Abstract
Tenofovir alafenamide (TAF), a novel prodrug of the NtRTI tenofovir (TFV), delivers TFV-diphosphate (TFV-DP) to target cells more efficiently than the current prodrug, tenofovir disoproxil fumarate (TDF), with a 90% reduction in TFV plasma exposure. TAF, within the fixed dose combination of elvitegravir /cobicistat / emtricitabine (FTC)/TAF (E/C/F/TAF), has been evaluated in one Phase 2 and two Phase 3 randomized, double-blinded studies in HIV-infected treatment-naive patients, comparing E/C/F/TAF to E/C/F/TDF. In these studies, the TAF-containing group demonstrated non-inferior efficacy to the TDF-containing comparator group with 91.9% of E/C/F/TAF patients having <50 copies/mL of HIV-1 RNA at week 48. An integrated resistance analysis across these three studies was conducted, including HIV-1 genotypic analysis at screening, and genotypic/phenotypic analysis for patients with HIV-1 RNA >400 copies/mL at virologic failure. Pre-existing primary resistance-associated mutations (RAMs) were observed at screening among the 1903 randomized and treated patients: 7.5% had NRTI-RAMs, 18.2% had NNRTI-RAMs, and 3.4% had primary PI-RAMs. Pre-treatment RAMs did not influence treatment response at Week 48. In the E/C/F/TAF group, resistance development was rare; seven patients (0.7%, 7/978) developed NRTI-RAMs, five of whom (0.5%, 5/978) also developed primary INSTI-RAMs. In the E/C/F/TDF group, resistance development was also rare; seven patients (0.8%, 7/925) developed NRTI-RAMs, four of whom (0.4%, 4/925) also developed primary INSTI-RAMs. An additional analysis by deep sequencing in virologic failures revealed minimal differences compared to population sequencing. Overall, resistance development was rare in E/C/F/TAF-treated patients, and the pattern of emergent mutations was similar to E/C/F/TDF.
Introduction
Historically, the three-drug regimens recommended for HIV-1 treatment involved the use of multiple pills taken several times a day. These complicated regimens consisting of first-generation molecules with a high pill burden resulted in reduced efficacy and increased emergence of drug resistance due to poor treatment adherence.Citation1 The first advancement towards regimen simplification was the development of once-daily fixed dose combination pills combining 2 nucleos(t)ide reverse transcriptase inhibitors (NRTI). These were followed by the first once-daily single tablet regimen (STR) that combined all three drugs needed to create a complete treatment regimen,Citation2 with additional regimens having been added to the available treatment options.Citation3 In addition to providing treatment simplification, the use of STRs for the treatment of HIV-1 infection has been associated with increased treatment adherence and increased virologic suppression compared to multi-pill regimens.Citation4 Two integrase strand transfer inhibitor (INSTI)-containing STRs – Stribild™ and Triumeq™ – are currently recommended as first-line regimens for the treatment of HIV-1 in antiretroviral (ARV) treatment-naive patientsCitation1; however, both have restrictions depending on patient characteristics. Consequently, there is a need to develop more broadly tolerated treatment regimens.
Tenofovir (TFV) is an HIV-1 NRTI that is currently included in four of the five DHHS recommended regimens as the prodrug TDF.Citation1 TFV as a parent drug has limited oral bioavailability due to limited cell permeability, which is driven by the polar nature of the phosphonate moiety.Citation5 TFV is currently administered as the prodrug TDF, which has been engineered to improve TFV oral bioavailability.Citation6 However, TDF is quickly metabolized to TFV upon administration by gut and serum esterases, requiring high plasma TFV exposure to obtain therapeutic effects in target cells.Citation7 Once inside the cells, TFV is converted to the active inhibitor tenofovir diphosphate (TFV-DP) by cellular kinases.Citation8,9 TFV is a substrate for the organic anion transporters OAT-1 and OAT-3, and as a consequence of the high TFV plasma concentration, TFV can accumulate in the kidney in certain conditions,Citation10 leading in some cases to renal impairment and reduced bone mineral density in patients.Citation11
Tenofovir alafenamide (TAF) is a novel investigational prodrug of TFV with improved metabolic stability compared to TDF.Citation12–15 Consequently, TAF is more efficiently delivered to target cells where it is metabolized into TFV by specific cellular enzymes, such as cathepsin A (CatA),Citation16 a ubiquitous enzyme enriched in the lymphoid tissues,Citation17 or by carboxy-esterase 1 (CES1).Citation18 Direct metabolism of TAF into TFV in target cells upon dosing with 25 mg of TAF in Phase 1 clinical studies was associated with an 86–91% reduced systemic TFV exposure compared to dosing with 300 mg TDF,Citation14,19 and resulted in higher TFV-DP concentrations in PBMCs with greater viral RNA reductions. The STR-containing E/C/F/TAF has the potential to offer a better safety profile and a higher inhibitory quotient (IQ) associated with higher intracellular levels of TFV-DP than the STR-containing E/C/F/TDF. Similar reductions in plasma levels of TFV were observed in Phase 2Citation19 and Phase 3Citation20 studies with E/C/F/TAF compared to the Phase 1 studies with TAF alone.Citation14, 19, 21 Furthermore, in Phase 3 studies, E/C/F/TAF treatment resulted in a significantly improved renal and bone safety profile compared to E/C/F/TDF.Citation20 Here we report the results of the integrated E/C/F/TAF resistance analysis combining the Phase 2 study GS-US-292–0102 and the two Phase 3 studies GS-US-292–0104 and GS-US-292–0111 in treatment-naive HIV-1 infected patients.
