Abstract
Background: Quantification of Circulating Tumor Cells (CTCs) as a prognostic marker in metastatic colorectal cancer (mCRC) has already been validated and approved for routine use. However, more than quantification, qualification or characterization of CTCs is gaining importance, since the genetic characterization of CTCs may reflect, in a real time fashion, genetic profile of the disease. Objective: To characterize KRAS mutations (codon 12 and 13) in CTCs from patients with mCRC and to compare with matched primary tumor. Additionally, correlate these mutations with clinical and pathological features of patients. Methods: Blood samples were collected from 26 patients with mCRC from the AC Camargo Cancer Center (São Paulo-Brazil). CTCs were isolated by ISET technology (Isolation by Size of Epithelial Tumors; Rarecells Diagnostics, France) and mutations analyzes were performed by pyrosequencing (QIAGEN). Results: KRAS mutation was detected in 7 of the 21 cases (33%) of samples from CTCs. In matched primary tumors, 9 of the 24 cases (37.5%) were found KRAS mutated. We observed that 5 of the 9 samples with KRAS mutation in their primary tumor had also KRAS mutation in CTCs, meaning a concordance of 71% of matched cases (P = 0.017). KRAS mutation neither on primary tumor nor in CTCs was associated with clinical-pathological parameters analyzed. Conclusion: Faced with a polyclonal disease like colorectal cancer, which is often treated with alternating and successive lines of chemotherapy, real time genetic characterization of CTCs, in a fast and feasible fashion, can provide important information to clinical management of metastatic patients. Although our cohort was limited, it was possible to show a high grade of concordance between primary tumor and CTCs, which suggests that CTCs can be used as surrogate of primary tumors in clinical practice, when the knowledge of mutation profile is necessary and the primary tumor is not available.
Abbreviations
| ASB-PCR | = | allele specific PCR with a blocking reagent |
| CEA | = | carcinoembryonic antigen |
| cftDNA | = | circulating free tumor DNA |
| CRC | = | colorectal cancer |
| CTCs | = | circulating tumor cells |
| EDTA | = | Ethylenediamine tetraacetic acid |
| EGFR | = | Endothelial Growth Factor Receptor |
| ESMO | = | European Society for Medical Oncology; ISET-Isolation by Size of Epithelial Tumors |
| JSCCR | = | Japanese Society for Cancer of the Colon and Rectum |
| mCRC | = | metastatic colorectal cancer |
| NCCN | = | National Comprehensive Cancer Network |
| PFS-progression | = | free survival |
| WT | = | wild type |
| MT | = | mutation |
| OS | = | overall survival. |
Introduction
The analysis of Circulating Tumor Cells (CTCs) in the era of personalized medicine is a promising tool for early detecting response to therapy. Nowadays, the identification of specific biomarkers that brings information to the target therapy selection is generally made with specimens collected at the moment of initial diagnosis. Frequently, the material is not available or is found insufficient for molecular analysis in advanced stages of the disease.Citation1 In addition, initial primary tumor specimens are not always representative of the metastasis that can occur many years after the resection of the primary tumor.Citation2 After primary tumor biopsy or surgical removal, many other mutations can be acquired and other specific abnormalities found in the primary tumor can be lost. In this way, the CTC analysis can provide a non-invasive way of evaluating a tumor genotype and better correlate with the dynamic process of tumor progression and resistance to chemotherapeutic drugs.Citation3
The development of target therapies has created a need for rapid and robust molecular characterization of cancer. KRAS gene codes for an essential protein involved in the activation of the pathway within the signaling cascade induced by the activation of the Endothelial Growth Factor Receptor (EGFR) gene. KRAS gene plays a central role in tumor development by regulating the expression of proteins that are involved in cell proliferation and survival, metastasis and angiogenesis.Citation4-6
In colorectal carcinoma, KRAS mutations are present in 30-40% of patients.Citation7 Activating mutations in KRAS are responsible to anti-EGFR therapy resistance (Cetuximab or Panitumumab) in metastatic colorectal cancer (mCRC). More recently, around 18% of patients with wild type KRAS - exon 2, were find to carry mutation in exon 3, 4 of KRAS gene and exon 2, 3, 4 of NRAS gene. Thus, only wild-type RAS (KRAS and NRAS) patients can receive anti-EGFR therapy for mCRC, according to Treatment Guidelines for CRC published by the National Comprehensive Cancer Network (NCCN), European Society for Medical Oncology (ESMO) and Japanese Society for Cancer of the Colon and Rectum (JSCCR).Citation8 Metastatic process is dynamic, so, longitudinal monitoring of CTCs can provide a noninvasive and real time evaluation of potential biomarkers related to resistance to specific target therapies.
