Abstract
Oncogene MYC is deregulated in many human cancers, especially in lymphoma. Previously, we showed that inauhzin (INZ) activates p53 and inhibits tumor growth. However, whether INZ could suppress cancer cell growth independently of p53 activity is still elusive. Here, we report that INZ(c), a second generation of INZ, suppresses c-Myc activity and thus inhibits growth of human lymphoma cells in a p53-independent manner. INZ(c) treatment decreased c-Myc expression at both mRNA and protein level, and suppressed c-Myc transcriptional activity in human Burkitt's lymphoma Raji cells with mutant p53. Also, we showed that overexpressing ectopic c-Myc rescues the inhibition of cell proliferation by INZ(c) in Raji cells, implicating c-Myc activity is targeted by INZ(c). Interestingly, the effect of INZ(c) on c-Myc expression was impaired by disrupting the targeting of c-Myc mRNA by miRNAs via knockdown of ribosomal protein (RP) L5, RPL11, or Ago2, a subunit of RISC complex, indicating that INZ(c) targets c-Myc via miRNA pathways. These results reveal a new mechanism that INZ(c) targets c-Myc activity in human lymphoma cells.
Abbreviations
| Dox | = | doxorubicin |
| FACS | = | Fluorescence-activated cell sorting |
| GTP | = | guanosine triphosphate |
| INZ | = | inauhzin |
| MTT | = | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| PBS | = | Phosphate Buffered Saline |
| PI | = | propidium iodide |
| q-RT-PCR | = | Real-time reverse transcription polymerase chain reaction |
| RISC | = | RNA-induced silencing complex |
| RP | = | ribosomal protein |
| UTR | = | untranslated region |
Introduction
c-Myc is an oncoprotein that transcriptionally regulates the expression of numerous genes involved in cell growth and proliferation.Citation1,2 Therefore, in normal cells c-Myc expression is highly regulated by multiple mechanisms, and deregulation of c-Myc is identified in many cancers.Citation3-9 As c-Myc activation has been reported to be required for a number of essential cellular processes, such as ribosome biogenesis, metabolic adaptation, cell survival, and cell division, which are vital for the growth of cancer cells, it is believed that inhibition of c-Myc activation could suppress cancer development.Citation1,10 Indeed, a number of studies have shown that tumor aggression and poor clinical outcome correlate with amplification of c-Myc by mutations or chromosomal abnormities in patients.Citation11-14 In addition, while growth factors are usually required for proliferation of normal cells, it has been reported that cancer cells with abnormal high level of c-Myc expression manage to grow and proliferate without growth factor stimulation, suggesting c-Myc could be a great therapeutic target for cancers with overexpression of c-Myc, such as lymphoma.Citation13,14,17-20
Previously, we identified a small molecule called Inauhzin (INZ) that could activate p53 by suppressing SIRT1 activity, leading to cell cycle arrest and apoptosis of cancer cells without introducing detectable DNA damage to cells.Citation15 Transcriptome analysis of cells treated with INZ showed that over 200 p53 targets were induced by INZ, suggesting INZ is indeed a p53 activator.Citation16 Also, we showed that INZ could activate p53 synergistically with Nutlin-3, an Mdm2 inhibitor, and sensitize cancer cells to chemotherapeutic drugs, such as cisplatin and doxorubicin.Citation17,18 In addition, in xenograft mouse model, INZ treatment reduced tumor size and promoted survival of tumor-bearing mice, suggesting INZ suppresses tumorigenesis and could be a potential drug for cancer therapy.Citation15 Very recently, we found that INZ can cause ribosomal stress by inhibiting cellular IMPDH2 activity and reducing cellular GTP level, consequently decreasing the level of nucleostemin,Citation19 a nucleolar GTP-binding protein important for rRNA processing.Citation20,21 This study provides a second mechanism for INZ activation of p53.Citation19 However, whether INZ could inhibit cancer development by targeting other oncoproteins remains unknown.
Here, we show that INZ(c), a second generation of INZ with better bioavailability and potency,Citation22 suppresses c-Myc expression and thus inhibits growth and proliferation of both of the human wild type p53-containing and deficient lymphoma cells. Interestingly, disrupting miRNA function in these cells abrogated the inhibitory effect of INZ on c-Myc expression, indicating that INZ(c) targets c-Myc via miRNAs. Hence, these findings demonstrate a new mechanism that INZ(c) could inhibit cancer cell growth by targeting c-Myc in a p53-independent fashion.