Methods
Study design
Studies GS-US-292-0104 (Study 104) and GS-US-292-0111 (Study 111) (ClinicalTrials.gov identifiers NCT01780506 and NCT01797445, respectively)Citation20 are two ongoing Phase 3, randomized, double-blind studies to evaluate the safety and efficacy of E/C/F/TAF vs. E/C/F/TDF in HIV-1 infected ARV treatment-naive adults. Each study enrolled over 860 patients randomized 1:1 to receive either E/C/F/TAF or E/C/F/TDF. Study GS-US-292-0102 (Study 102) (ClinicalTrials.gov identifier NCT 01497899)Citation19 was a Phase 2 study that evaluated E/C/F/TAF vs. E/C/F/TDF in a smaller patient population (170 patients randomized 2:1 to either E/C/F/TAF or E/C/F/TDF). All studies were stratified based on screening HIV-1 RNA values (≤ or >100,000 copies/mL), as well as geography and screening CD4 cell counts (Studies 104 and 111 only). The primary endpoint was the proportion of patients with HIV-1 RNA <50 copies/mL at week 48 (Studies 104 and 111) or week 24 (Study 102) using the FDA snapshot algorithm.
Screening genotype
HIV-1 genotyping of the protease (PR), reverse transcriptase (RT), and integrase (IN; Studies 104 and 111 only) segments of the pol gene was conducted at screening for all prospective patients to assess for the presence of pre-existing resistance. Genotypic testing was carried out at Monogram Biosciences (South San Francisco, CA, USA), and sensitivity to EVG, FTC, and TFV (delivered as TDF or TAF) were determined using the Monogram Biosciences algorithm. Only patients with genotypic sensitivity to all study drugs were allowed to enroll in the studies.
Resistance analysis criteria
Patients with either (1) suboptimal virologic response (HIV-1 RNA reduction of less than 1 log10 from baseline and HIV-1 RNA ≥50 copies/mL (Studies 104 and 111) or 400 copies/mL (Study 102) at the Week 8 visit, followed with HIV-1 RNA ≥400 copies/mL at the next scheduled or unscheduled subsequent visit), (2) virologic rebound (rebound in HIV-1 RNA above 50 copies/mL (Studies 104 and 111) or 400 copies/mL (Study 102), followed with HIV-1 RNA ≥400 copies/mL at the next scheduled or unscheduled subsequent visit; or two subsequent visits with HIV-1 RNA greater than 1 log10 above the nadir), or (3) HIV-1 RNA ≥400 copies/mL at their last visit while maintaining the study drug regimen (or within 72 h after interruption or discontinuation of study drugs), were considered virologic failures and included in resistance analyses.
