Abstract
Uveal melanoma (UM) represents approximately 5–6% of all melanoma diagnoses and up to 50% of patients succumb to their disease. Although several methods are available, accurate diagnosis is not always easily feasible because of potential accidents (e.g., intraocular hemorrhage). Based on the assumption that the profile of circulating miRNAs is often altered in human cancers, we verified whether UM patients showed different vitreous humor (VH) or serum miRNA profiles with respect to healthy controls. By using TaqMan Low Density Arrays, we analyzed 754 miRNAs from VH, vitreal exosomes, and serum of 6 UM patients and 6 healthy donors: our data demonstrated that the UM VH profile was unique and only partially overlapping with that from serum of the same patients. Whereas, 90% of miRNAs were shared between VH and vitreal exosomes, and their alterations in UM were statistically overlapped with those of VH and vitreal exosomes, suggesting that VH alterations could result from exosomal dysregulation. We report 32 miRNAs differentially expressed in UM patients in at least 2 different types of samples analyzed. We validated these data on an independent cohort of 12 UM patients. Most alterations were common to VH and vitreal exosomes (e.g., upregulation of miR-21,-34 a,-146a). Interestingly, miR-146a was upregulated in the serum of UM patients, as well as in serum exosomes. Upregulation of miR-21 and miR-146a was also detected in formalin-fixed, paraffin-embedded UM, suggesting that VH or serum alterations in UM could be the consequence of disregulation arising from tumoral cells. Our findings suggest the possibility to detect in VH and serum of UM patients “diagnostic” miRNAs released by the affected eye: based on this, miR-146a could be considered a potential circulating marker of UM.
Abbreviations
| CM | = | cutaneous melanoma |
| Ct | = | cycle threshold |
| DCts | = | delta cycle thresholds |
| DE | = | Differentially Expressed |
| DLS | = | dynamic light scattering |
| FFPE | = | formalin-fixed paraffin-embedded |
| RQ | = | relative quantities |
| SAM | = | Significance Analysis of Microarrays |
| UM | = | uveal melanoma |
| TLDA | = | TaqMan Low Density Array |
| VH | = | vitreous humor. |
Introduction
Uveal melanoma (UM) is the most common primary intraocular malignancy in adults, with about 2000 new cases diagnosed each year in the United States.Citation1 This corresponds to an incidence of 6 cases per million, compared to 153.5 cases per million for cutaneous melanoma (CM).Citation2 UM is distinct from CM because of its very strong propensity to metastasize to the liver. Because of the absence of ocular lymphatic drainage, UM does not spread to regional lymph nodes except in rare cases through conjunctival lymphatics or direct invasion of conjunctiva.Citation3 UM may be located at any site in the uveal tract, with choroid and ciliary body being more frequent locations than the iris; it threatens not only the visual function but also the patient's life. It has been estimated that even with early diagnosis, appropriate treatment and close follow-up, 40–50% of patients with UM die from metastatic disease; the liver is involved in up to 90% of individuals and the median survival is 4–5 months.Citation4 New diagnostic and therapeutic approaches have led to increased eye preservation rates, even though patients' survival has not been improved.Citation5 Most UM cases are correctly diagnosed through ophthalmoscopy, ultrasonography, fundus fluorescein angiography, indocyanine green angiography and magnetic resonance imaging.Citation6 However, clinical presentation, size of lesion, opacity of refractive media, intraocular hemorrhage and other factors may cause false negative or positive results. Recently, the characterization of serum or plasma miRNAs as novel biomarkers has represented a new approach for diagnostic minimally invasive screening.Citation7-9 Moreover, several recent studies have shown that circulating miRNAs fulfil a number of criteria as ideal biomarkers: accessibility through non-invasive methods, high degree of specificity and sensitivity, ability to differentiate pathologies, long half-life within samples, rapid and accurate detection.Citation7-9 Several studies have identified miRNAs in lipid vesicles secreted by cells.Citation10,11 Among these, exosomes produced by cancer cells can contribute to the horizontal propagation of oncogenic miRNAs and their associated transforming phenotype among subsets of cancer cells.Citation12,13 These secreted miRNAs may play a pivotal role as signaling molecules in physiological and pathological events. Besides the possibility of assessing concentrations of circulating miRNAs in serum or plasma, the quantification of such markers in other body fluids is another interesting possibility. In fact, in our previous work we showed that the expression of circulating miRNAs in Vitreous Humor (VH) is altered in different eye pathologies, including UM.Citation14 The amount of miRNAs that cancer cells secrete in vitro and in vivo is altered compared to their physiological counterpart. Based on this, the aim of this work was to detect potential dysregulations of circulating miRNA expression in VH, vitreal exosomes and serum of UM patients, with respect to healthy donors. Our findings should contribute to improve the diagnosis of UM and to shed light on mechanisms favoring the extracellular spreading of oncogenic signals from this tumor.