Results
INZ(c) suppresses c-Myc expression in a p53-independent manner
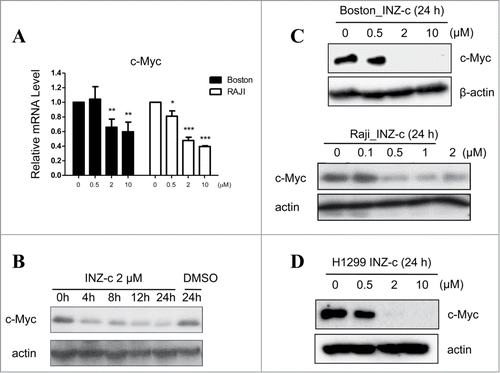
Previously, we reported that INZ and its analogs activate p53 and thus inhibit cancer cell growth.Citation15-18,22 However, whether INZ could function to suppress the expression of c-Myc and inhibit lymphoma cell growth remains unknown. To this end, we treated human lymphoma Boston cells with INZ(c), an INZ analog with better potency.Citation22 As shown in , INZ(c) treatment dramatically decreased c-Myc mRNA level in a dose dependent fashion. As p53 had been reported to suppress c-Myc expression via miRNAs,Citation23 we next tested whether INZ(c) could suppress c-Myc expression in human lymphoma Raji cells without active p53 to see if p53 is dispensable for this regulation. As shown in , INZ(c) also inhibited the expression of c-Myc mRNA in Raji cells, indicating the targeting of c-Myc by INZ(c) is independent of p53 activity in cells. In addition, our results showed that c-Myc protein expression is suppressed dramatically by INZ(c) in Boston (with wild-type p53), Raji (with mutant p53), and H1299 (without p53) cells (). These findings suggest that INZ(c) suppresses c-Myc expression in a p53-independent manner.
Figure 1. INZ(c) inhibits c-Myc expression. (A) INZ(c) treatment decreases c-Myc mRNA level. H1299 cells were treated with various concentrations of INZ(c) for 24 h. c-Myc mRNA was determined by q-RT-PCR. (B) INZ(c) reduces c-Myc protein level. H1299 cells were treated with 2 μM INZ(c) for 0, 4, 8, 12 or 24 h. (C) Suppression of c-Myc expression by INZ(c) is independent of p53 activity. Boston and RAJI cells were treated with various concentrations of Inauhzin-C for 24 h. Cell lysates were prepared and subjected to Western blotting for c-Myc. (D) H1299 cells were treated with various concentrations of Inauhzin-C for 24 h. Western blotting was conducted to determine expression of c-Myc.
INZ(c) suppresses growth of lymphoma cells
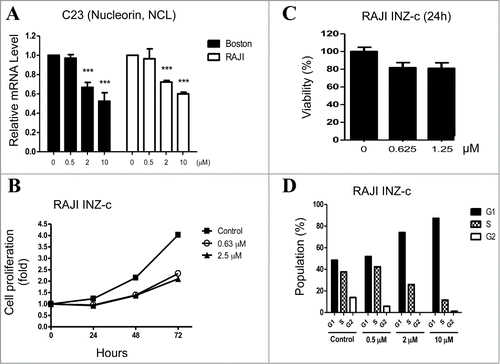
As c-Myc is deregulated in many cancers especially in lymphoma,Citation3-9 we next tested whether INZ(c) could inhibit lymphoma cell growth by targeting c-Myc. c-Myc transcriptionally regulates the expression of a large number of genes and promotes cell proliferation.Citation1,2 To determine whether c-Myc transcriptional activity is suppressed by INZ(c), we first carried out q-RT-PCR to test the effect of INZ(c) on the expression of C23, a known c-Myc target.Citation24 As expected, INZ(c) treatment decreased C23 mRNA level significantly in both Boston and Raji cells (), indicating INZ(c) indeed suppresses c-Myc transcriptional activity. Since c-Myc transcriptional activity is important for proliferation of cancer cells,Citation1,2 we speculated that INZ(c) might inhibit cell growth by targeting c-Myc. As shown in , a low concentration (0.63 μM) of INZ(c) dramatically inhibited the proliferation of Raji cells. This result was confirmed by the MTT assay that showed INZ(c) suppresses cell viability of Raji cells too (). To further validate these results, we carried out FACS analysis and showed that INZ(c) treatment decreases the number of cells in S phase and arrests cells at G1 phase dose-dependently (), which is in line with literature showing c-Myc activity is required for cells to enter S phase.Citation25 These results show that INZ(c) inhibits the growth of lymphoma cells.
Figure 2. INZ(c) inhibits c-Myc transcriptional activity and suppresses cell growth. (A) INZ(c) suppresses c-Myc transcription activity. Total RNAs were isolated from Boston and Raji cells treated with indicated concentrations of INZ(c) for 24 hours. C23 mRNA level was dertermined by q-RT-PCR. Data represent means ± SD. (B) INZ(c) inhibits cell Proliferation. Proliferation assay was carried out for Raji cells treated with 0, 0.63, or 2.5 μINZ(c) at indicated time points. Data represent means ± SD. (C) INZ(c) decreases viability of Raji cells. Raji cells were treated with indicated concentrations of INZ(c) and then subjected to cell viability assay. Data represent means ± SD. (D) INZ(c) decreases the number of cells in S phase. Raji cells were treated with indicated concentration of INZ(c) and subjected to flow cytometer for cell cycle analysis.
