Abstract
Multiple juxtacrine and paracrine interactions occur between cancer cells and non-cancer cells of the tumor microenvironment (TME) that direct tumor progression. Cancer Associated Fibroblasts (CAFs) are an integral component of the TME, and the majority of breast tumor stroma is comprised of CAFs. Heterotypic interactions between cancer cells and non-cancer cells of the TME occur via soluble agents, including cytokines, hormones, growth factors, and secreted microRNAs. We previously identified a microRNA signature indicative of hyperactive MAPK signaling (hMAPK-miRNA signature) that significantly associated with reduced recurrence-free and overall survival. Here we report that the hMAPK-miRNA signature associates with a high metric of stromal cell infiltrate, and we investigate the role of microRNAs, particularly hMAPK-microRNAs, secreted by CAFs on estrogen receptor (ER) expression in breast cancer cells. ER-positive MCF-7/ltE2- cells were treated with conditioned media (CM) from CAFs derived from breast cancers of different PAM50 subtypes (CAFBAS, CAFHER2, and CAFLA). CAF CM isolated specifically from ER-negative primary breast tumors led to ER repression in vitro. Nanoparticle tracking analysis and transmission electron microscopy confirmed the presence of CAF-secreted exosomes in CM and the uptake of these exosomes by the ER+ MCF-7/ltE2- cells. Differentially expressed microRNAs in CAF CM as well as in MCF-7/ltE2- cells treated with this CM were identified. Knockdown of miR-221/222 in CAFBAS resulted in knockdown of miR221/222 levels in the conditioned media and the CM from CAFBAS; miR221/222 knockdown rescued ER repression in ER-positive cell lines treated with CAFBAS-CM. Collectively, our results demonstrate that CAF-secreted microRNAs are directly involved in ER-repression, and may contribute to the MAPK-induced ER repression in breast cancer cells.
Abbreviations
| CAFs | = | cancer associated fibroblasts |
| CM | = | conditioned media |
| ER | = | estrogen receptor |
| hMAPK | = | hyperactive MAPK |
| MAPK | = | mitogen activated protein kinase |
| MCF-7/ltE2- | = | MCF-7 cell line long-term conditioned for growth in estrogen depleted conditions |
| miRNA | = | microRNA |
| NSC | = | non-silencing control |
| RT-PCR | = | real time polymerase chain reaction |
| TME | = | tumor microenvironment. |
Introduction
While understanding the genetic and epigenetic changes that occur within breast cancer cells has identified novel therapeutic targets and biomarkers for disease progression, over the past decade an accumulating body of evidence has clearly established the significance of surrounding stromal cells (also known as the tumor microenvironment (TME)) in promoting breast cancer progression.Citation1-4 Cross-talk between the TME and cancer cells is known to play a major role in driving breast cancer progression and poor clinical outcome.Citation1,5-11 Cancer associated fibroblasts (CAFs) form an essential component of the TME, and the majority of TME in many cancer types, including breast cancer, is comprised mainly of CAFs.Citation12-14 There is increasing evidence highlighting the role of CAFs in various processes that range from tumor initiation, progression, epithelial to mesenchymal transition (EMT), tumor invasion, angiogenesis, and metastasis, as well as conferring drug resistance to cancer cells.Citation5,15-20 CAFs secrete soluble factors including cytokines, MMPs, and growth factors which are major mechanisms by which CAFs promote tumor progression.Citation15,16,19,21-27 Many of these growth factors and cytokines activate the growth factor dependent-MAPK signaling in breast tumor cells Citation17,27-33, which has been well-established as a major pathway activated in breast cancer.