Results
Characterization of VH exosomes
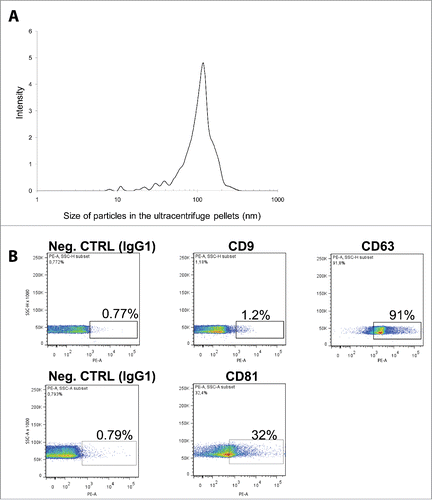
After exosome isolation, the size of pelleted particles was determined through dynamic light scattering (DLS) using a Zetasizer Nano. The results show that the pellet consisted of particles with an average size of 100 nm in diameter, consistent with the characteristic size range of exosomes (). By using flow cytometry we found that the isolated nano-particles were positive for at least one of the canonical exosome markers CD9, CD63 and CD81 (). Take together, these 2 analysis confirm that our samples were enriched in nano-vesicles.
Figure 1. Characterization of VH exosomes. (A) Average particle size in VH exosome samples was determined by dynamic light scattering. Y-axes: signal intensity (%); X-axes: size of particles (nm). (B) Flow cytometry detection of surface molecules on nanoparticles isolated from VH samples. The exosomes were bound to aldehyde-sulfate latex beads conjugated with anti-CD9, anti-CD63 or anti-CD81 antibodies and analyzed by flow cytometry. The antibodies were compared with their appropriate isotype control IgG1.
Comparison of miRNA profiles from VH, VH exosomes and serum
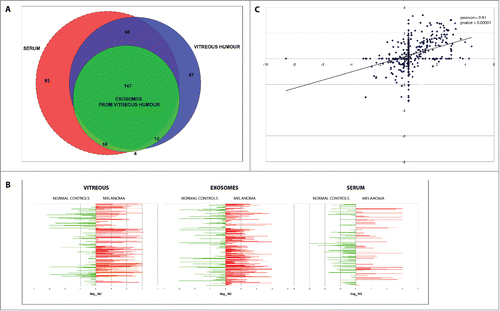
Using TaqMan Low Density Array (TLDA) technology, we determined the profiles of 754 miRNAs in VH, VH exosomes and serum from 6 patients affected by UM and 6 unaffected controls. For each patient, we compared the sets of miRNAs in VH (274 miRNAs detected), VH exosomes (179 miRNAs) and serum (324 miRNAs) (). The three biological matrices shared 147 miRNA species; specifically, about 90% of exosomal miRNAs were also detected in VH, but only 66% of serum miRNAs were present also in VH. Interestingly, about 13% of VH miRNAs were not detected in exosomes and serum. This suggests that the VH miRNA profile is unique and only partially overlaps that of serum; moreover, a fraction of circulating miRNAs in VH may also derive from other sources than exosomes (i.e., from other microvesicles, complexes comprising RNA binding proteins, or apoptotic bodies).
Figure 2. Comparison of miRNAs found in VH, VH exosomes and serum of UM and healthy controls. (A) Venn diagrams showing the overlap between miRNA sets found in different types of samples. (B) Quantitative representation of miRNA different expression between UM patients and controls in VH, VH exosomes, serum. (C) Correlation between RQs from VH and its exosomes: x-axis represents the −log10 of RQ of vitreal miRNAs in UM patients with respect to normal controls; y-axis represents the −log10 of RQ of exosomal miRNAs in UM patients with respect to normal controls.
miRNA alterations in UM VH and exosomes significantly overlap
The comparison of miRNA profiles in VH, exosomes from VH and serum from UM patients and normal controls showed a strong dysregulation of pathological samples () (miRNA list and expression values used to create are reported in supplementary file 1). Notably, the correlation coefficient (Pearson) of Relative Quantities (RQs) between VH and VH exosomes, VH and serum, VH exosomes and serum showed a significant positive correlation (p = 0.41; p value <0.00001) for RQs from VH vs VH exosomes only (). These data showed that a consistent overlapping exists between miRNA expression alterations in VH of UM patients and those in VH exosomes. This suggests that the major source of miRNA alterations in VH could be an altered cargo from exosomes circulating inside the eye chamber. On the contrary, miRNA dysregulations in serum of UM patients did not mirror the alterations of VH miRNAs.