Resistance analyses
Plasma samples from virologic failure timepoints with HIV-1 RNA ≥400 copies/mL were tested for the potential emergence of genotypic and phenotypic resistance to study drugs using the PR/RT PhenoSenseGT™ and IN GeneSeq and PhenoSense™ assaysCitation22 at Monogram Biosciences. The list of resistance-associated mutations (RAMs; see Supplementary Digital Content Table 1) to HIV-1 inhibitors utilized is based on International AIDS Society-USA guidelines (IAS-USA) with some modifications.Citation23, 24
Deep sequencing of HIV-1 PR/RT and IN
Deep sequencing (minority variant) analyses were conducted at Seq-IT (Kaiserslautern, Germany). HIV-1 RNA was extracted from plasma of HIV-1 infected patients using QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany). For plasma samples with viral loads <1,000 copies/mL an additional centrifugation step was introduced. Up to 2 mL plasma of low viral load samples were centrifuged at 17,000 × g for 1 h at 4 °C. Supernatant was discarded down to 140 μL. Pellet was resuspended in the remaining 140 μL. One-step RT-PCR was done using the SuperScript™ III One-Step RT-PCR System with Platinum® Taq High Fidelity (Life Technologies, Darmstadt, Germany) and primers GAAGAAATGATGACAGCATGTCAGGG (HIV-1NL4–3 nt 1819 to 1844), TAATTTATCTACTTGTTCATTTCCTCCAAT (HIV-1NL4–3 nt 4173 to 4202) for PR/RT and ATTGGAGGAAATGAACAAGT (HIV-1NL4–3 nt 4173 to 4192), ATCCTGTCTACYTGCCACACAA (HIV-1NL4–3 nt 5066 to 5087) for IN. Nested PCR for PR/RT was done using Platinum® Taq High Fidelity enzyme (Life Technologies) and primers AGACAGGCTAATTTTTTAGGGA (HIV-1NL4–3 nt 2074 to 2095), ATGGYTCTTGATAAATTTGATATGTCC (HIV-1NL4–3 nt 3585 to 3559). All amplicons were purified using Agencourt AMPure XP PCR Purification beads on a BioMek NX workstation (Beckman Coulter, Krefeld, Germany), quantified fluorometrically on a FluoStar Optima (BMG Labtech, Ortenberg, Germany) using Quant-iT Picogreen dsDNA reagent (Life Technologies) and sample-specific amplicons were pooled equimolarly for library preparation. The sequencing library was prepared with the Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s description and sequenced using a MiSeq Benchtop Sequencer (Illumina) generating paired-end reads of 2×250 bp length with a calculated coverage of 10,000 to 20,000 reads.
Analysis of deep sequencing data was performed using a pipeline where sequences in the form of FASTQ files were assembled and aligned via a multi-step method. Baseline samples were assembled by the following steps: Contigs from paired-end reads were assembled from FASTQ using VICUNA,Citation25 and were then merged to generate an assembly sequence based on an alignment to the HIV reference sequence HIV-1 NL4-3 (Genbank Accession AF324493). Prior to alignment, next-generation sequencing (NGS) data were evaluated based on quality scores. Any reads with phred score <15 were trimmed, and reads consisting of <50 nucleotides in length after trimming were excluded. These reads were aligned using MOSAIK v1.1.0017Citation26 to the sample’s corresponding baseline assembly sequence. All aligned reads were translated in-frame and variants were tabulated.
Results
Pre-treatment genotypic analysis
Genotyping of the PR/RT (Studies 102, 104, and 111) and IN (Studies 104 and 111) genes was conducted at the screening visit to assess for the presence of pre-existing RAMs that could adversely affect treatment response to E/C/F/TAF or E/C/F/TDF. Drug sensitivity was established on the basis of the Monogram Biosciences report and proprietary resistance algorithm. Consistent with enrollment criteria, all patients showed sensitivity to EVG, FTC, and TFV at screening except for one patient with possible EVG resistance and two patients with possible TDF resistance, all of whom were randomized to receive E/C/F/TDF treatment. The patient with possible EVG resistance at baseline had the low-level polymorphism T97A mutation in IN as a mixture with wild-type (T97T/A). That patient experienced full virologic suppression with undetectable HIV-1 RNA through 96 weeks while receiving E/C/F/TDF. The two patients with possible TDF resistance at baseline (one with RT mutations A62V, L210T, and K219R; and one with RT mutations M41L, L210W, and T215D) both experienced HIV-1 RNA suppression to undetectable levels at the time of early discontinuation from the study after up to three weeks of E/C/F/TDF treatment.
Patients entered the studies with varying levels of pre-existing background RAMs. NNRTI-associated RAMs were the most prevalent with 18.2% of patients carrying one or more of these mutations (Table ); the K103N/S mutation was found in 5.7% of patients. NRTI-associated RAMs were observed in 7.5% of patients, with the majority being the 3TC-resistance-associatedCitation27 polymorphism V118I (4.7%). Thymidine-analog associated mutations (TAMs) were detected in 2.3% of patients. Primary PI-associated RAMs were observed in 3.4% of patients. The distribution of pre-existing RAMs was comparable between treatment groups. Patients had predominantly HIV-1 subtype B (87%), followed by subtype AE (6.7%), complex subtype (1.5%), and subtypes AG and C (each 1.4%). Other HIV-1 subtypes (A, A1, A2, BC, BF, D, F, F1, and G) were found in less than 1% of patients. The majority of AE patients were from southeast Asia, where subtype AE is the predominant HIV-1 strain. The HIV-1 subtype distribution was comparable between treatment groups (not shown).
