenomics has revolutionized the field of gene annotation by discovering post-translational protein modifications and protein cleavage [Citation11]. Thus, proteomics has great potential to identify novel biomarkers, modified host and viral proteins, vaccines, and pharmacological targets against COVID-19 and SARS-CoV-2 mutants. For instance, proteomics-based studies (e.g. ultra-high-throughput clinical proteomics involving sequential window acquisition of all theoretical mass spectra – a specific variant of the data-independent acquisition method that supports the quantitative analysis of peptides composed of up to 9000 proteins with high accuracy and consistency – and quantitative proteomic analysis using the BoxCar method) have identified host immune response and associated factors to be responsible for the different phenotypes of COVID-19 (asymptomatic, symptomatic, mild to moderate, and severe) patients, changes in immunometabolic reprogramming among various immune cells controlling their immune function, and the development of specific biomarkers for COVID-19 () [Citation12–14].
3. Proteomics-based approaches for understanding COVID-19 and drug discovery
Humans have more than 20,000 genes that encode different proteins, including 1000,000 proteoforms, such as splice variants and proteins with post-translational modifications. Proteomics-based approaches, such as liquid chromatography and tandem mass spectrometry (LC-MS/MS), are crucial for identifying and characterizing the human proteome [Citation15]. Proteomic analysis (LC-MS/MS) of SARS-CoV-2-infected host cells has helped to identify the translatome and proteome infection profile of the virus, showing that SARS-CoV-2 infection changes the central cellular pathways, including splicing, translation, protein metabolism, carbon metabolism, and nucleic acid metabolism [Citation16]. LC-MS/MS can also identify the genetic basis of resistance and susceptibility to COVID-19, such as HIV-1 infection [Citation17,Citation18]. In addition, the occurrence of cytokine storm in patients raises the severity of COVID-19, which can be predicted by analyzing the de-mono-ADP-ribosylation of STAT1 by SARS-CoV-2 non-structural protein (Nsp) 3, as this process is the putative cause of severe disease in most patients [Citation19]. Another study has also identified host proteome-based criteria for COVID-19-associated cytokine storms [Citation20].
Proteomic analysis of SARS-CoV-2 has also identified that its genomic mRNA encodes four structural proteins: S, envelope (E), membrane (M), and nucleocapsid (N). Sixteen Nsps (Nsp1–Nsp16) of SARS-CoV-2 play a crucial role in viral replication and escape from the host immune response. LC-MS/MS has been used to study the protein-protein interactions (PPIs) between the host and virus to elucidate the host immune response and potential drug targets [Citation21]. Hence, targeting these proteins may lead to better therapeutic approaches against COVID-19 [Citation22]. For example, Nsp15 may serve as a crucial antiviral target for COVID-19. Furthermore, LC-MS/MS-based quantitative proteomics has identified the broad-spectrum antiviral effect of the antiparasitic drug, ivermectin, for treating COVID-19 [Citation23].
The pro-drug, EIDD-2801 (also called molnupiravir or MK-4482), derived from the ribonucleoside analog β-D-N4–hydroxycytidine, exerts antiviral effects by acting as a competitive alternate substrate for the viral RNA polymerase. This allows its incorporation into the viral RNA, thereby accumulating mutations in the viral RNA genome and inducing error catastrophe [Citation24]. EIDD-2801 exerts both SARS-CoV-2 infection prophylaxis and treatment effects through the oral route of administration [Citation25]. The drug is presently undergoing clinical trial phases 2 and 3. Additionally, multivalent nanobodies or variable domains of heavy-chain-only Abs have been designed by studying the detailed structures of their epitopes and binding modes to the S protein of SARS-CoV-2 as well as the mechanisms by which the virus fuses to the cell membrane through the S protein [Citation26]. These multivalent nanobodies block SARS-CoV-2 infection and suppress their mutational escape by targeting two independent epitopes [Citation26]. The deep learning approach to predict and design a multi-epitope vaccine (DeepVacPred) combines in silico immunoinformatics and deep neural network strategies. The DeepVacPred computational framework has predicted several potential vaccine subunits from the available SARS-CoV-2 spike protein sequence [Citation27]. Moreover, in silico analysis of the immune system protein interactome associated with SARS-CoV-2 infection and severity has revealed several novel therapeutic targets for drug repurposing against COVID-19 [Citation28]. Hence, proteomics combined with bioinformatics may serve as a novel tool for understanding COVID-19 pathogenesis and designing therapeutics and vaccines.
4. Conclusion
Future studies are crucial in the field of drug discovery and vaccine design for the treatment and prevention of COVID-19. Although we have developed vaccines for COVID-19, novel antiviral agents and better vaccine candidates are still needed. For example, the identification and isolation of naturally circulating attenuated SARS-CoV-2 variants may serve as another potential vaccine candidate [Citation29]. In addition, vaccines for short-term immune response (6–12 months) are currently in demand due to the urgency of preventing additional cases of COVID-19 worldwide; however, we still need a vaccine for long-term immunity. Hence, proteomics with other omics techniques (e.g. genomics, metabolomics, lipidomics, and phenomics) are potential solutions for the development of antivirals, potent immunomodulatory agents, and vaccines.