miR-146a is upregulated in VH, exosomes from VH, serum and exosomes from serum of UM patients
By applying Significance Analysis of Microarrays (SAM) statistical method to identify Differentially Expressed (DE) miRNAs, we performed 3 different comparisons between DCts (Delta Cycle Thresholds) obtained from TLDAs: (i) VH miRNAs from UM patients vs VH miRNAs from control donors; ii) exosomal miRNAs from VH of UM patients vs exosomal miRNAs from VH of control donors; iii) serum miRNAs of UM patients vs serum miRNAs of control donors. Specifically, we detected 32 circulating miRNAs differentially expressed in at least 2 different types of samples (). Most dysregulations were observed in VH and VH exosomes and were consistent with the general trend of expression changes (e.g., miR-21, miR-34a, miR-126), as already shown by Pearson correlations. Conversely, we detected few miRNA alterations in serum of UM patients and they were nearly always discordant with those observed in VH or its exosomes and found that miR-146a was upregulated in all types of samples. As we considered the data on miR-146a potentially interesting for their biological consistence and for diagnostic purposes, we validated these data on an independent cohort of 12 UM patients. We performed single TaqMan assays for miR-146a and observed a statistically significant upregulation in UM patients with respect to normal controls in VH, VH exosomes, serum and exosomes from serum (). We also statistically validated the expression of miR-21, miR-34a, miR-618 in the same cohort of patients, as reported in. Notably, miR-618 was downregulated in VH but upregulated in VH exosomes and serum (), suggesting that part of the expression profile observed in VH might not derive from the exosomes.
Figure 3. Single TaqMan assays for miR-21, miR-34a, miR-146a, miR-618. Box plots representing the expression of: (A) miR-146a, (B) miR-21, (C) miR-34a, (D) miR-618, analyzed by single TaqMan assay on whole vitreous humor, exosomes from vitreous (ExoVitr.), whole serum, or exosomes from serum (ExoSer.) from an independent cohort of 12 patients. y-axis represents the –ΔCt of miRNAs in UM patients with respect to normal controls. Statistical significance was evaluated by the Wilcoxon rank sum test (p-value < 0.05).
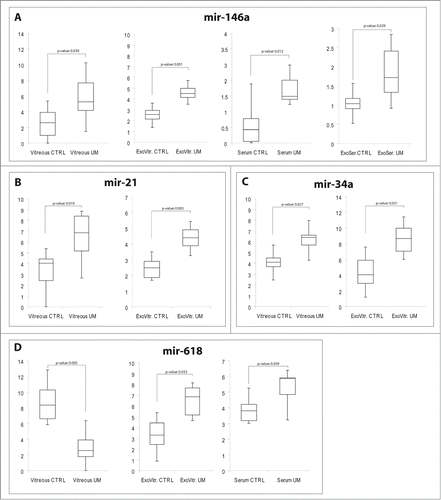
Table 2. Demographics, tumor parameters, time treatment/enucleation in UM
Table 1. DE miRNAs in UM patients
miR-21, miR-34a and miR-146a are upregulated in FFPE uveal melanoma specimens
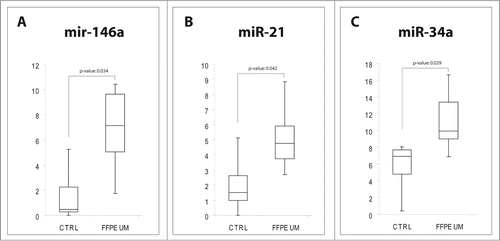
To verify whether the miRNA alterations observed in eye chamber fluid and serum from UM patients were the result of specific miRNA secretion or the consequence of molecular dysregulations arising from tumoral cells, we performed single TaqMan assays for miR-21, miR-34a and miR-146a on 12 formalin-fixed, paraffin-embedded (FFPE) UM samples (epithelioid cells) and compared them to choroidal melanocytes from 5 unaffected eyes. Real Time PCR analysis showed that miR-21, miR-34a and miR-146a were statistically upregulated in UM cells (), as already shown for VH and VH exosomes.
Figure 4. MiRNAs expression in FFPE UM specimens. Box plots representing the expression of: (A) miR-146a, (B) miR-21, (C) miR-34a, analyzed by single TaqMan assay on paraffin-embedded UM compared to healthy choroidal melanocytes. y-axis represents the −ΔCt of miRNAs in UM patients with respect to normal controls. Statistical significance was evaluated by the Wilcoxon rank sum test (p-value < 0.05).