Overexpressing ectopic c-Myc rescues the inhibitory effect of INZ(c) on the growth of lymphoma cells
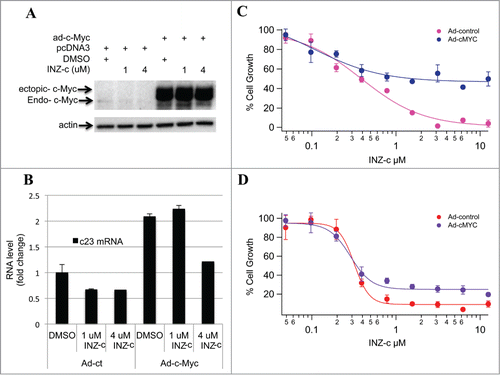
To investigate whether ectopic c-Myc could rescue the effect of INZ(c) on cell growth, we treated cells with INZ(c) after overexpressing c-Myc () and found that c-Myc transcriptional activity is restored to normal level after INZ(c) treatment (). It is rational to assume that the inhibitory effect of INZ(c) on cell growth should be rescued by overexpressing c-Myc, if the inhibition of cell growth were due to the decreasing of c-Myc activity by INZ(c). Indeed, as shown in , restoration of c-Myc expression in both Boston and Raji cells significantly impaired the inhibition of cell growth by INZ(c) treatment, suggesting INZ(c) suppresses lymphoma cell growth by targeting c-Myc.
Figure 3. Ectopic c-Myc rescues the inhibitory effect of INZ(c) on cell growth. (A) H1299 cells were treated with ad-c-Myc as indicated and treated with INZ(c) for 24 h. Cell lysates were prepared and subjected to Western blotting for c-Myc and actin. (B) Overexpression of c-Myc rescues the inhibitory effect of INZ(c) on c-Myc activity. Cells were treated with ad-c-Myc as indicated and incubated with INZ(c) for 24 h. Real-time PCR was performed to determine c23 mRNA. (C) and (D) Ectopic c-Myc rescues the inhibitory effect of INZ(c) on cell growth. Boston (C) and Raji (D) Cells were treated with ad-c-Myc as indicated and incubated with INZ(c) for 24 hours. Cells were then harvested and subject to WST cell growth assay. Values represent means ± SD (n = 3).
INZ(c) targets c-Myc via miRNAs
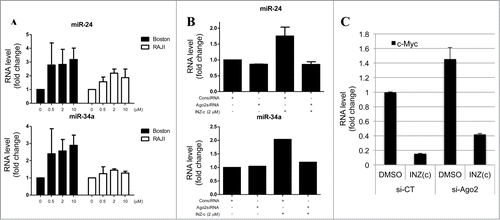
Interestingly, we found that while INZ(c) suppresses the expression of endogenous c-Myc, the level of ectopic c-Myc expressed by transfecting cells with recombination DNA is not affected by INZ(c) treatments (), suggesting that INZ(c) might target c-Myc via miRNAs, as recombinant DNA does not have the 3′ UTR where most miRNAs target. To test this hypothesis, we treated both Boston and Raji cells with INZ(c), and checked the expression of miR-24 and miR-34a, which have been reported to target c-Myc.Citation26,27 Intriguingly, both miR-24 and miR-34a were induced by INZ(c) treatment dose-dependently (), indicating that INZ(c) might target c-Myc via regulating the functions of miRNAs. Next, we knocked down Ago2, a component of RISC complex that is vital for miRNA processing and function,Citation28-30 to see whether disrupting miRNA functions in cells could affect the regulation of c-Myc by INZ(c). As shown in , the induction of miR-24 and miR-34a by INZ(c) was abrogated by Ago2 knockdown, which is consistent with literature showing that knocking down RISC complex could affect the level of mature miRNAs.Citation31,32 Interestingly, Ago2 knockdown impaired the targeting of c-Myc by INZ(c) as shown in , suggesting INZ(c) regulates c-Myc expression via the miRNA pathway.
Figure 4. Inauhzin-C targets c-Myc via miRNAs. (A) INZ(c) increases miR-24 and miR-34a level. Boston and Raji cells were treated with INZ(c) for 24 hours. RNA was isolated and subjected to q-RT-PCR to determine miR-24 and miR-34a level. (B) Ago2 knockdown abrogates the induction of miR-24 and miR-34a by INZ(c). Cells were treated with INZ(c) after incubating with Ago2 siRNA for 48 hours. Real-time PCR was performed to determine the level of miR-24 and miR-34a. (C) Ago2 knockdown impairs the inhibitory effect of INZ(c) on c-Myc expression. H1299 cells treated with indicated siRNAs and drugs were harvested for q-RT-PCR assay. Data are presented as means ± SD.