Table 1 Integrated baseline genotypic analysis
Treatment response per baseline virological category
The impact of pre-existing RAMs and HIV-1 subtype on treatment outcomes was assessed for all patients (Table ). The proportions of patients with treatment success at Week 48 in the presence or absence of RAMs (NRTI-R, TAMs, NNRTI-R, K103N/S, and PI-R), or with HIV-1 subtype B vs. non-B were compared using Fisher’s exact test. Proportions of patients with treatment success at week 48 per treatment group for each virological category were also analyzed using Fisher’s exact test. No statistical significance was found in any of the comparisons tested (p > 0.05), indicating that treatment response was not affected by the presence of pre-existing RAMs or the patient’s HIV-1 subtype.
Table 2 Impact of pre-existing RAMs and HIV-1 subtype on treatment outcomes
Resistance analysis population
A large majority of patients in both treatment groups achieved treatment success at Week 48 (>90%). In the E/C/F/TAF group, 899 of 978 patients (91.9%) were treatment successes, while the remainder had either no virologic data (NVD; 41/978, 4.2%) or virologic failure (VF; 38/978, 3.9%). The resistance analysis population (RAP) included 2 patients with treatment success, 2 patients with NVD, and 15 patients with VF for a total of 19 patients. The 23 remaining VF patients did not qualify for resistance analyses due to HIV RNA being below the lower limit of performance for the resistance assays (<400 copies/mL) at the time of failure. Five of the 19 RAP patients had resuppression of HIV-1 RNA to <50 copies/mL while maintaining study drugs after resistance testing was conducted, and were excluded from the final-RAP. Therefore, the final-RAP in the E/C/F/TAF group consisted of 14 patients. None of the patients excluded from the final-RAP had emergent resistance to study drugs (data not shown). In the E/C/F/TDF group, 835 of 925 patients (90.3%) were treatment successes, while the remainder was either in the NVD (49/925, 5.3%) or VF category (41/925, 4.4%). The RAP included 5 patients with treatment success, 1 patient with NVD, and 16 patients with VF for a total of 22 patients. The 25 remaining VF patients did not qualify for resistance analyses due to HIV RNA being below 400 copies/mL at the time of failure. Six of the 22 RAP patients had resuppression of HIV-1 RNA to <50 copies/mL while maintaining study drugs after resistance testing was conducted, and were excluded from the final-RAP. Therefore, the final-RAP in the E/C/F/TDF group consisted of 16 patients. None of the patients excluded from the final-RAP had emergent resistance to study drugs (data not shown).
Emergence of resistance by week 48 in final-RAP patients
In the E/C/F/TAF group, genotypic data were obtained for 14/14 (RT) and 11/14 (IN) final-RAP patients, and in the E/C/F/TDF group, 15/16 (RT) and 13/16 (IN) final-RAP patients (Table ). A similar proportion of patients developed primary resistance mutations in each treatment group, with 7 of 978 patients (0.7%) in the E/C/F/TAF group and 7 of 925 patients (0.8%) in the E/C/F/TDF group (Fig. ). The FTC primary resistance mutation M184V/I in RT was detected in all patients developing primary resistance from both treatment groups (7 patients in each group). The presence of M184V/I was associated with phenotypic resistance to FTC except in two patients where the mutation was detected as a mixture with wild-type (M184 M/V) (Table ). Of note, the M184V mutation was detected in one patient at Week 48 with HIV-1 RNA of 73 copies/mL (patient 24, Table ). That patient became undetectable at the next visit, and remained suppressed for up to seven months prior to being lost to follow-up. In addition to M184V/I, primary INSTI resistance was detected in 5/7 and 4/7 patients in the E/C/F/TAF and E/C/F/TDF groups, respectively, with similar patterns of INSTI mutations for both groups (Table ). Emergence of INSTI-R mutations was associated with phenotypic resistance to EVG and cross-resistance to RAL in most patients (Table ). Primary resistance to TAF or TDF was detected in addition to M184V/I in 1/7 and 3/7 patients in the E/C/F/TAF and E/C/F/TDF groups, respectively. One E/C/F/TAF patient and two E/C/F/TDF patients developed the K65R RT mutation in addition to M184V/I and INSTI-R (N155H, Q148R, and E92Q, respectively). No phenotypic resistance to TAF or TDF was detected in the two patients with available data; however, cross-resistance to ABC was detected in both cases. Lastly, one E/C/F/TDF patient developed a K70K/E mutation mixture with wild-type in addition to M184M/V, with no phenotypic resistance to TAF/TDF. Overall, emergence of resistance occurred in <1% of patients in each treatment group. No other unique substitutions in the RT or IN genes were observed in more than 1 final-RAP patient.