We previously established that hyperactivation of ERK1/2 MAPK in breast cancer cells leads to repression of estrogen receptor (ER),Citation34 and we identified a hyperactive MAPK (hMAPK) gene signature that correlates with ER-negative breast cancers.Citation35 Furthermore, we demonstrated the importance of MAPK activation in establishing the ER-negative breast tumor phenotype, as inhibition of ERK1/2 MAPK leads to re-expression of ER in multiple ER-negative breast cancer cell lines and in primary cultures established from ER-negative breast tumors.Citation36,37
Aberrant mitogenic signaling also affects expression of microRNAs (miRNAs).Citation38 We have recently identified a novel, patient-derived miRNA signature indicative of hMAPK signaling in breast cancer.Citation38 This signature identifies breast cancers with poor clinical outcomes, including decreased recurrence-free and disease-specific survival; moreover, ER+ breast cancers exhibiting this hMAPK-miRNA signature had reduced recurrence-free and disease-specific survival with hormone therapy.Citation38 Several of the hMAPK-miRNAs have been shown to target various proteins like ER, EMT transcription factors, markers of luminality, stemness, and ECM.Citation39-43
miRNAs are small, 19-22 nucleotide long, non-coding RNAs, are often found to be differentially regulated in cancer, and are known to play multifaceted roles in tumorigenesis.Citation44-47 miRNAs are known to have both oncogenic and tumor suppression functions.Citation45 In breast cancer, miRNA expression associates with tumor grade, proliferation, and the major molecular subtypes.Citation38,48-52 It is now well known that changes in the level of miRNAs can occur by interaction between cancer cells and stromal cells via paracrine secreted cytokines/growth factors or direct secretion of miRNAs.Citation45 The hMAPK-miRNA signature was developed from primary breast cancer specimens that are composed of a tumor cell population that contains breast cancer cells and also containing infiltrating stromal cells. This suggests that there may be significant stromal contribution to these hMAPK-miRNAs, as the presence of non-tumor stromal cells has been shown to strongly impact gene expression signatures.Citation53-57
Our results show that the hMAPK-miRNA signature significantly associates with tumors that have a high stromal score, as defined by Yoshihara et. al.Citation54 We demonstrate that expression of a number of the miRNAs in our hMAPK-miRNA signature are more highly expressed in CAFs than in cultured dissociated tumor cells. Treatment of ER+ breast cancer cells with conditioned media (CM) from CAFs represses both ER protein and mRNA expression. Importantly, we show that this ER repression is specific to CM isolated from CAFs from ER-negative breast tumors (CAF21H2N and CAF23BAS), and not seen in CM isolated from CAFs obtained from ER+-positive tumors (CAF19LA). The effect of ER repression at protein, mRNA and ER 3'UTR luciferase activity was more significant by conditioned media from CAF23BAS when compared to CAF19LA alone. We found that miR-221 and miR-222 were overexpressed in CM from CAF23BAS vs CAF19LA, as determined by Nanostring analysis. miR-221/222 are part of the hMAPK-miRNA signature and known to repress ER expression.Citation42 To determine the role of miR-221/222 in CAF mediated ER repression, we performed knockdown of these miRNAs in CAF23BAS and were able to partially rescue the CAF23BAS mediated ER repression. Our results indicate that CAF secreted hMAPK-miRNAs miR-221/222 can influence the ER-negative phenotype of breast cancer cells via paracrine interactions within the tumor microenvironment and that this miRNA-mediated repression of ER is specific to CM from CAFs obtained from basal breast cancers.
Results
Correlation of hMAPK-microRNA signature with tumor stroma and CAFs
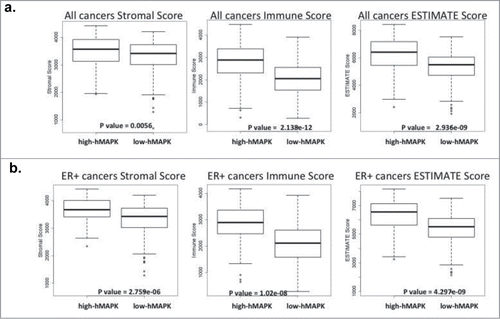
Gene expression profiling of tumor samples have helped better classify tumors and understand their biology. Tumor samples are not only composed of tumor cells but also consist of infiltrating stromal and immune cells. It is now clear that the associated stromal and immune cells can influence gene expression and miRNA expression profiles of tumor samples.Citation45,53-57 Using breast cancer specimens, we developed an hMAPK-miRNA signature that associates with poor survival.Citation38 Since the breast cancer specimens in patient datasets are composed of tumor cells and associated stroma, we wanted to investigate the connection between hMAPK-miRNAs and stroma. We employed the method developed by Yoshihara et al.Citation54 to calculate stromal and immune scores for breast cancers from the TCGA and METABRIC data sets; these scores provide an indication for the proportion of immune and stromal infiltrate of a tumor sample. According to this method, a higher stromal score or immune score indicates a higher stromal or immune cell infiltrate in the tumor, and the ESTIMATE (estimation of stromal and immune cells in malignant tumor tissue) score represents the combination of stromal and immune scores. Breast tumors from the TCGA and METABRIC datasets classified as high hMAPK-miRNA according to our signature are significantly enriched for tumors with higher stromal, immune, and ESTIMATE scores, when compared to breast tumors classified as low hMAPK-miRNAs (). Because CAFs make up a large portion of breast tumor stromaCitation12-14 and have been shown to contribute to gene and miRNA expression of tumor cells,Citation45,53-57 we hypothesized that CAFs could similarly contribute to the hMAPK-miRNA signature.