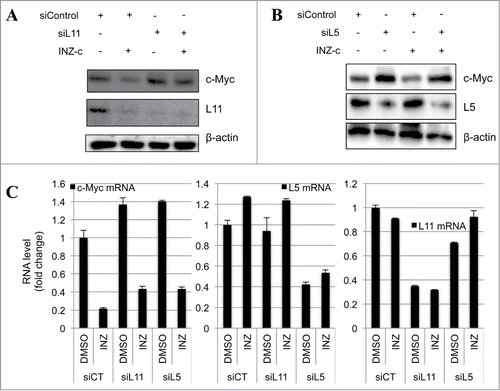
RPL5 and RPL11 are required for the targeting of c-Myc by INZ(c)
Recently, both our lab and another group have reported that RPL5 and RPL11 can recruit a miRNA complex that binds to and inhibits c-Myc mRNA.Citation33,34 To further confirm that INZ(c) regulates c-Myc expression through a miRNA pathway, we next tested whether RPL5 and RPL11 are required for this regulation. To this end, RPL5 and RPL11 were knocked down in cells, and the expressions of c-Myc protein and mRNA were determined by western blot and q-RT-PCR, respectively. As expected, knocking down either RPL11 or RPL5 rescued the targeting of c-Myc by INZ(c) at both protein and mRNA levels (), confirming that the RPL11/RPL5-miRNA axis is indispensible for the inhibitory effect of INZ(c) on c-Myc expression. Taken together, our results reveal a new pathway that INZ(c) utilizes to target c-Myc expression and inhibit cancer cell growth.
Figure 5. RPL11 and RPL5 are required for the targeting of c-Myc by INZ(c). (A) and (B) Knocking down RPL11 (A) and RPL5 (B) rescues the suppression of c-Myc protein expression by INZ(c). H1299 cells transfected with indicated plasmids and harvested for WB analysis to check the expression of c-Myc, RPL11, RPL5, and actin. (C) Knocking down RPL11 or RPL5 impairs the inhibitory effect of INZ(c) on c-Myc mRNA expression. H1299 cells treated with indicated drugs and siRNAs were harvested and subjected to qRT-PCR analysis.
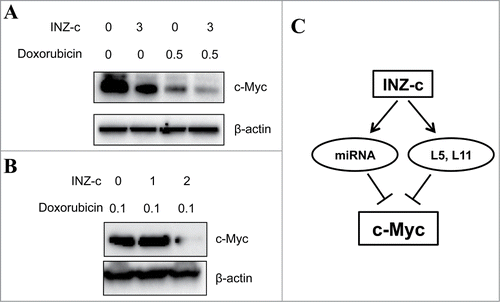
INZ(c) cooperatively suppresses c-Myc expression with doxorubicin
We previously showed that INZ does not introduce noticeable DNA damageCitation15 and could sensitize p53-dependent cytotoxicity and tumor suppression of chemotherapeutic agents, such as doxorubicin (Dox).Citation17 Therefore, we next checked whether INZ(c) could cooperatively target c-Myc expression with doxorubicin, as doxorubicin had been shown to suppress c-Myc expression.Citation35 To test this hypothesis, we treated cells with INZ(c) alone, Dox alone, or both of them, and observed that INZ(c) cooperatively inhibited c-Myc expression with Dox (), suggesting INZ(c) could sensitize cytotoxicity of Dox by targeting c-Myc expression. These results implicate that INZ(c) could be used to decrease the dose of chemotherapeutic drugs and reduce DNA damage to normal tissues during chemotherapy.
Figure 6. INZ(c) cooperatively decreases c-Myc expression with doxorubicin. (A) H1299 cells were treated both INZ(c) and Doxorubicin for 24 h and subjected to Western blotting for c-Myc and actin. (B) Raji cells were treated both INZ(c) and Doxorubicin for 24 h and subjected to Western blotting for c-Myc and actin. (C) A schematic model for the targeting of c-Myc by INZ(c).
Discussion
c-Myc deregulation is highly associated with lymphoma development, which suggests that targeting c-Myc could be a good therapeutic strategy for the management of lymphoma patients.Citation13,14,17-20 Previously we showed that INZ activates p53 and functions to suppress tumorigenesis without introducing DNA damage to cells.Citation15-18,22 However, whether INZ(c) could inhibit cancer cell growth independently of p53 is still unknown. In this study, we identified that INZ(c) suppresses c-Myc expression and inhibits cell growth and proliferation of lymphoma cells via a miRNA pathway. First, we showed that INZ(c) suppresses the expression of c-Myc at both mRNA and protein levels in lymphoma cells with or without wild type p53 (). Consistently, we found that INZ(c) treatment decreases c-Myc transcriptional activity and suppresses growth of lymphoma cells (). Also, intriguingly, putting back ectopic c-Myc into lymphoma cells rescued the inhibitory effect of INZ(c) on cell growth, suggesting that the c-Myc pathway is indeed targeted by INZ(c) (). In addition, we found that the targeting of c-Myc by miRNA is indispensable for the effect of INZ(c) on c-Myc expression, for knocking down Ago2 or ribosomal proteins, RPL5 and RPL11, which had been shown to facilitate the targeting of c-Myc by miRNAs,Citation28,29,33,34 impaired the regulation of c-Myc by INZ(c) (). Finally, we uncovered that INZ(c) and doxorubicin act together to repress c-Myc expression cooperatively (). These results demonstrate that INZ(c) could suppress lymphoma cell growth independent of p53 by inhibiting c-Myc expression ().