Table 3 Week 48 genotypic resistance results
Figure 1 Integrated analysis: RAP inclusion criteria and genotypic results through week 48.
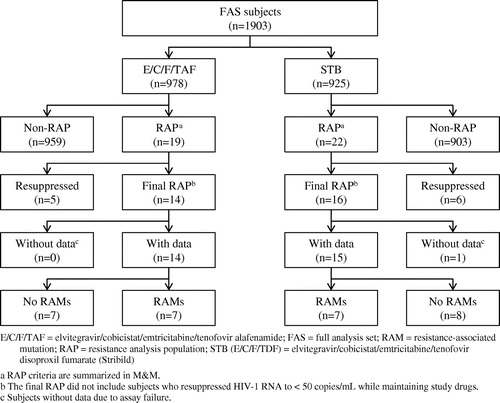
Table 4 Week 48 phenotypic resistance results
Deep sequencing analyses
Deep sequencing analyses were conducted to investigate the potential presence of minority variants harboring resistance mutations not previously detected by population sequencing. Deep sequencing was performed for all RAP patients in Studies 104 and 111 (16 and 19 patients in the E/C/F/TAF and E/C/F/TDF groups, respectively), with data available for 14 and 16 patients in the E/C/F/TAF and E/C/F/TDF groups, respectively. Additionally, deep sequencing was performed for non-RAP virologic failure patients who had low-level HIV viremia (HIV-1 RNA between 50 and 400 copies/mL at confirmatory or last visit past week 8), adding 13 and 17 patients to the analysis set in the E/C/F/TAF and E/C/F/TDF groups, with data available for 9 and 17 patients, respectively. Among the RAP patients previously analyzed by population sequencing, deep sequencing identified RAMs in 1 additional patient in the E/C/F/TAF group (Table ). This patient had no RAMs detected by population sequencing, but had M184V/I detected by deep sequencing (20%) at virologic failure. No other RAP patients had additional resistance detected by deep sequencing, including the five subjects with IN genotypic assay failure by population sequencing (see Table ). No deep sequencing data were obtained for the subject with dual RT and IN population sequencing assay failure. Among patients with low-level viremia not included in the protocol-defined RAP, 1 patient had Q148R detected by deep sequencing (2%) in the E/C/F/TAF group and 3 patients in the E/C/F/TDF group had resistance mutations detected: M184V/I (71%), M184I (99%), and Q148H (10%). Deep sequencing assay failure in 4 of the 13 subjects with low-level viremia tested from the E/C/F/TAF group may have limited resistance detection in these four subjects.
Table 5 Patients with resistance by population and deep sequencing analyses
Discussion
Studies 102, 104, and 111 are the first randomized, double-blind clinical studies to evaluate the use of E/C/F/TAF in treatment-naive patients.Citation19, 20 Safety and efficacy through Week 48 demonstrated that E/C/F/TAF was not inferior to E/C/F/TDF, with safety benefits in bone mineral density and renal biomarkers for the E/C/F/TAF groups. Across the two large Phase 3 studies, rates of virologic suppression reached over 92%, representing the highest rates of suppression at Week 48 observed in randomized studies conducted in HIV-1 infected treatment-naive patients to date.Citation20 Correspondingly, resistance emergence was rare, with similar rates of resistance emergence for the E/C/F/TAF (0.7%) and the E/C/F/TDF groups (0.8%).
Across these three studies comparing E/C/F/TAF with E/C/F/TDF, the distribution of pre-existing RAMs and HIV-1 subtype was comparable across treatment groups, and had no impact on treatment outcomes. Across all patients entering these studies, pre-existing RAMs (PI, NNRTI, NRTI) were observed in 3–18% of patients. The presence of these mutations in treatment-naive patients suggests that they represent transmitted drug-resistant (TDR) viruses. The rate of TDRs observed in these studies is comparable to other recent studies,Citation28 suggesting that TDR is still a major issue in HIV infected patients, and supports the need for the development of new HIV-1 treatment regimens. Importantly, the presence of these pre-existing RAMs had no impact on E/C/F/TAF response rates, suggesting that E/C/F/TAF is a viable regimen for the vast majority of treatment-naive patients.