Discussion
The discovery of circulating miRNAs suggested the presence of new mediators of gene regulation. In fact, while most miRNAs are found intracellularly, a large number of miRNAs has also been detected outside cells and in various body fluids.Citation15,16 The function of these circulating miRNAs remains not completely understood. One of the most intriguing hypothesis is that extracellular miRNAs may work as mediators of cell-cell communication: some miRNAs are specifically secreted by donor cells to be transferred to recipient cells.Citation17,18 Recent studies have shown that secreted miRNAs are packaged in specific structures to protect them against RNase digestion, which would naturally occur in serum and other body fluids. MiRNAs would be shielded from degradation by being packaged in lipid vesicles (i.e., exosomes, microvesicles), or by being included in complexes with RNA-binding proteins.Citation19-21 Exosomes can be transferred from one cell to another, and their cargo can work in the new cellular environment. Given their biological importance, it is not surprising that the expression of circulating miRNA is frequently dysregulated in human cancer. There is some evidence suggesting that circulating miRNAs can contribute to tumorigenesis by modulating oncogenic or tumor suppressor pathways.Citation22,10 Several studies have compared the expression profiles of miRNAs in serum or plasma across a variety of tumors to identify cancer-specific expression patterns: data obtained suggested that circulating miRNAs expression could be used to discriminate disease samples, demonstrating their potential use as blood-based diagnostic cancer markers.Citation23-26
Circulating miRNAs in VH of UM patients mostly derive from VH exosomes
In this study, we explored the possibility that in UM patients there could be alterations of circulating miRNAs in VH and serum. We found expression alterations for several miRNAs in VH of UM patients compared to healthy donors: this demonstrated that the presence of tumor growth and their infiltrating different layers of the eye could affect physiological miRNA secretion. We also observed in the vitreous the presence of vesicles with an average size of 100 nm and characterized by the expression of exosomes-specific tetraspanins on their surface. The possibility that these vesicles could be microvesicles or melanosomes was excluded, since both these particles are significantly larger than 100 nm (500 – 1000 nm) and lack specific tetraspanins on their surface.Citation27-29 The expression profiles of miRNAs carried by these nanoparticles were statistically correlated to those observed in total VH. These data suggested that dysregulation of circulating miRNA in VH of UM patients could be mostly caused by alterations of the molecular content of VH exosomes. We cannot exclude the presence of other miRNA molecular shuttles in VH (e.g., other types of nano-vesicles or protein complexes), which might contribute to cell-cell communication: indeed, we found that the amount of some miRNAs dysregulated in VH was unaltered in exosomes, while other miRNAs showed an opposite behavior (e.g., miR-618 downregulated in UM VH, but upregulated in its exosomes). It is well known that many tumors have a remarkable ability to mold their stromal environment to their own advantage: exosomes from cancer cells can contribute to the horizontal propagation of oncogenic miRNAs among subsets of cancer cells or immuno-suppressive miRNAs to negatively regulate the immune system.Citation30,31 We hypothesize that in UM the cell source of this altered secretion could be transformed melanocytes. It has already been reported for CM that melanocytes are able to produce exosomes and that melanoma-derived exosomes have unique miRNA expression signatures, compared to exosomes from normal melanocytes.Citation32 Melanocytes from UM could secrete oncogenic exosomes and affect their tumor microenvironment, but they also would flow inside the vitreal chamber helped by retinal detachment, commonly occurring in UM patients. Among miRNAs contemporarily dysregulated in VH and VH exosomes, we detected the upregulation of miR-21, miR-34a, and miR-130a and the downregulation of miR-149. This kind of alteration for these miRNAs was also found in exosomes from CM compared to normal melanocytes.Citation32 On the other hand, we found that miR-146a was upregulated and let-7c was downregulated in both VH and VH exosomes (see Table 1); while Xiao et al. reported an opposite regulation of these 2 miRNAs in exosomes from CM.Citation32 These observations agree with molecular data showing that CM and UM differ in some aspects, although they are histopathologically similar.Citation33
Upregulation of miR-146a in serum of UM patients could derive from melanocytes through exosome secretion
By comparing miRNA serum profiles of UM patients with those of healthy donors, we pinpointed some DE miRNAs. Notably, VH had significantly more DE miRNAs compared to serum and the 2 profiles were statistically unrelated: this would suggest that serum miRNA alterations were not a direct consequence of their dysregulation in the vitreal chamber. However, we found that miR-146a, which is overexpressed in VH and its exosomes, was also upregulated in the serum and exosomes from UM patients. In a previous study, we showed upregulation of miR-146a in VH of UM patients;Citation14 and recently, Achberger et al. demonstrated that levels of miR-146a were higher in plasma and CD3+, CD56+ and CD15+ cells from UM patients compared to controls.Citation34 MiR-146a is considered an immune miRNA with a potential immune-suppressive role: it is a mediator of inflammation and it is involved in NK cell development and function; its overexpression in NK cells inhibits proliferation and induces apoptosis.Citation35-37 It is interesting to note that miR-146a was reported to be involved in inflammatory pathways related to diabetic retinopathy in in vitro and in vivo studies.Citation38 We found that miR-146a was also upregulated in FFPE UM specimens, suggesting its potential role in neoplastic melanocytes. miR-146a is highly upregulated by oncogenic BRAF and NRAS in CM. Expression of miR-146a increases the ability of human CM cells to proliferate in culture and form tumors in mice.Citation39 Although BRAF and NRAS mutations are uncommon in UM, the oncogenic activation of the MAPK cascade is very frequent, mainly driven by GNAQ mutations.Citation40 Moreover, miR-146a is a target of MITF (Microphthalmia-associated Transcription Factor), a proto-oncogenic transcription factor acting as a master regulator of melanocyte development, function and survival; it may also be implicated in choroidal melanoma pigmentation and proliferation.Citation41-43 Based on these data, miR-146a could have an important regulatory role in the survival of melanocytes from UM. Our data suggest that transformed melanocytes in the eyes of UM patients may change the expression of circulating miRNAs in the vitreous humor by dysregulation of the miRNA cargo of secreted exosomes. Some circulating miRNA alterations mirrored the expression differences observed in ex-vivo UM samples (e.g.,, upregulation of miR-21, miR-146a). The secretion of these oncogenic exosomes would contribute to release cancer signals in the surrounding microenvironment. Exosomes could pass through tumor blood vessels and flow into the blood. In other words, circulating miRNAs found in systemic circulation could derive from UM cells through exosome secretion: indeed, also purified exosomes from serum of UM patients showed an upregulation of miR-146a. Based on this, upregulation of miR-146a in serum of UM patients could be a long range effect of exosome secretion from cancerous melanocytes and represents an innovative non-invasive diagnostic biomarker for UM. However, we do not exclude some other sources of circulating miR-146a in UM serum, i.e, RNA binding protein complexes or other nano-vesicles.