Both overexpression of c-Myc and disruption of p53 normal activity are highly associated with human cancers.Citation36-38 It is therefore logical to assume that developing drugs targeting both pathways simultaneously might provide better outcomes for cancer therapy. Interestingly, a number of proteins, including ARF, RPL11, and RPL5,Citation24,33,34,39-48 had been identified to target both c-Myc and p53 pathways in cells, suggesting that screening drugs that target these 2 pathways is possible. Recently, we also identified IMPDH2 as another target of INZ, and inhibiting cellular IMPDH2 activity led to the decrease of cellular GTP and nucleostemin levels, consequently causing ribosomal stress that leads to p53 activation.Citation19 Consistent with this recent work,Citation19 our study presented here also demonstrated the requirement of RPL11 and RPL5 for INZ(c) inactivation of c-Myc (), suggesting that INZ might also inactivate c-Myc by inducing ribosomal stress. This study together with our previously and recently published dataCitation15-19,22 show that INZ(c) can not only activate p53 and suppress tumorigenesis of cancers with wild type p53, but also repress c-Myc expression and inhibit growth of cancer cells with overexpression of c-Myc.
Cancer cells often become resistant to DNA damaging agents during chemotherapy, especially when tumor cells are treated with high concentrations of drugs.Citation49-52 One of the mechanisms underlying chemoresistance is that cancer cells manage to promote c-Myc expression after exposure to chemotherapeutic drugs, indicating elevated c-Myc expression might be at least partially responsible for chemoresistance of cancers.Citation53-55 Here we showed that INZ(c) represses c-Myc expression via miRNA pathways, providing a potential drug to inhibit or delay drug resistance of cancer cells during chemotherapy. Further studies on the effect of INZ(c) on the growth of drug-resistant cancer cells would provide more information about how INZ(c) functions to facilitate or re-sensitize drug-resistant cancer cells to chemotherapeutic agents.
Materials and Methods
Cell culture
Boston and RAJI cells were maintained in RPMI1640 medium supplemented 10% fetal bovine serum (FBS), 50 U/ml penicillin and 0.1 mg/ml streptomycin at 37°C in a 5% CO2-humidified atmosphere. H1299 and MCF7 cells were maintained in DMEM medium supplemented 10% fetal bovine serum (FBS), 50 U/ml penicillin and 0.1 mg/ml streptomycin at 37°C in a 5% CO2-humidified atmosphere. Adenoviruses to overexpress full-length c-Myc or control GFP were described previously.Citation24,56
Cytotoxicity assay
Cytotoxic effects of Inauhzin-c Boston and Raji cells were evaluated by cell counting kit (Dojindo Molecular Technologies Inc.., Gaithersburg, Maryland). Cells were seeded onto 96-well micro plates at a density of 2 × 104 cells per well and exposed to various concentrations of Inauhzin-c for 24 h. The cells were incubated WST-8 at a final concentration of 10% to each well and incubate for 2 h. Optical density (OD) was measured using a micro plate reader (Molecular Device, SpectraMax M5e) at 450 nm. Cell viability was calculated as a percentage of viable cells in drug-treated group versus untreated control by following equation.
Cell viability (%) = [OD (Drug)−OD (Blank)]/[OD (Control)−OD (Blank)]×100
Western blot analysis
Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM EDTA, 1 mM Na3VO4, 1 mMNaF, protease inhibitors cocktail). The extracts were incubated on ice for 30 min and supernatants were collected by centrifugation at 14,000g at 4ºC. Proteins were separated by electrophoresis on 10-15% SDS-PAGE gel and transferred onto membrane with transfer buffer (25 mMTris, 250 mM glycine, 15% methanol) at 15V, 1.0A for 35 min. The membrane was blocked in 5% nonfat skim milk, and probed with primary antibodies for c-Myc (Abcam, Cambridge, United Kingdom), Cyclin D1, Cyclin E, and LC3A/B (Cell Signaling Tech., Danvers, MA), SIRT1 and RPS14 (Santa Cruz Biotechnologies, Santa Cruz, CA), RPL5 and RPL11 antibodies have been described.Citation24,44
Cell cycle analysis
Cell cycle analysis was performed by PI staining. Boston and Raji cells were treated with Inauhzin-c for 24 h, collected and fixed in 75% ethanol. The cells were then incubated at 37°C with 0.1% RNaseA in PBS for 30 min and suspended in PBS containing 25 μg/ml PtdIns for 30 min at room temperature. The stained cells were analyzed for DNA content in FACSCalibur (Becton Dickinson, Franklin Lakes, NJ) using the Cell Quest program (Becton Dickinson, Franklin Lakes, NJ).