In patients who developed resistance across both treatment groups, M184V/I was always detected by population sequencing, either with or without additional NRTI and/or INSTI RAMs, which is typical for most prescribed NRTI-containing regimens.Citation29 As has been previously observed with other FTC or 3TC-containing treatment regimens, M184V appears to be one of the first RAMs to emerge during cART treatment in patients who develop resistance, with mutations to other drug classes typically emerging with additional time on treatment.Citation30 Further supporting this observation, deep sequencing results in this study demonstrated development of M184V/I in three additional patients without INSTI RAMs. These data, along with data from other clinical studies, support performing genotypic testing on patients immediately after viral load rebound in order to reduce the potential for development of multi-drug resistant viruses. A comparative review of recent clinical studies highlighted differences in clinical trial management and discussed advantages/disadvantages of earlier genotypic testing on resistance rates in clinical practice.Citation31
Despite the fact that TFV plasma exposure has been shown to be consistently reduced by ~90% when using TAF over TDF in E/C/F/TAF clinical studies,Citation14, 19, 20 the rate of resistance observed in E/C/F/TAF-treated patients in these studies was similar to that in E/C/F/TDF-treated patients. This may be due to the fact that there is no reduction of intracellular TFV-DP (TFV active entity) in patients treated with TAF.Citation19 Thus, there is still strong selective pressure for TFV resistance. In spite of this, only one case of TAF resistance was observed. This patient had low treatment adherence by returned pill counts (subject was discontinued from study by the investigator for non-compliance) which may have contributed to the virologic failure and subsequent development of resistance.
Experimental resistance studiesCitation32 using non-clinical drug concentrations have shown that TAF and TFV have qualitatively the same resistance profile in vitro. This was expected since both TAF and TFV lead to the same active moiety, TFV-DP. However, the higher intracellular levels of TFV-DP observed with TAF in the clinic could predict higher levels of virologic suppression or reduced levels of resistance, as the TFV IQ is increased via TAF delivery. In support of this hypothesis, the Phase 1 data demonstrated that TAF 25 mg had higher potency than TDF 300 mg.Citation14 Yet, in the context of an STR, it is more challenging to determine the contribution of each drug to the overall potency of the ARV regimen, especially with the fast viral load decrease observed with INSTI-containing regimens, as seen in these Phase 2 and Phase 3 studies.
Finally, 30 patients with low-level viremia, not included in the protocol-defined RAP, were analyzed for the presence of low-level variants by deep sequencing analysis. A minority of these patients (8/30) either maintained low-level viremia after their analysis timepoints or had no follow-up, including one E/C/F/TDF subject with resistance detected (M184V 37% and M184I 34%). The remainder of these patients (22/30) showed a transient rebound in HIV-1 RNA that was subsequently resuppressed <50 copies/mL, including three patients for whom resistance was detected. Two of these three patients had an INSTI-R mutation at position Q148 that was detected at low-level (one E/C/F/TDF patient with Q148H 10%, and one E/C/F/TAF patient with Q148R 2%), and one E/C/F/TDF patient had the M184I RT mutation that was detected at >99%. This indicates that the presence of these resistance mutations did not preclude resuppression of HIV-1 RNA to undetectable level in these patients. Similarly, one E/C/F/TDF patient in the RAP was able to resuppress HIV RNA to undetectable level in the presence of M184V at 100%.
In summary, the resistance analyses conducted across the one Phase 2 and two Phase 3 studies of E/C/F/TAF vs. E/C/F/TDF in treatment-naive patients demonstrate that E/C/F/TAF has a similar resistance profile to E/C/F/TDF, with <1% resistance observed in both treatment groups. In addition, the patterns of emergent resistance observed in the E/C/F/TAF-treated patients were similar to the E/C/F/TDF-treated patients.
Funding
These studies were sponsored by Gilead Sciences. All authors are employees and stockholders of Gilead Sciences.
Acknowledgements
The authors would like to thank the patients and investigators who participated in these studies, the E/C/F/TAF project team for their contributions to this analysis, as well as Silvia Chang, Krishna Chodavarapu and Martin Daeumer for the deep sequencing analyses.