In this sense, our study had the objective to characterize KRAS mutations on CTCs isolated by ISET from patients with mCRC and to compare KRAS results with primary tumors. We also tried to correlate KRAS mutations with clinical and pathological features of patients with mCRC.
Results
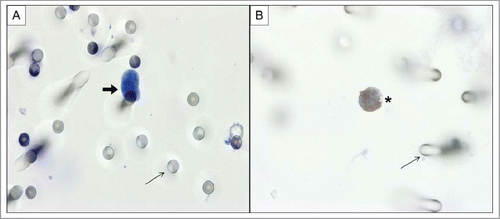
CTCs were identified in 23/26 patients, with a median CTC count of 2 CTCs/mL (range 0-14 CTCs/mL). shows CTCs isolated by ISET and residual leukocytes from a patient with mCRC. Three patients had no detectable CTC.
Figure 1. (A) CTC (thick arrow) isolated by ISET from patient with mCRC stained with hematoxylin-eosin. (B) Residual leukocyte (asterisk), without any CTC visualized on the ISET membrane. Spiked cells were stained with antibody against CD45 to identify leukocytes, visualized with DAB (3,3′- Diaminobenzidine) and counterstained with haematoxylin. Images were taken using a light microscope (Axioskop 40; Carl Zeiss Meditec, Jena, Germany) coupled to a digital camera (Sony Cyber-Shot Dsc-s75; Sony, Tokyo, Japan) at 60× magnification. Fine arrows represent pores of ISET membrane.
We evaluated the KRAS (codons 12 and 13, exon 2) in CTCs samples obtained from 23/26 patients with mCRC. Then, we matched their results with the results of KRAS mutations from primary tumors obtained from medical records.
In 5 samples of the 23 patients there was no PCR amplification. We found KRAS mutation in 7 samples of CTCs (7/21) (33%). All mutations of KRAS in CTC were found in codon 12.
In matched primary tumors, we observed KRAS mutation in 9 cases (9/24) (37.5%). In six primary tumors samples, KRAS mutation was observed in codon 12 and in 3 samples the mutation was observed in codon 13. There was concordance between KRAS mutations in CTCs and primary tumor in 5 samples (71%; P = 0.017) (patients 2, 7, 13, 14 and 20) (). Additionally, KRAS mutations in CTCs were found in 2 patients that were KRAS wild type (wt) in matched primary tumor (patients 4 and 22). shows mutational status of KRAS in CTCs samples and primary tumors.
Table 1. Status mutation of KRAS (codon 12/13) and BRAF (V600E) genes, tumor site, CTC count, in CTC and primary tumor of patients with mCRC
Table 2. Correlation between clinical-pathological parameters and KRAS mutations in CTC and primary tumors in patients with mCRC
We observed KRAS wt in 3 cases without counting CTC, reflecting the presence of leukocytes in samples, as observed by immunocytochemistry with anti-CD45 ().
The KRAS status and its association with clinical-pathological parameters are shown in . KRAS mutation was not associated with the clinical-pathological parameters analyzed (histological grade, lymph node status, diagnosis, tumor progression, carcinoembryonic antigen (CEA). The progression-free survival (PFS) was not statistically different between patients with KRAS mutation and KRAS wild type neither on primary tumor samples nor on CTC's samples (P = 0.143 and P = 0.185, respectively). We did not find association between PFS and CTC's counts (P = 0.194), histological grade (P =0.381) and CEA level (P = 1.000).