5. Expert Opinion
COVID-19 has emerged as one of the biggest infectious disease pandemic of the 21st century since its first origin in the China in December 2019. We currently do not have a potent antiviral agent for the SARS-CoV-2, the causative agent for the COVID-19. However, with the use of proteomics-based approaches along with other molecular biology techniques we have successfully developed different vaccines to prevent further spread of the infection. But the emergence of different mutants of the SARS-CoV-2 in many countries and their spread to other nations has also compelled researchers to think about vaccination strategies. This is because some mutants have shown resistance against different vaccines-derived immune response. Hence, different proteomics-based approaches in combination with bioinformatics, metabolomics, lipidomics, and other molecular biology techniques are required to track mutant viruses that can be used to design potent vaccine for long lasting immunity and design novel antiviral to directly target the virus.
Declaration of interest
The author has no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Additional information
Funding
References
- Faulkner N, Ng KW, Wu M, et al. Reduced antibody cross-reactivity following infection with B.1.1.7 than with parental SARS-CoV-2 strains. bioRxiv. 2021;2021.2003.2001.433314.
- Zhang W, Davis BD, Chen SS, et al. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA. 2021;325:1324.
- Li Q, Wu J, Nie J, et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294.e1289.
- Weisblum Y, Schmidt F, Zhang F, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312.
- Liu Z, VanBlargan LA, Bloyet L-M, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29(3):477-488.
- Mahase E. Covid-19: novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ. 2021;372:n296.
- Cele S, Gazy I, Jackson L, et al. Escape of SARS-CoV-2 501Y.V2 variants from neutralization by convalescent plasma. medRxiv. 2021. DOI: 10.1038/s41586-021-03471-w.
- McCullough PA, Kelly RJ, Ruocco G, et al. Pathophysiological basis and rationale for early outpatient treatment of SARS-CoV-2 (COVID-19) infection. Am J Med. 2021;134(1):16–22.
- McCullough PA. Favipiravir and the need for early ambulatory treatment of SARS-CoV-2 infection (COVID-19). Antimicrob Agents Chemother. 2020;64(12):e02017–02020.
- McCullough PA, Alexander PE, Armstrong R, et al. Multifaceted highly targeted sequential multidrug treatment of early ambulatory high-risk SARS-CoV-2 infection (COVID-19). Rev Cardiovasc Med. 2020;21(4):517–530.
- Gupta N, Tanner S, Jaitly N, et al. Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 2007;17(9):1362–1377.
- Grenga L, Armengaud J. Proteomics in the COVID-19 battlefield: first semester check-up. PROTEOMICS. 2021;21(1):2000198.
- Messner CB, Demichev V, Wendisch D, et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020;11(1):11–24.e14.
- Park J, Kim H, Kim SY, et al. In-depth blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non-severe COVID-19 patients. Sci Rep. 2020;10(1):22418.
- Chapman JD, Goodlett DR, Masselon CD. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom Rev. 2014;33(6):452–470.
- Bojkova D, Klann K, Koch B, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583(7816):469–472.
- Chakravarty S. COVID-19: The effect of host genetic variations on host–virus interactions. J Proteome Res. 2021;20(1):139–153.
- Kumar V, Prakash O, Manpreet S, et al. Genetic basis of HIV-1 resistance and susceptibility: an approach to understand correlation between human genes and HIV-1 infection. Indian J Exp Biol. 2006;44(9):683–692.
- Claverie J-M, Putative A. Role of de-Mono-ADP-ribosylation of STAT1 by the SARS-CoV-2 Nsp3 protein in the cytokine storm syndrome of COVID-19. Viruses. 2020;12(6):646.
- Caricchio R, Gallucci M, Dass C, et al. Preliminary predictive criteria for COVID-19 cytokine storm. Ann Rheum Dis. 2021;80(1):88–95.
- Lachén-Montes M, Corrales FJ, Fernández-Irigoyen J, et al. Proteomics insights into the molecular basis of SARS-CoV-2 infection: what we can learn from the human olfactory axis. Front Microbiol. 2020;11(2101). DOI:10.3389/fmicb.2020.02101
- Kumar V. Understanding the complexities of SARS-CoV2 infection and its immunology: a road to immune-based therapeutics. Int Immunopharmacol. 2020;88(106980). DOI: 10.1016/j.intimp.2020.106980.
- Li N, Zhao L, Zhan X. Quantitative proteomics reveals a broad-spectrum antiviral property of ivermectin, benefiting for COVID-19 treatment. J Cell Physiol. 2021;236(4):2959–2975.
- Sheahan TP, Sims AC, Zhou S, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12(541):eabb5883.
- Wahl A, Gralinski LE, Johnson CE, et al. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591(7850):451–457.
- Koenig P-A, Das H, Liu H, et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371(6530):eabe6230.
- Yang Z, Bogdan P, Nazarian S. An in silico deep learning approach to multi-epitope vaccine design: a SARS-CoV-2 case study. Sci Rep. 2021;11(1):3238.
- López-Cortés A, Guevara-Ramírez P, Kyriakidis NC, et al. In silico analyses of immune system protein interactome network, single-cell RNA sequencing of human tissues, and artificial neural networks reveal potential therapeutic targets for drug repurposing against COVID-19. Front Pharmacol. 2021;12(63). DOI:10.3389/fphar.2021.598925.
- Armengaud J, Delaunay-Moisan A, Thuret JY, et al. The importance of naturally attenuated SARS-CoV-2in the fight against COVID-19. Environ Microbiol. 2020;22(6):1997–2000.