Reverse transcription (RT) and quantitative (q) PCR analysis
Total RNA was isolated from cells using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. A reverse transcription kit (Promega, Fitchburg, WI) was used to construct the template cDNA. Quantitative PCR (qPCR) was conducted using SYBR green mis according to the manufacturer's protocol (Bio-Rad, Hercules, CA). Primers for RPL5 and RPL11 have been previously described.Citation24,56
RNA interference and plasmids
The siRNA pool against RPL11, RPL5, RPS14 and Ago2 (Santa Cruz Biotechnology) were purchased. 40 nM of siRNAs were introduced into cells using Turbofect siRNA transfection reagent. Cells were incubated 48 h and then treated by Inauhzin-c for 24 h. After treated, cells were harvested.
Reverse transcriptase-polymerase chain reaction and quantitative real-time PCR analysis
RT and Q-PCR for mRNAs were done by using the methods described previously.Citation44,56 Briefly, quantitative real-time PCR was performed on an ABI 7300 real-time PCR system (Applied Biosystems) using SYBR Green Mix (Applied Biosystems). Relative gene expression was calculated using the C method, following the manufacturer's instruction. All reactions were carried out in triplicate.
Knockdown of the endogenous mRNAs
siRNAs for RPL5 and RPL11 were described previously.Citation24,44 siRNA for Ago2 was purchased from Santa Cruz Biotechnology. Transfection of siRNAs was performed the same as that of normal siRNA as described previouslyCitation57 by using siLentFectTM Lipid (Bio-Rad), following the manufacturer's protocol.
Statistical Analysis
All data were presented as means ± standard deviation (S.D). Statistical significance was verified by Student's t-test using Sigmaplot software (Systat Software Inc., San Jose, CA).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by the Lady Leukemia League fund in Metairie, LA, and NIH-NCI grants CA095441, and CA172468 to H.L. SHK was in part supported by the Korea government grant [MEST] (No. 2012-0005755).
References
- Dang C, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res 2009; 15:6479-83; PMID:19861459; http://dx.doi.org/10.1158/1078-0432.CCR-09-0889
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012; 151:68-79; PMID:23021216; http://dx.doi.org/10.1016/j.cell.2012.08.033
- Verbeek S, van Lohuizen M, van der Valk M, Domen J, Kraal G, Berns A. Mice bearing the E mu-myc and E mu-pim-1 transgenes develop pre-B-cell leukemia prenatally. Mol Cell Biol 1991; 11:1176-9; PMID:1990273
- Soucek L, Whitfield J, Martins C, Finch A, Murphy D, Sodir N, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature 2008; 455:679-83; PMID:18716624; http://dx.doi.org/10.1038/nature07260
- Sodir N, Swigart L, Karnezis A, Hanahan D, Evan G, Soucek L. Endogenous Myc maintains the tumor microenvironment. Genes Dev 2011; 25:907-16; PMID:21478273; http://dx.doi.org/10.1101/gad.2038411
- Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell 1999; 3:565-77; PMID:10360173; http://dx.doi.org/10.1016/S1097-2765(00)80350-0
- Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 2002; 297:102-4; PMID:12098700; http://dx.doi.org/10.1126/science.1071489
- Flores I, Murphy D, Swigart L, Knies U, Evan G. Defining the temporal requirements for Myc in the progression and maintenance of skin neoplasia. Oncogene 2004; 23:5923-30; PMID:15208685; http://dx.doi.org/10.1038/sj.onc.1207796
- Felsher D, Bishop J. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell 1999; 4:199-207; PMID:10488335; http://dx.doi.org/10.1016/S1097-2765(00)80367-6
- Kim J, Chu J, Shen X, Wang J, Orkin S. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 2008; 132:1049-61; PMID:18358816; http://dx.doi.org/10.1016/j.cell.2008.02.039
- Wright J, Brown S, Cole M. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol 2010; 30:1411-20; PMID:20065031; http://dx.doi.org/10.1128/MCB.01384-09
- Pomerantz M, Ahmadiyeh N, Jia L, Herman P, Verzi M, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 2009; 41:882-4; PMID:19561607; http://dx.doi.org/10.1038/ng.403