Conclusions
Careful examination by experienced clinicians remains the most important test to establish the presence of ocular melanoma: often, distinguishing a small UM from a nevus can be very difficult. Ancillary diagnostic testing, including fluorescein angiography and ultrasonography, can be used to establish and confirm the diagnosis.Citation44,45 However, effective molecular diagnostic markers for this tumor have not been described. Although a very low metabolic exchange exists between systemic circulation and VH, the vitreal chamber can be considered a nearly closed biological compartment. Accordingly, if surrounding tumoral cells are able to secrete specific miRNAs, they could be detected in VH, as shown in this paper. VH has already been demonstrated to be a very useful tool for analyzing the patho-physiological events that take place in the retina of diabetic patients.Citation46 However, data shown in this paper also suggest the possibility to screen the blood of UM patients to find miRNAs released by the affected eyes. Further multicentric studies on larger cohorts of patients and on other neoplastic diseases will be needed to verify the effective diagnostic and discriminatory power of miR-146a and other circulating miRNAs in uveal melanoma.
Materials and Methods
This study was performed at the University of Catania, Italy. All patients were selected between October 2012 and April 2014. Our research followed the tenets of the Declaration of Helsinki: informed consent was obtained from all patients after explaining the nature and possible consequences of the study. Following diagnosis of UM, enucleation was performed at the Eye Clinic of the University of Catania. Eligible patients underwent enucleation because of large basal tumor diameter and height; other methods were considered unlikely to conserve the eye and useful vision without causing excessive morbidity.Citation47 At enrolment, all patients were free of metastases and other cancers. Age at diagnosis, gender, location of the tumor, cell type, largest diameter, thickness, extrascleral extension, and pathological TNM stage were retrieved from clinical files, pathology reports and histopathological samples (). VH from 6 cornea donors was used as controls: all of them were Caucasian; 3 males, 3 females; mean age = 57.5; years range = 37 – 67). We excluded from our study patients with systemic disease, other ocular diseases, and previous ocular surgical procedures.
VH sampling
After enucleation, the unfixed eyes underwent VH sampling by scleral puncture in an area far from the melanoma, marked with ink by the surgeon. A 20-gauge needle and a 2 mL syringe were used. One mL of VH was collected with careful suction; non transparent, bloody samples were not analyzed. Samples were stored at −80°C until analysis.
Serum sampling
All enrolled patients underwent fasting venous blood sampling. We analyzed serum from 12 healthy donors as controls. Blood samples were obtained by vein puncture using dry vacutainer tubes (BD Biosciences, Italy). The samples were processed for serum isolation within 2 h from withdrawal. Whole blood was left to stand for 30′ at 20°C before being centrifuged at 3000 rpm for 15′ at 4°C. Serum was divided into aliquots, and stored at −80°C until analysis.
Tumor samples and clinical-pathological data
The pathology reports of UM patients who had their eye enucleated were retrieved at the Unit of Anatomical Pathology, Department G.F. Ingrassia, University of Catania, Catania, Italy. Cases of iris melanoma, with incomplete patient records or without representative tumor tissue in paraffin blocks, were excluded. Twelve patients who underwent enucleation were selected for miRNA analysis (). Healthy controls were choroidal melanocytes from 5 unaffected eyes. All the control subjects were Caucasian: 3 males, 2 females; mean age = 43.8; years range = 33 – 54.
RNA isolation from VH, serum and miRNA profiling by TaqMan Low Density Array
VH and serum samples were centrifuged at 2000 rpm for 10' to pellet any circulating cells or debris. miRNAs were extracted from 400 μl of vitreous and serum samples by using Qiagen miRNeasy mini kit (Qiagen, GmbH, Hilden, Germany), according to Qiagen supplementary protocol for purification of small RNAs from serum and plasma, and finally eluted in 40 μl of elution buffer.Citation14 RNAs were quantified by fluorometry and spectrophotometry. To profile the transcriptome of 754 miRNAs on TLDA, 30 ng of serum or VH RNAs were retrotranscribed and pre-amplified, according to the manufacturer's instructions. Pre-amplified products were loaded onto TLDAs, TaqMan Human MicroRNA Array v3.0 A and B (Applied Biosystems│Life Technologies™ Monza, Italy). PCRs on TLDAs were performed on a 7900 HT Fast Real Time PCR System (Applied Biosystem│Life Technologies™ Monza, Italy). Results were validated by single TaqMan assays (Applied Biosystems│Life Technologies™Monza, Italy) using the same amount of RNAs, according to the manufacturer's instructions.