We observed that 9 of 23 patients analyzed here showed disease progression after inclusion in our study (all of them were included after a diagnosis of metastasis or protocol change and the blood was collected before first or line second line of chemotherapy). In 4/9 patients we could not assess the KRAS status in CTC because there was no PCR amplification. In 3/9 patients we observed KRAS mutation in CTCs and 2 of them died in 3.18 and 9.18 months after inclusion in study. In we show status of KRAS mutation (codon 12/13) gene in primary tumors and in CTCs in these 9 patients that showed disease progression after inclusion in our study, as also types of treatment.
Table 3. Status of KRAS mutation (codon 12/13) gene in primary tumors and in CTCs, forms of treatment and time to progression after CTC analysis of 9 mCRC patients that showed disease progression after inclusion in our study
Discussion
Approximately half of patients with mCRC carry mutations in KRAS and NRAS genes, and theses mutations result in resistance to anti-EGFR therapy. Although, 50% of patients with wild type RAS present better response to Cetuximab and Panitumumab therapy, not all patients benefit equally from these specific and expensive drugs. Other biomarkers have already been linked to drug resistance, for example PI3KCA, PTEN, Amphiregulin, Epiregulin.Citation9 Several studies have shown that CTC detection is an important prognostic factor in patients with mCRC.Citation10-14 High levels of CTCs are correlated with worse prognosis in breast cancer and their decrease after treatment are related to better survival.Citation15 More recently, liquid biopsy technology, such as CTCs, have shown to be able to provide real time information about the tumor biology. Genotyping CTCs may be an early manner to detect mutations that may have therapeutic implication.Citation10,16-18
Here, we performed analysis of KRAS mutations in CTCs samples and KRAS status in matched primary tumor of patients with mCRC. In matched cases, we found a concordance of KRAS status between CTC and primary tumors of 71%. Similar results were also found by other groups.Citation7,19-21 Fabbri et al.,Citation19 using a dielectrophoresis-based device, had analyzed KRAS mutation (exon 2), and observed 50% of concordance between KRAS mutation in CTCs and matched primary tumor of mCRC. Mostert et al.Citation7 found 5 of the 42 (12%) mCRC patients with KRAS mutation (codon 12/13) in CTCs by nested ASB-PCR. This group also observed that 4 of the 5 patients analyzed had the same KRAS mutation in CTC and their metastasis tissue. Raimondi et al.Citation21 investigated KRAS mutation in CTCs and tumor tissue of CRC patients and observed 29% of correlation of KRAS mutation between CTC and tumor tissue matched. Here, we also observed that 33% of CTCs samples showed KRAS mutation. This result is similar to previous findings published by Gasch et al.,Citation20 that verified the presence of mutation in KRAS in 1 of the 5 CTCs of mCRC patient studied and the same mutation was observed in primary tumor.
Concerning this level of agreement found by our group and othersCitation7,19,20 between CTCs and primary tumors, it has been hypothesized that the development of resistance to EGFR therapy is caused by rare cells with KRAS mutations that pre exist at low levels in mCRC with apparent KRAS wt.Citation22 Diaz et al.Citation22 tested this hypothesis by analyzing 24 patients with mCRC, all of them with KRAS wt in primary tumor and resistant to anti- EGFR therapy. They showed by circulating free tumor DNA (cftDNA) that 9 of the 24 (38%) patients showed KRAS mutation in their serum. These authors suggested that KRAS mutations were present in subclones before the initiation of treatment. In this way, this mutation could be a mediator of acquired resistance to EGFR therapy. It is expected that cells containing KRAS mutation would be released into the circulation being possible to detect by non-invasive manner, as cftDNACitation22 or CTC technique.Citation18 Although our study had fewer numbers of patients, we could observe CTCs containing KRAS mutations in 2 patients whose primary tumors were KRAS wt. As described by Diaz et al.,Citation22 our result suggests that these patients could develop resistance to anti-EGFR treatment by KRAS mutation present in subclones in primary tumor.