Figure 1. hMAPK miRNA signature associates with stromal, immune and estimate score. (A) TCGA breast cancer gene expression data set comparing hMAPK-miRNA signature to stromal, immune and estimate score. (B) ER+ breast cancer gene expression TCGA dataset comparing hMAPK-miRNA signature to stromal, immune and estimate score.
Media conditioned by CAFs derived from different breast tumor subtypes elicits distinct effects on ER repression in ER-positive breast cancer cells
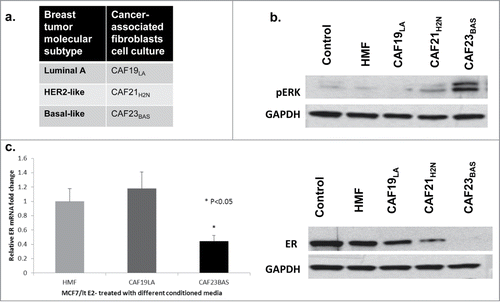
We previously reported that breast tumors from basal-like and HER2 molecular subtypes are enriched for the hMAPK-miRNA signature.Citation38 Because CAFs are known to secrete factors that can stimulate growth factor signaling pathways in breast cancer cells, and hyperactivation of growth factor pathways in breast cancer contributes to repression of ER and development of hormone independent growth, we hypothesized that CAFs derived from different subtypes of breast cancer would differentially impact ER biology in ER-positive breast cancer cells. To determine the in vitro effect of CAF interactions with ER+ breast cancer cells, MCF-7/ltE2- cells were treated with conditioned media collected from CAFs obtained from different breast cancer molecular subtypes () and normal human mammary fibroblasts at various time points. Conditioned media from CAF21H2N and CAF23BAS transiently activated MAPK signaling (20min). CM from CAF23BAS resulted in sustained MAPK activation and repression of ER (4hrs) in MCF-7/ltE2- cells (). Conditioned media from additional luminal and basal-like CAFs, CAF8LA and CAF31BAS respectively (Fig. S1A), produced similar results. These CAFs were confirmed using fibroblasts markers (Fig. S1B and 1C) and their ability to not form soft agar colonies (Fig. S1D). CM from CAF31BAS elicited a more sustained MAPK activation (20min) and repression of ER (4h) in MCF-7/ltE2- cells than CM from CAF8LA (Fig. S1E). ER gene expression was analyzed in total RNA isolated from MCF-7/ltE2- cells treated with different CAF-CM at various time points. CM from CAF19LA did not cause repression of ER mRNA, while conditioned media from CAF23BAS led to significant repression in ER mRNA (). Similar results were obtained by treating parental MCF-7 cells with CAF conditioned media (data not shown); we employed the MCF-7/ltE2- cell line model because the differences in ER protein expression were more pronounced than in parental MCF-7 cells, due to the enhanced expression of ER in the MCF-7/ltE2- cells.