RNA isolation and RT-PCR from formalin-fixed, paraffin-embedded samples
For miRNA extraction, 8 sections of 20 μm each were cut from FFPE samples (eyes) of choroidal melanocytes from 12 UMs on a RM2245 microtome (Leica, Bannockburn, IL, USA). Sections were transferred to glass slides, and tumor tissue was isolated by hand with a scalpel. We also analyzed FFPE from 5 unaffected eyes. RNAs were extracted by using a Recover All Total Nucleic Acid Isolation Kit (Ambion), following the manufacturer's protocol. The expression of miRNAs from FFPE samples was analyzed by TaqMan MicroRNA Assay, as previously specified.
Exosome isolation and characterization
Exosomes were extracted from VH or serum supernatants by centrifugation at 300 g to pellet cell debris, and then at 16500 g for 30′, followed by filtration through a 0.2 μm filter. The final supernatant was ultracentrifuged at 120000 g on a Beckman L8–70M ultracentrifuge in a SW28 rotor for 70′. Exosome pellets were resuspended in 300 μl PBS for FACS analysis or directly lysed for RNA isolation by adding 350 μl of Qiazol and following Qiagen miRNeasy mini kit protocol. Exosomes were analyzed by: (1) Zetasizer Nano ZS (Malvern Instruments, UK); (2) flow cytometry for size determination and surface marker characterization, as previously reported.Citation48 Aldehyde/sulfate latex beads (Invitrogen, Sweden) (140000) were incubated with 200 μl of vitreous exosomes at 37˚C for 30′ and then at 4˚C for 16 h on a rotator apparatus. Following centrifugation at 4000 g for 10′, pellets were resuspended in 100 μl PBS; 20 μl of 1 M glycine were added to block unspecific binding sites at 20°C for 30′. After one wash with PBS and 1% FBS, exosome-coated beads were incubated with PE-conjugated CD9, CD63 or CD81 antibodies or isotype controls (BD Biosciences) for 60' at 4˚C. For FACS analysis, samples were washed and resuspended in 200 μl PBS/FBS and analyzed with FACSCantoII (Becton Dickinson, San Diego CA, USA) and FlowJo software (TreeStar).
Analysis of miRNAs expression data
To obtain an accurate miRNA profiling, we used the global median normalization method. Similar to microarray analysis, cycle threshold (Ct) values were normalized to the median Ct of the arrays for each sample.Citation47 By computing the Pearson correlation between the Ct medians of each array and the Ct of each miRNA, we identified miRNAs that showed an expression profile close to the median of TLDAs, i.e. miR-320 for VH and serum samples and snRNA U6 for vitreal and serum exosomes. Accordingly, miR-320 and snRNA U6 were used as reference genes for validation by single TaqMan assays in VH, serum and exosomal samples. Expression fold changes were calculated by the 2−ΔΔCT method. DE miRNAs were identified by Significance Analysis of Microarrays (SAM), computed by Multi experiment viewer v4.8.1, by applying a 2-class unpaired test among ΔCts and using a p-value based on 100 permutations; imputation engine: K-nearest neighbors (10 neighbors); false discovery rate <0.05 was used as correction for multiple comparisons. The Wilcoxon signed-rank test (p < 0.05) was applied to statistically evaluate the expression differences between UM patients and healthy controls by single TaqMan validation assays in VH, serum, exosomes and FFPE samples.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental File 1
Download MS Excel (69.5 KB)Acknowledgments
We thank Micron Foundation for the financial support to the scientific project, believing in the improvement of the quality of life in the context where Micron operates. We thank Scientific Bureau of the University of Catania for language support.