In favor of dynamic process that evolves tumor progression, in 2 patients from our study, we found differences in KRAS mutation in CTCs samples (mutation in codon 12) and matched primary tumors (mutation in codon 13). This result suggests that more than one subclone could be present in tumor. Other reports also found discrepancy of concordance between types of KRAS mutations in CTCs and primary tumor or metastasis tissue. Again, it can be explained by the cellular heterogeneity found in epithelial malignancies, which can be reflected by the heterogeneity found in CTCs.Citation7,19,22,23 Another explanation for our findings is the parallel progression model, which suggests that it may be possible to have various metastatic subclones which are disseminated early during disease development and remain dormant for years.Citation24
Additionally, we tried to correlate our findings with clinical-pathological characteristics of patients. It is presumed that patients with KRAS mutations are more likely to develop progression,Citation12 but we could not associate this mutation with progression or another clinical-pathological feature and progression-free survival (PFS) in our CTC samples neither on primary tumors. The lack of association may be due to the small number of cases analyzed and the relatively poor frequency of KRAS mutation in both, CTCs (n = 7) and primary tumors (n = 9). Another group, recently, showed that CTCs count and KRAS status in tumor were independent prognostic factors for patients with mCRC treated with monoclonal antibody (bevacizumab plus chemotherapy). In this study, patients with less CTC's count (<3 CTCs/7.5mL) and KRAS wt in tumor had higher PFS and overall survival (OS) than patients with more CTCs (>3 CTCs/7.5mL) and KRAS mutation in tumor (P = 0.001 and P = 0.004, respectively).Citation17
The presence of heterogeneous cell population in the blood may be an obstacle to the detection of gene mutation in CTCs. We can note that the leukocyte contamination of CTCs sample and their retention on membrane surface could camouflage the results. We observed than in 3 cases without CTC score, KRAS wt has been observed, reflecting the presence of other cells, in addition to the CTC, in the ISET membrane. Maybe, a viable alternative is to make laser microdissection of ISET membranes. In the case of KRAS, which is detected in 30–40% of patients with CRC,Citation7 we believe that the laser microdissection is not necessary and contamination with leukocytes is not sufficient to disrupt the reading. The pyrosequencing is a sensitive technique, being able to identify the mutation in a few cells. Several studies have shown its sensitivity.Citation25,26 However, we should give attention to leucocyte contamination when a qualitative method of sequencing is been employed.
Our results show that molecular characterization of CTCs, such as KRAS status, are relevant, as they show that these cells can be used as “liquid biopsy,” since samples from primary tumors are not always available to make molecular analysis. Moreover, CTCs can be a useful tool to determine real time molecular profile of mCRC patients in order to characterize the evolution of a determined biomarker responsible for resistance. Here, we used pyrosequencing, after isolating CTCs by ISET technology. ISET has been used for research purpose only, but it underwent technical.Citation27,28 and clinical validation.Citation29 In addition, this technique has been show to be extremely sensible and specific as cells with malignant phenotype were found only in patients with cancer and never in healthy volunteers when tested on ISET (reviewed by Chinen et al, 2014).Citation30 Thus, we suggest that the method described here using CTCs isolated by ISET can be used for the most common mutations, as it is faster, cheaper and practical. However, despite its feasibility, molecular analysis after CTCs isolation by ISET, must be tested in a larger cohort of patients in order to validate our findings.
In conclusion, we demonstrated that CTC can be characterized based on molecular profile and genetic analysis. Moreover, for mCRC, there are discrepancies among KRAS mutation both, in CTCs and primary tumor. The hypothesis that this disease presents dynamic changes from the point of view of molecular abnormalities is reinforced by our data.
Materials and Methods
Patients
This study included 26 patients with mCRC obtained from the A C Camargo Cancer Center, São Paulo-Brazil, between February and November (2013). It was approved by local Ethics Committee (study nº 1708/12). Patients with stage IV CRC were recruited at Clinical Oncology Department. The majority of patients (18/26) (69.2%) had received previous therapy (first line chemotherapy) and were receiving monoclonal therapy with anti-EGFR antibody (17/26) (63.4%) before the blood collection for CTC analysis. The most common site of metastasis was liver (8/26) (30.8%), followed by liver and lung (7/26) (29.6%)
Evaluation of tumor response to therapy followed the radiologic criteria of RECIST (version 1.1).Citation31 Clinical-pathological characteristics of patients are summarized in .