- Meyer N, Penn L. Reflecting on 25 years with MYC. Nat Rev Cancer 2008; 8:976-90; PMID:19029958; http://dx.doi.org/10.1038/nrc2231
- Eilers M, Eisenman R. Myc's broad reach. Genes Dev 2008; 22:2755-66; PMID:18923074; http://dx.doi.org/10.1101/gad.1712408
- Zhang Q, Zeng SX, Zhang Y, Zhang Y, Ding D, Ye Q, Meroueh SO, Lu H. A small molecule Inauhzin inhibits SIRT1 activity and suppresses tumour growth through activation of p53. EMBO Mol Med 2012; 4:298-312; PMID:22331558; http://dx.doi.org/10.1002/emmm.201100211
- Liao JM, Zeng SX, Zhou X, Lu H. Global effect of inauhzin on human p53-responsive transcriptome. PloS One 2012; 7:e52172; PMID:23284922; http://dx.doi.org/10.1371/journal.pone.0052172
- Zhang Y, Zhang Q, Zeng SX, Hao Q, Lu H. Inauhzin sensitizes p53-dependent cytotoxicity and tumor suppression of chemotherapeutic agents. Neoplasia 2013; 15:523-34; PMID:23633924
- Zhang Y, Zhang Q, Zeng SX, Zhang Y, Mayo LD, Lu H. Inauhzin and Nutlin3 synergistically activate p53 and suppress tumor growth. Cancer Biol Ther 2012; 13:915-24; PMID:22785205; http://dx.doi.org/10.4161/cbt.20844
- Zhang Q, Zhou X, Wu R, Mosley A, Zeng SX, Xing Z, Lu H. The role of IMP dehydrogenase 2 in Inauhzin-induced ribosomal stress. Elife 2014; 3; e03077; PMID:25347121
- Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol 2008; 28:4365-76; PMID:18426907; http://dx.doi.org/10.1128/MCB.01662-07
- Lo D, Dai MS, Sun XX, Zeng SX, Lu H. Ubiquitin- and MDM2 E3 ligase-independent proteasomal turnover of nucleostemin in response to GTP depletion. J Biol Chem 2012; 287:10013-20; PMID:22318725; http://dx.doi.org/10.1074/jbc.M111.335141
- Zhang Q, Ding D, Zeng SX, Ye QZ, Lu H. Structure and activity analysis of Inauhzin analogs as novel antitumor compounds that induce p53 and inhibit cell growth. PloS One 2012; 7:e46294; PMID:23115626; http://dx.doi.org/10.1371/journal.pone.0046294
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A 2009; 106:3207-12; PMID:19202062; http://dx.doi.org/10.1073/pnas.0808042106
- Dai MS, Arnold H, Sun XX, Sears R, Lu H. Inhibition of c-Myc activity by ribosomal protein L11. EMBO J 2007; 26:3332-45; PMID:17599065; http://dx.doi.org/10.1038/sj.emboj.7601776
- Heikkila R, Schwab G, Wickstrom E, Loke SL, Pluznik DH, Watt R, Neckers LM. A c-myc antisense oligodeoxynucleotide inhibits entry into S phase but not progress from G0 to G1. Nature 1987; 328:445-9; PMID:3302722; http://dx.doi.org/10.1038/328445a0
- Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund AH. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differe 2010; 17:236-45; PMID:19696787; http://dx.doi.org/10.1038/cdd.2009.109
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to "seedless" 3'UTR microRNA recognition elements. Mol Cell 2009; 35:610-25; PMID:19748357; http://dx.doi.org/10.1016/j.molcel.2009.08.020
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005; 436:740-4; PMID:15973356; http://dx.doi.org/10.1038/nature03868
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 2005; 6:961-7; PMID:16142218; http://dx.doi.org/10.1038/sj.embor.7400509
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005; 123:607-20; PMID:16271386; http://dx.doi.org/10.1016/j.cell.2005.08.044
- Gerin I, Bommer GT, McCoin CS, Sousa KM, Krishnan V, MacDougald OA. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab 2010; 299:E198-206; PMID:20484008
- Huang V, Zheng J, Qi Z, Wang J, Place RF, Yu J, Li H, Li LC. Ago1 Interacts with RNA polymerase II and binds to the promoters of actively transcribed genes in human cancer cells. PLoS Genet 2013; 9:e1003821; PMID:24086155; http://dx.doi.org/10.1371/journal.pgen.1003821
- Challagundla KB, Sun XX, Zhang X, DeVine T, Zhang Q, Sears RC, Dai MS. Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol Cell Biol 2011; 31:4007-21; PMID:21807902; http://dx.doi.org/10.1128/MCB.05810-11
- Liao JM, Zhou X, Gatignol A, Lu H. Ribosomal proteins L5 and L11 co-operatively inactivate c-Myc via RNA-induced silencing complex. Oncogene 2014; 33:4916-23; PMID:24141778; http://dx.doi.org/10.1038/onc.2013.430