References
- Materin MA, Faries M, Kluger HM. Molecular alternations in uveal melanoma. Curr Probl Cancer 2011; 35(4):211-24; PMID:21911184; http://dx.doi.org/10.1016/j.currproblcancer.2011.07.004
- McLaughlin CC, Wu XC, Jemal A, Martin HJ, Roche LM, Chen VW. Incidence of noncutaneous melanomas in the U.S. 2005; Cancer. 103:1000-7; PMID:15651058
- Dithmar S, Diaz CE, Grossniklaus HE. Intraocular melanoma spread to regional lymph nodes: report of two cases. Retina 2000; 20:76-9; PMID:10696752; http://dx.doi.org/10.1097/00006982-200001000-00014
- Kujala E1, Mäkitie T, Kivelä T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci 2003; 44(11):4651-9; PMID:14578381; http://dx.doi.org/10.1167/iovs.03-0538
- Singh AD, Topham A. Survival rates with uveal melanoma in the United States:1973-1997. Ophthalmology 2003; 110(5):962-5; PMID:12750098; http://dx.doi.org/10.1016/S0161-6420(03)00077-0
- Yang W, Hu S, Wang J, Wang L, Zheng B. Color Doppler imaging diagnosis of intra-ocular tumor. Chin Med J (Engl) 1997; 110(9):664-6; PMID:9642319
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18(10):997-1006; PMID:18766170; http://dx.doi.org/10.1038/cr.2008.282
- Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, et al. Serum microRNAs are promising novel biomarkers. PLoS One 2008; 3(9):e3148; PMID:18773077; http://dx.doi.org/10.1371/journal.pone.0003148
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008; 105(30):10513-8; PMID:18663219; http://dx.doi.org/10.1073/pnas.0804549105
- Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010; 101(10):2087-92; PMID:20624164; http://dx.doi.org/10.1111/j.1349-7006.2010.01650.x
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9(6):654-9; PMID:17486113; http://dx.doi.org/10.1038/ncb1596
- Corrado C, Raimondo S, Chiesi A, Ciccia F, De Leo G, Alessandro R. Exosomes as intercellular signaling organelles involved in health and disease: basic science and clinical applications. Int J Mol Sci 2013; 14(3):5338-66; PMID:23466882; http://dx.doi.org/10.3390/ijms14035338
- Hendrix A, Westbroek W, Bracke M, De Wever O. An ex(o)citing machinery for invasive tumor growth. Cancer Res 2010; 70(23):9533-7; PMID:21098711; http://dx.doi.org/10.1158/0008-5472.CAN-10-3248
- Ragusa M, Caltabiano R, Russo A, Puzzo L, Avitabile T, Longo A, Toro MD, Di Pietro C, Purrello M, Reibaldi M. MicroRNAs in vitreus humor from patients with ocular diseases. Mol Vis 2013; 19:430-40; PMID:23441115
- Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, Dahlsveen IK. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013; 59(1):S1-6; PMID:23036329; http://dx.doi.org/10.1016/j.ymeth.2012.09.015
- Pacifici M, Delbue S, Kadri F, Peruzzi F. Cerebrospinal fluid MicroRNA profiling using quantitative real time PCR. J Vis Exp 2014; (83):e51172; PMID:24514260
- Kosaka N, Yoshioka Y, Hagiwara K, Tominaga N, Katsuda T, Ochiya T. Trash or Treasure: extracellular microRNAs and cell-to-cell communication. Front Genet 2013; 4:173; PMID:24046777; http://dx.doi.org/10.3389/fgene.2013.00173
- Shah MY, Calin GA. The mix of two worlds: non-coding RNAs and hormones. Nucleic Acid Ther 2013; 23(1):2-8; PMID:23051203
- Boon RA, Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol 2013; 33(2):186-92; PMID:23325475; http://dx.doi.org/10.1161/ATVBAHA.112.300139
- Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011; 39(16):7223-33; PMID:21609964; http://dx.doi.org/10.1093/nar/gkr254
- Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol 2012; 23(2):91-7; PMID:22418571; http://dx.doi.org/10.1097/MOL.0b013e328350a425
- Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev 2013 23(1):3-11; PMID:23465882; http://dx.doi.org/10.1016/j.gde.2013.01.004
- Fortunato O, Boeri M, Verri C, Conte D, Mensah M, Suatoni P, Pastorino U, Sozzi G. Assessment of circulating microRNAs in plasma of lung cancer patients. Molecules 2014; 19(3):3038-54; PMID:24619302; http://dx.doi.org/10.3390/molecules19033038
- Ganepola GA, Rutledge JR, Suman P, Yiengpruksawan A, Chang DH. Novel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancer. World J Gastrointest Oncol 2014; 6(1):22-33; PMID:24578785; http://dx.doi.org/10.4251/wjgo.v6.i1.22
- Mar-Aguilar F, Rodríguez-Padilla C, Reséndez-Pérez D. Use of serum-circulating miRNA profiling for the identification of breast cancer biomarkers. Methods Mol Biol 2014; 1165:71-80; PMID:24839019; http://dx.doi.org/10.1007/978-1-4939-0856-1_6
- Wang J, Huang SK, Zhao M, Yang M, Zhong JL, Gu YY, Peng H, Che YQ, Huang CZ. Identification of a circulating microRNA signature for colorectal cancer detection. PLoS One 2014; 9(4):e87451; PMID:24709885; http://dx.doi.org/10.1371/journal.pone.0087451