Table 4. Clinical and pathological characteristics of patients with mCCR
CTCs were isolated by ISET technology (Isolation by Size of Epithelial Tumors; Rarecells Diagnostics, Paris, France). Blood samples (8mL) from patients were collected in EDTA tubes, stored at room temperature for up to 4 hours under homogenization and processed by ISET according to manufacturer's instructions. ISET methodology isolates intact CTCs from blood through direct filtration without using antibodies, as it exploits the larger size of tumor cells as compared with leukocytes. Recently, we reported the advantage of ISET, as compared with a “marker-related” approach, in showing progression of a metastatic lung cancer before the progression became evident by imaging.Citation32 ISET uses a polycarbonate membrane with 8-μm-diameter cylindrical pores. We used procedures of this platform and CTCs definition criteria according to previous detailed reports.Citation33 ISET membrane spots were cut and submitted to immunocytochemistry with anti-CD45 antibody (1:100; clone 2B11 + PD7/26, Dako™), a surface leukocyte marker, in order to differentiate them from the CTCs. For CTCs counting, 8 spots from the ISET filter were used (corresponding to 8 mL of blood), after staining with hematoxylin and eosin. Counting was made per mL of blood. Cells were considered as CTCs if they were negative for CD45, have presented with hypercromatic nucleus, irregular shape, high cytoplasm nucleus ratio (>0.5) and nuclear size ≥12 µm. Images of results obtained with this technique were taken using a light microscope (Research System Microscope BX61– Olympus, Tokyo, Japan) coupled to a digital camera (SC100–Olympus, Tokyo, Japan) at 40× and 60× magnification.
Although ISET has passed through technical,Citation27,28 and clinical validation,Citation32,34 we made our own tests of its sensitivity and specificity, as described previously by Chinen et al., (2014).Citation30 Briefly, we spiked different numbers of cells HCT 116 (colorectal carcinoma cell line) in 1 ml of blood from healthy subject. We made the dilutions in a counting chamber before spiking the cells. Then, we filtered the blood by using the ISET. We made the experiment with 3 concentrations, tested in triplicate, in one ml of blood: 50, 100 and 150 cells. Each spot was stained with antibody against CD45 to visualize leucocytes, counterstained with haematoxilin and read in a light microscope. We found a mean number of HCT 116 cells closely consistent with the number of cells we added to the blood before ISET.
Paraffin-embedded primary tumor samples from the same patients included for CTCs´ analysis were evaluated for KRAS mutation by Anatomic Pathology Department, as the analysis of KRAS status is standard practice in AC Camargo Cancer Center.
Analysis of Kras mutation by pyrosequencing
We performed DNA extraction of CTC in polycarbonate membrane obtained by ISET technique (after quantification of CTCs) by QIAamp DNA Micro Kit (Qiagen, Valencia, CA USA). The spot's membrane (4 spots per patient) were cut in small pieces and transferred into the lyses buffer and proteinase K supplied by Kit and the extractions were conducted according to protocol provided by the manufacturer. For analysis of primary tumor, total DNA was isolated from formalin-fixed paraffin embedded tissue with QIAamp DNA Micro Kit (Qiagen, Valencia, CA USA). All pyrosequencing reactions included DNA sample without mutation of KRAS gene, supplied by kit. We could not perform analyses of KRAS gene in metastasis samples from our patients because the majority of them were not available.
PCR amplification and pyrosequencing targeted for KRAS codon 12 and 13 were done by PyroMark Q24 KRAS (24), version 2.0, version 2.0 (QIAGEN, Valencia, CA USA), respectively, using the instrument PyroMark Q96 (QIAGEN, Valencia, CA USA).