- Frenzel A, Zirath H, Vita M, Albihn A, Henriksson MA. Identification of cytotoxic drugs that selectively target tumor cells with MYC overexpression. PloS One 2011; 6:e27988; PMID:22132187; http://dx.doi.org/10.1371/journal.pone.0027988
- Cole MD. The myc oncogene: its role in transformation and differentiation. Ann Rev Genet 1986; 20:361-84; PMID:3028245; http://dx.doi.org/10.1146/annurev.ge.20.120186.002045
- Dai MS, Jin Y, Gallegos JR, Lu H. Balance of Yin and Yang: ubiquitylation-mediated regulation of p53 and c-Myc. Neoplasia 2006; 8:630-44; PMID:16925946; http://dx.doi.org/10.1593/neo.06334
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000; 408:307-10; PMID:11099028; http://dx.doi.org/10.1038/35042675
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 1998; 92:725-34; PMID:9529249; http://dx.doi.org/10.1016/S0092-8674(00)81401-4
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol 1999; 1:20-6; PMID:10559859; http://dx.doi.org/10.1038/8991
- Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J 1999; 18:22-7; PMID:9878046; http://dx.doi.org/10.1093/emboj/18.1.22
- Datta A, Nag A, Pan W, Hay N, Gartel AL, Colamonici O, Mori Y, Raychaudhuri P. Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. J Biol Chem 2004; 279:36698-707; PMID:15199070; http://dx.doi.org/10.1074/jbc.M312305200
- Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature 2004; 431:712-7; PMID:15361884; http://dx.doi.org/10.1038/nature02958
- Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem 2004; 279:44475-82; PMID:15308643; http://dx.doi.org/10.1074/jbc.M403722200
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol 2004; 24:7654-68; PMID:15314173; http://dx.doi.org/10.1128/MCB.24.17.7654-7668.2004
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 2003; 23:8902-12; PMID:14612427; http://dx.doi.org/10.1128/MCB.23.23.8902-8912.2003
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 2003; 3:577-87; PMID:12842086; http://dx.doi.org/10.1016/S1535-6108(03)00134-X
- Jin A, Itahana K, O'Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol 2004; 24:7669-80; PMID:15314174; http://dx.doi.org/10.1128/MCB.24.17.7669-7680.2004
- Andrews PA, Howell SB. Cellular pharmacology of cisplatin: perspectives on mechanisms of acquired resistance. Cancer cells 1990; 2:35-43; PMID:2204382
- Kashani-Sabet M, Lu Y, Leong L, Haedicke K, Scanlon KJ. Differential oncogene amplification in tumor cells from a patient treated with cisplatin and 5-fluorouracil. Eur J Cancer 1990; 26:383-90; PMID:2141497; http://dx.doi.org/10.1016/0277-5379(90)90238-O
- Twentyman PR, Wright KA, Rhodes T. Radiation response of human lung cancer cells with inherent and acquired resistance to cisplatin. Int J Radiat Oncol Biol Phys 1991; 20:217-20; PMID:1846846; http://dx.doi.org/10.1016/0360-3016(91)90093-J
- Christen RD, Jekunen AP, Jones JA, Thiebaut F, Shalinsky DR, Howell SB. In vitro modulation of cisplatin accumulation in human ovarian carcinoma cells by pharmacologic alteration of microtubules. J Clin Invest 1993; 92:431-40; PMID:8100837; http://dx.doi.org/10.1172/JCI116585
- Niimi S, Nakagawa K, Yokota J, Tsunokawa Y, Nishio K, Terashima Y, Shibuya M, Terada M, Saijo N. Resistance to anticancer drugs in NIH3T3 cells transfected with c-myc and/or c-H-ras genes. Br J Cancer 1991; 63:237-41; PMID:1997100; http://dx.doi.org/10.1038/bjc.1991.56
- Sklar MD, Prochownik EV. Modulation of cis-platinum resistance in Friend erythroleukemia cells by c-myc. Cancer Res 1991; 51:2118-23; PMID:2009531
- Mizutani Y, Fukumoto M, Bonavida B, Yoshida O. Enhancement of sensitivity of urinary bladder tumor cells to cisplatin by c-myc antisense oligonucleotide. Cancer 1994; 74:2546-54; PMID:7923012; http://dx.doi.org/10.1002/1097-0142(19941101)74:9%3c2546::AID-CNCR2820740924%3e3.0.CO;2-Y
- Liao JM, Lu H. Autoregulatory suppression of c-Myc by miR-185-3p. J Biol Chem 2011; 286:33901-9; PMID:21832077; http://dx.doi.org/10.1074/jbc.M111.262030
- Sun XX, Dai MS, Lu H. Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. J Biol Chem 2008; 283:12387-92; PMID:18305114; http://dx.doi.org/10.1074/jbc.M801387200