- Ando H, Niki Y, Ito M, Akiyama K, Matsui MS, Yarosh DB, Ichihashi M. Melanosomes are transferred from melanocytes to keratinocytes through the processes of packaging, release, uptake, and dispersion. J Invest Dermatol 2012; 132(4):1222-9; PMID:22189785; http://dx.doi.org/10.1038/jid.2011.413
- Crescitelli R, Lässer C, Szabó TG, Kittel A, Eldh M, Dianzani I, Buzás EI, Lötvall J. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles 2013; 12:2; PMID:24223256
- Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borràs FE, Breakefield X, Budnik V, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol 2012; 10(12):e1001450; PMID:23271954; http://dx.doi.org/10.1371/journal.pbio.1001450
- Filipazzi P, Bürdek M, Villa A, Rivoltini L, Huber V. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin Cancer Biol 2012; 22(4):342-9; PMID:22369922; http://dx.doi.org/10.1016/j.semcancer.2012.02.005
- Yang C, Robbins PD. The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol 2011; 2011:842849; PMID:22190973
- Xiao D, Ohlendorf J, Chen Y, Taylor DD, Rai SN, Waigel S, Zacharias W, Hao H, McMasters KM. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS One 2012; 7(10):e46874; PMID:23056502; http://dx.doi.org/10.1371/journal.pone.0046874
- Gaudi S, Messina JL. Molecular bases of cutaneous and uveal melanomas. Patholog Res Int 2011; 2011:159421; PMID:21876842
- Achberger S, Aldrich W, Tubbs R, Crabb JW, Singh AD, Triozzi PL. Circulating immune cell and microRNA in patients with uveal melanoma developing metastatic disease. Mol Immunol 2014; 58(2):182-6; PMID:24370793; http://dx.doi.org/10.1016/j.molimm.2013.11.018
- Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, et al. MiR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med 2011; 208(6):1189-201; PMID:21555486; http://dx.doi.org/10.1084/jem.20101823
- Paik JH, Jang JY, Jeon YK, Kim WY, Kim TM, Heo DS, Kim CW. MicroRNA-146a downregulates NFκB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin Cancer Res 2011; 17(14):4761-71; PMID:21610143; http://dx.doi.org/10.1158/1078-0432.CCR-11-0494
- Sheedy FJ, O'Neill LA. Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann Rheum Dis 2008; 67 3:iii50-5; PMID:19022814
- Mastropasqua R, Toto L, Cipollone F, Santovito D, Carpineto P, Mastropasqua L. Role of microRNAs in the modulation of diabetic retinopathy. Prog Retin Eye Res 2014; 43C:92-107; PMID:25128741; http://dx.doi.org/10.1016/j.preteyeres.2014.07.003
- Forloni M, Dogra SK, Dong Y, Conte D Jr, Ou J, Zhu LJ, Deng A, Mahalingam M, Green MR, Wajapeyee N. miR-146a promotes the initiation and progression of melanoma by activating Notch signaling. Elife 2014; 3:e01460; PMID:24550252; http://dx.doi.org/10.7554/eLife.01460
- Onken MD, Worley LA, Long MD, Duan S, Council ML, Bowcock AM, Harbour JW. Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest Ophthalmol Vis Sci 2008; 49(12):5230-4; PMID:18719078; http://dx.doi.org/10.1167/iovs.08-2145
- Chen X, Wang J, Shen H, Lu J, Li C, Hu DN, Dong XD, Yan D, Tu L. Epigenetics, microRNAs, and carcinogenesis: functional role of microRNA-137 in uveal melanoma. Invest Ophthalmol Vis Sci 2011; 52(3):1193-9; PMID:21051724; http://dx.doi.org/10.1167/iovs.10-5272
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev 2008; 22(22):3172-83; PMID:19056895; http://dx.doi.org/10.1101/gad.1706508
- Yajima I, Kumasaka MY, Thang ND, Goto Y, Takeda K, Iida M, Ohgami N, Tamura H, Yamanoshita O, Kawamoto Y, et al. Molecular Network Associated with MITF in Skin Melanoma Development and Progression. J Skin Cancer 2011; 2011:730170; PMID:22046555; http://dx.doi.org/10.1155/2011/730170
- Field MG, Harbour JW. Recent developments in prognostic and predictive testing in uveal melanoma. Curr Opin Ophthalmol 2014; 25(3):234-9; PMID:24713608; http://dx.doi.org/10.1097/ICU.0000000000000051
- Rashid AB, Grossniklaus HE. Clinical, pathologic, and imaging features and biological markers of uveal melanoma. Methods Mol Biol 2014; 1102:397-425; PMID:24258990; http://dx.doi.org/10.1007/978-1-62703-727-3_21
- Simó-Servat O, Hernández C, Simó R. Usefulness of the vitreous fluid analysis in the translational research of diabetic retinopathy. Mediators Inflamm 2012; 2012:872978
- Damato B, Lecuona K. Conservation of eyes with choroidal melanoma by a multimodality approach to treatment: an audit of 1632 patients. Ophthalmology 2004; 111(5):977-83; PMID:15121377; http://dx.doi.org/10.1016/j.ophtha.2003.09.028
- Ragusa M, Statello L, Maugeri M, Barbagallo C, Passanisi R, Alhamdani MS, Li Destri G, Cappellani A, Barbagallo D, Scalia M, et al. Highly skewed distribution of miRNAs and proteins between colorectal cancer cells and their exosomes following Cetuximab treatment: biomolecular, genetic and translational implications. Oncoscience 2014; 1(2):132-157; PMID:25594007