Additional information
Funding
Notes
Presented in part at the International HIV Drug Resistance Workshop, Feb 21–22, 2015, Seattle, USA, at the 2015 Conference on Retroviruses and Opportunistic Infections (CROI), Feb 23–26, Seattle, USA, and at the European AIDS Clinical Society (EACS), Oct 21–24, 2015, Barcelona, Spain
References
- Department for Health and Human Services (DHHS). Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents. Developed by the DHHS Panel on antiretroviral guidelines for adults and adolescents – A Working Group of the Office of AIDS Research Advisory Council (OARAC). http://aidsinfo.nih.gov/guidelines on 5/11/2015. Last Updated 08 April 2015.
- Deeks ED, Perry CM. Efavirenz/emtricitabine/tenofovir disoproxil fumarate single-tablet regimen (Atripla®). Drugs. 2010;70:2315–2338.10.2165/11203800-000000000-00000
- Aldir I, Horta A, Serrado M. Single-tablet regimens in HIV: does it really make a difference? Curr Med Res Opin. 2013;30:89–97.
- Hanna DB, Hessol NA, Golub ET, et al. Increase in single-tablet regimen use and associated improvements in adherence-related outcomes in HIV-infected women. J Acquir Immune Defic Syndr. 2014;65:587–596.10.1097/QAI.0000000000000082
- Shaw J-P, Sueoka CM, Oliyai R, et al. Metabolism and pharmacokinetics of novel oral prodrugs of 9-[(R)-2-(phosphonomethoxy)propyl]adenine (PMPA) in dogs. Pharm Res. 1997;14:1824–1829.10.1023/A:1012108719462
- Arimilli M, Kim C, Bischofberger N. Synthesis, in vitro biological evaluation and oral bioavailability of 9-[2-(phosphonomethoxy)propyl]adenine (PMPA) prodrugs. Antivir Chem Chemother. 1997;8:557–564.
- Naesens L, Bischofberger N, Augustijns P, et al. Antiretroviral efficacy and pharmacokinetics of oral bis(isopropyloxycarbonyloxymethyl)-9(2-phosphonylmethoxypropyl) adenine in mice. Antimicrob Agents Chemother. 1998;42:1568–1573.
- Suo Z, Johnson KA. Selective inhibition of HIV-1 reverse transcriptase by an antiviral inhibitor, (R)-9-(2-Phosphonylmethoxypropyl) adenine. J Biol Chem. 1998;273:27250–27258.10.1074/jbc.273.42.27250
- Robbins BL, Greenhaw JJ, Connelly MC, Fridland A. Metabolic pathways for activation of the antiviral agent 9-(2-phosphonylmethoxyethyl)adenine in human lymphoid cells. Antimicrob Agents Chemother. 1995;39:2304–2308.10.1128/AAC.39.10.2304
- Kohler JJ, Hosseini SH, Green E, et al. Tenofovir renal proximal tubular toxicity is regulated by OAT1 and MRP4 transporters. Lab Invest. 2011;91:852–858.10.1038/labinvest.2011.48
- Rodriguez-Nóvoa S, Alvarez E, Labarga P, Soriano V. Renal toxicity associated with tenofovir use. Expert Opin Drug Saf. 2010;9:545–559.10.1517/14740331003627458
- Lee WA, He G-X, Eisenberg E, et al. Selective intracellular activation of a novel prodrug of the human immunodeficiency virus reverse transcriptase inhibitor tenofovir leads to preferential distribution and accumulation in lymphatic tissue. Antimicrob Agents Chemother. 2005;49:1898–1906.10.1128/AAC.49.5.1898-1906.2005
- Eisenberg EJ, He G-X, Lee WA. Metabolism Of Gs-7340, A novel phenyl monophosphoramidate intracellular prodrug of PMPA, in blood. Nucleosides Nucleotides Nucleic Acids. 2001;20:1091–1098.10.1081/NCN-100002496
- Ruane PJ, DeJesus E, Berger D, et al. Antiviral activity, safety, and pharmacokinetics/pharmacodynamics of tenofovir alafenamide as 10-day monotherapy in HIV-1-positive adults. J Acquir Immune Defic Syndr. 2013;63:449–455.10.1097/QAI.0b013e3182965d45
- Callebaut C, Stepan G, Tian Y, Miller MD. In vitro virology profile of tenofovir alafenamide, a novel oral prodrug of tenofovir with improved antiviral activity compared to tenofovir disoproxil fumarate. Antimicrob Agents Chemother 2015;59:5909–5916.