The results from KRAS status in primary tumor samples were retrieved from medical records. KRAS analysis is currently made in mCRC patients as approved test to help in choice of anti-EGFR therapies.Citation35 The evaluation of KRAS status in these samples was performed by the same methods described above for CTCs, by the Anatomic Pathology Department of the AC Camargo Cancer Center.
Statistical analysis
Association between mutational status of KRAS and clinical and pathological parameters were assessed by the X2-test. The progression free survival (PFS) graphics were constructed according to the Kaplan-Meier statistical method and the difference between curves was analyzed by log-rank method.
All p-values were on 2-tailed statistical analysis and p value ≤0.05 was considered as statistically significant. Statistical analysis were performed using the SPSS 15.0 statistical package (SPSS Inc., Chicago, IL USA).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Brazil for financial support.
References
- Yang MJ, Chiu HH, Wang HM, Yen LC, Tsao DA, Hsiao CP, Chen YF, Wang JY, Lin SR. Enhancing detection of circulating tumor cells with activating KRAS oncogene in patients with colorectal cancer by weighted chemiluminescent membrane array method. Ann Surg Oncol 2010; 17(2):624-33; PMID:19937133; http://dx.doi.org/10.1245/s10434-009-0831-8
- Yen LC, Yeh YS, Chen CW, Wang HM, Tsai HL, Lu CY, Chang YT, Chu KS, Lin SR, Wang JY. Detection of KRAS oncogene in peripheral blood as a predictor of the response to cetuximab plus chemotherapy in patients with metastatic colorectal cancer. Clin Cancer Res 2009; 15(13):4508-13; PMID:19549774; http://dx.doi.org/10.1158/1078-0432.CCR-08-3179
- Maheswaran S, Haber DA. Circulating tumor cells: a window into cancer biology and metastasis. Current Opin Genet Dev 2010; 20:96-9; PMID:20071161; http://dx.doi.org/10.1016/j.gde.2009.12.002
- Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008; 359(17):1757-65; PMID:18946061; http://dx.doi.org/10.1056/NEJMoa0804385
- Ramos FJ, Macarulla T, Capdevila J, Elez E, Tabernero J. Understanding the predictive role of K-ras for epidermal growth factor receptor-argeted therapies in colorectal cancer. Clin Colorectal Cancer 2008; 7 Suppl 2:S52-7; PMID:19064407; http://dx.doi.org/10.3816/CCC.2008.s.008
- Phipps AI, Buchanan DD, Makar KW, Win AK, Baron JA, Lindor NM, Potter JD, Newcomb PA. KRAS-mutation status in relation to colorectal cancer survival: the joint impact of correlated tumour markers. Br J Cancer 2013; 108(8):1757-64; PMID:23511557; http://dx.doi.org/10.1038/bjc.2013.118
- Mostert B, Jiang Y, Sieuwerts AM, Wang H, Bolt-de Vries J, Biermann K, Kraan J, Lalmahomed Z, van Galen A, de Weerd V, et al. KRAS and BRAF mutation status in circulating colorectal tumor cells and their correlation with primary and metastatic tumor tissue. Int J Cancer 2013; 133(1):130-41; PMID:23233388; http://dx.doi.org/10.1002/ijc.27987
- Okada.K, Kimura T, Mikamo H, Kasahara K, Seki M, Takakura S, Tokimatsu I, Ohmagari N,Takahashi Y, Matsumoto K, et al. Clinical practice guidelines for therapeutic drug monitoring of arbekacin: a consensus review of the Japanese Society of Chemotherapy and the Japanese Society of Therapeutic Drug Monitoring. J Infect Chemother 2014; 20(1):1-5; PMID:2448616; http://dx.doi.org/10.1016/j.jiac.2013.08.008
- De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010; 11(8):753-62; PMID:20619739; http://dx.doi.org/10.1016/S1470-2045(10)70130-3
- Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008; 26(19):3213-21; PMID:18591556; http://dx.doi.org/10.1200/JCO.2007.15.8923
- Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse MA, Mitchell E, Miller MC, et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann Oncol 2009 Jul; 20(7):1223-9; PMID:19282466; http://dx.doi.org/10.1093/annonc/mdn786
- Sastre J, Maestro ML, Gómez-España A, Rivera F, Valladares M, Massuti B, Benavides M, Gallén M, Marcuello E, Abad A, et al. Circulating tumor cell count is a prognostic factor in metastatic colorectal cancer patients receiving first-line chemotherapy plus bevacizumab: a Spanish Cooperative Group for the Treatment of Digestive Tumors study. Oncologist. 2012; 17(7):947-55; PMID:22643538; http://dx.doi.org/10.1634/theoncologist.2012-0048
- Pierga JY, Hajage D, Bachelot T, Delaloge S, Brain E, Campone M, Diéras V, Rolland E, Mignot L, Mathiot C, et al. High independent prognostic and predictive value of circulating tumor cells compared with serum tumor markers in a large prospective trial in first-line chemotherapy for metastatic breast câncer patients. Ann Oncol. 2012; 23(3):618-24; PMID:21642515; http://dx.doi.org/10.1093/annonc/mdr263
- de Albuquerque A, Kubisch I, Stölzel U, Ernst D, Boese-Landgraf J, Breier G, Stamminger G, Fersis N, Kaul S. Prognostic and predictive value of circulating tumor cell analysis in colorectal cancer patients. J Transl Med 2012; 10:222; PMID:23146106; http://dx.doi.org/10.1186/1479-5876-10-222
- Cristofanilli M, Fortina P. Circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013; 369(1):93; PMID:23822789; http://dx.doi.org/10.1056/NEJMc1306040
- Rahbari NN, Aigner M, Thorlund K, Mollberg N, Motschall E, Jensen K, Diener MK, Büchler MW, Koch M, Weitz J. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology. 2010; 138(5):1714-26; PMID:20100481; http://dx.doi.org/10.1053/j.gastro.2010.01.008
- Sastre J, Vidaurreta M, Gómez A, Rivera F, Massutí B, López MR, Abad A, Gallen M, Benavides M, Aranda E, et al. Spanish Cooperative Group for the Treatment of Digestive Tumors. Prognostic value of the combination of circulating tumor cells plus KRAS in patients with metastatic colorectal cancer treated with chemotherapy plus bevacizumab. Clin Colorectal Cancer 2013; 12(4):280-6; PMID:24012456; http://dx.doi.org/10.1016/j.clcc.2013.06.001
- Steinert G, Schölch S, Niemietz T, Iwata N, García SA, Behrens B, Voigt A, Kloor M, Benner A, Bork U, et al. Immune escape and survival mechanisms in circulating tumor cells of colorectal cancer. Cancer Res 2014; 74(6):1694-704; PMID:24599131; http://dx.doi.org/10.1158/0008-5472.CAN-13-1885
- Fabbri F, Carloni S, Zoli W, Ulivi P, Gallerani G, Fici P, Chiadini E, Passardi A, Frassineti GL, Ragazzini A, et al. Detection and recovery of circulating colon cancer cells using a dielectrophoresis-based device: KRAS mutation status in pure CTCs. Cancer Lett 2013; 335(1):225-31; PMID:23419522; http://dx.doi.org/10.1016/j.canlet.2013.02.015
- Gasch C, Bauernhofer T, Pichler M, Langer-Freitag S, Reeh M, Seifert AM, Mauermann O, Izbicki JR, Pantel K, Riethdorf S. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin Chem 2013; 59(1):252-60; PMID:23136247; http://dx.doi.org/10.1373/clinchem.2012.188557
- Raimondi C, Nicolazzo C, Gradilone A, Giannini G, De Falco E, Chimenti I, Varriale E, Hauch S, Plappert L, Cortesi E, Gazzaniga P. Circulating tumor cells: exploring intratumor heterogeneity of colorectal cancer. Cancer Biol Ther 2014; 15(5):496-503; PMID:24521660; http://dx.doi.org/10.4161/cbt.28020
- Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012; 486(7404):537-40; PMID:22722843
- Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res 2010; 16(3):790-9; PMID:20103678; http://dx.doi.org/10.1158/1078-0432.CCR-09-2446
- Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer 2009; 9: 302-12; PMID:19308069; http://dx.doi.org/10.1038/nrc2627
- Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn 2005 Aug; 7(3):413-21; PMID:16049314; http://dx.doi.org/10.1016/S1525-1578(10)60571-5
- Sha J, Liang G, Pan J, Xuan H, Ping P, Li D, Bo J, Liu D, Shen W, Liu W, et al. Application of pyrosequencing technique for improved detection of K-Ras mutation in formalin-fixed and paraffin-embedded prostate carcinoma tissues in Chinese patients. Clin Chim Acta. 2012; 413(19-20):1532-5; PMID:22732508; http://dx.doi.org/10.1016/j.cca.2012.06.008
- Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schütze K, Capron F, Franco D, Pazzagli M, Vekemans M, et al. Isolation by size of epithelial tumor cells : a new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol 2000; 156(1):57-63; PMID:10623654; http://dx.doi.org/10.1016/S0002-9440(10)64706-2
- Krebs MG, Hou JM, Sloane R, Lancashire L, Priest L, Nonaka D, Ward TH, Backen A, Clack G, Hughes A, et al Analysis of circulating tumor cells in patients with non-small cell lung cancer using epitelial marker-dependent and -independent approaches. J Thorac Oncol 2012; 7(2):306-15; PMID:22173704; http://dx.doi.org/10.1097/JTO.0b013e31823c5c16
- Hofman V, Long E, Ilie M, Bonnetaud C, Vignaud JM, Fléjou JF, Lantuejoul S, Piaton E, Mourad N, Butori C, et al. Morphological analysis of circulating tumour cells in patients undergoing surgery for non-small cell lung carcinoma using the isolation by size of epithelial tumour cell (ISET) method. Cytopathology 2012; 23(1):30-8; PMID:21210876; http://dx.doi.org/10.1111/j.1365-2303.2010.00835.x
- Chinen LTD, Mello CAL, Abdallah EA, Ocea LMM, Buim ME, Breve NM, Junior JLG, Fanelli, MF, Paterlini-Brechót P. Isolation, detection, and immunomorphological characterization of circulating tumor cells (CTCs) from patients with different types of sarcoma using isolation by size of tumor cells: a window on sarcoma-cell invasion. Onco Targets Ther 2014; 7:1609-17; PMID:25258541; http://dx.doi.org/10.2147/OTT.S62349
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45:228-47; PMID:19097774; http://dx.doi.org/10.1016/j.ejca.2008.10.026
- Chinen LT, de Carvalho FM, Rocha BM, Aguiar CM, Abdallah EA, Campanha D, Mingues NB, de Oliveira TB, Maciel MS, Cervantes GM, et al. Cytokeratin-based CTC counting unrelated to clinical follow up. J Thorac Dis 2013; 5(5):593-9; PMID:24255771
- Khoja L, Backen A, Sloane R, Menasce L, Ryder D, Krebs M, Board R, Clack G, Hughes A, Blackhall F, et al. A pilot study to explore circulating tumour cells in pancreatic cancer as a novel biomarker. Br J Cancer 2012; 106(3):508-16; PMID:22187035; http://dx.doi.org/10.1038/bjc.2011.545
- Hofman V, Ilie M, Long E, Selva E, Bonnetaud C, Molina T, Vénissac N, Mouroux J, Vielh P, Hofman P. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay TM and the isolation by size of epithelial tumor cell method. Int J Cancer 2011c; 129:1651-60; PMID:21128227; http://dx.doi.org/10.1002/ijc.25819
- Tougeron D, Cortes U, Ferru A, Villalva C, Silvain C, Tourani JM, Levillain P, Karayan-Tapon L. Epidermal growth factor receptor (EGFR) and KRAS mutations during chemotherapy plus anti-EGFR monoclonal antibody treatment in metastatic colorectal cancer. Cancer Chemother Pharmacol 2013; 72(2):397-403; PMID:23765179; http://dx.doi.org/10.1007/s00280-013-2211-0