- Reich M, Spindler KD, Burret M, Kalbacher H, Boehm BO, Burster T. Cathepsin A is expressed in primary human antigen-presenting cells. Immunol. Lett. 2010;128:143–147.10.1016/j.imlet.2009.11.010
- Birkus G, Kutty N, He GX, et al. Activation of 9-[(R)-2-[[(S)-[[(S)-1-(Isopropoxycarbonyl)ethyl]amino] phenoxyphosphinyl]-methoxy]propyl]adenine (GS-7340) and other tenofovir phosphonoamidate prodrugs by human proteases. Mol Pharmacol. 2008;74:92–100.10.1124/mol.108.045526
- Murakami E, Wang T, Park Y, et al. Implications of efficient hepatic delivery by tenofovir alafenamide (GS-7340) for hepatitis B virus therapy. Antimicrob Agents Chemother. 2015;59:3563–3569.10.1128/AAC.00128-15
- Sax PE, Zolopa A, Brar I, et al. Tenofovir alafenamide vs. tenofovir disoproxil fumarate in single tablet regimens for initial HIV-1 therapy: a randomized phase 2 study. J Acquir Immune Defic Syndr. 2014;67:52–58.10.1097/QAI.0000000000000225
- Sax PE, Wohl D, Yin MT, et al. Tenofovir alafenamide versus tenofovir disoproxil fumarate, coformulated with elvitegravir, cobicistat, and emtricitabine, for initial treatment of HIV-1 infection: two randomised, double-blind, phase 3, non-inferiority trials. Lancet. 2015;385:2606–2615.
- Markowitz M, Zolopa A, Squires K, et al. Phase I/II study of the pharmacokinetics, safety and antiretroviral activity of tenofovir alafenamide, a new prodrug of the HIV reverse transcriptase inhibitor tenofovir, in HIV-infected adults. J Antimicrob Chemother. 2014;69:1362–1369.10.1093/jac/dkt532
- Petropoulos CJ, Parkin NT, Limoli KL, et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–928.10.1128/AAC.44.4.920-928.2000
- Varghese V, Liu TF, Rhee SY, et al. HIV-1 integrase sequence variability in antiretroviral naive patients and in triple-class experienced patients subsequently treated with raltegravir. AIDS Res Hum Retroviruses. 2010;26:1323–1326.10.1089/aid.2010.0123
- Wensing AM, Calvez V, Gunthard HF, et al. 2014 update of the drug resistance mutations in HIV-1. Topics in antiviral med. 2014;22:642–650.
- Yang X, Charlebois P, Gnerre S, et al. De novo assembly of highly diverse viral populations. BMC Genomics. 2012;13:475.10.1186/1471-2164-13-475
- Lee WP, Stromberg MP, Ward A, Stewart C, Garrison EP, Marth GT. MOSAIK: a hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS ONE. 2014;9:e90581.10.1371/journal.pone.0090581
- Hertogs K, Bloor S, De Vroey V, et al. A novel human immunodeficiency virus type 1 reverse transcriptase mutational pattern confers phenotypic lamivudine resistance in the absence of mutation 184 V. Antimicrob Agents Chemother. 2000;44:568–573.10.1128/AAC.44.3.568-573.2000
- Taniguchi T, Nurutdinova D, Grubb JR, et al. Transmitted drug-resistant HIV type 1 remains prevalent and impacts virologic outcomes despite genotype-guided antiretroviral therapy. AIDS Res Hum Retroviruses. 2012;28:259–264.10.1089/aid.2011.0022
- Gupta R, Hill A, Sawyer AW, Pillay D. Emergence of drug resistance in HIV Type 1–infected patients after receipt of first‐line highly active antiretroviral therapy: a systematic review of clinical trials. Clinical Infectious Diseases. 2008;47:712–722.10.1086/592125
- Messou E, Chaix ML, Gabillard D, et al. Increasing rate of TAMs and etravirine resistance in HIV-1-infected adults between 12 and 24 months of treatment: the VOLTART cohort study in Cote d’Ivoire, West Africa. J Acquir Immune Defic Syndr 2013;64:211–219.10.1097/QAI.0b013e3182a009e4
- White KL, Raffi F, Miller MD. Resistance analyses of integrase strand transfer inhibitors within phase 3 clinical trials of treatment-naive patients. Viruses. 2014;6:2858–2879.10.3390/v6072858
- Margot NA, Johnson A, Miller MD, Callebaut C. Characterization of HIV-1 resistance to tenofovir alafenamide In Vitro. Antimicrob Agents Chemother. 2015;59:5917–5924.10.1128/AAC.01151-15