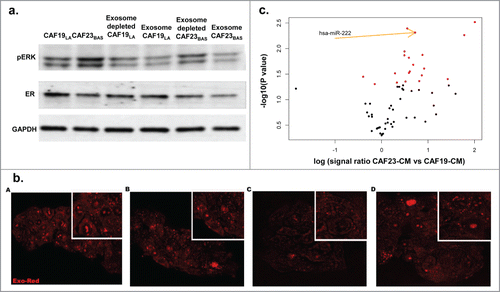
Figure 2. CAF23BAS conditioned media leads to estrogen receptor repression. (A) Table showing the 3 different types of cancer associated fibroblasts (CAFs) cell cultures isolated from 3 different primary breast tumors. (B) Western blot data showing ER (4h) and pERK (20min) expression when MCF-7/lt E2- cells are treated with different CAF-CM. GAPDH used as loading control. (C) qPCR data showing ER mRNA expression when treated with different CAF-CM.
miRNA-dependent repression of ER in ER+ cancer cells treated with CM from basal-like CAFs
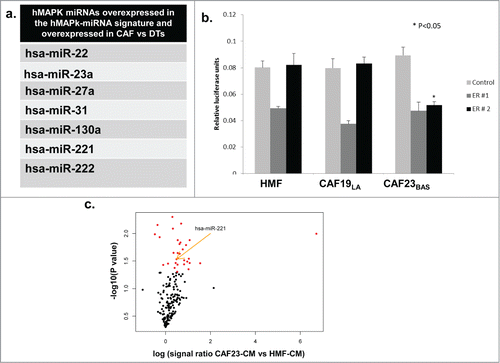
In order to address if CAFs could be the source of over-expressed miRNAs in the hMAPK-miRNA signature we performed miRNA expression analysis on CAFs and dissociated primary human breast tumor cells (DTs) using TaqMan® Array Human MicroRNA A+B Cards Set v2.0 panel. (CAFs and DTs are previously describedCitation34). We observed that several miRNAs overexpressed in the hMAPK-miRNA signature were significantly overexpressed in CAFs compared to dissociated tumor cells (). Several of these hMAPK-miRNAs overexpressed in CAFs compared to DTs are also known to target ER, (including miR-22, miR-221 and miR-222).Citation38,42 We treated the hyperactive MAPK cell line ca-Raf/MCF-7Citation37,58 with MAPK signaling inhibitor U0126 and found that hMAPK miR221/222 levels went down with the decrease in MAPK signaling. The decrease in hMAPK miR221/222 levels upon U0126 treatment was comparable to antagmiR knockdown of miR221/222 (Fig. S2A). To confirm that hMAPK-miR221/222 plays a role in the MAPK-mediated ER repression, we transfected the ca-Raf/MCF-7 cells with ER 3'UTR Dual Glo® luciferase reporter constructs or a control vector. ER 3'UTR Dual Glo® vectors contains firefly luciferase (LucF) under 3'UTR control of the full ER 3'UTR (ER 3'UTR is split among 2 vectors [ER # 1 and ER # 2] due to its large size), while the LucF in the control vector contains a non-targeted 3'UTR (Fig. S2B). We also observed an increase in the ER 3'UTR luciferase activity in ca-Raf/MCF-7 cell lines upon knockdown of miR221/222 (). Thus MAPK-regulated expression of miR221/222 participates in MAPK-mediated ER repression. To investigate if the ER repression effect of CAF-CM involved miRNA regulation, MCF-7/ltE2 cells were transfected with ER 3'UTR Dual Glo® luciferase reporter constructs or control vector, and then treated with CM from HMF vs CAFs. CAF23BAS–CM repressed ER 3'UTR luciferase activity, while CM from other CAFs did not and furthermore, the repression was with the ER 3'UTR#2 sequence containing miR221/222 sites (); indicating that CAF23BAS-CM impacts regulation of the miRNA regulatory elements present in the latter portion of the ER 3'UTR. Moreover, when the ER+ MCF-7/ltE2- cells were treated with different CAF CM we found using nCounter® Human miRNA Expression Assays that expression of several miRNAs, including miR221/222, within MCF-7/ltE2- was increased upon treatment with CAF23BAS CM and not HMF CM (). This shows that CAF-CM elicits differential ER repression by short-term expression of miRNAs upon interaction with tumor cells.
Figure 3. hMAPK miRNA 221/222 are involved in CAF mediated ER repression. (A) Table showing list of hMAPK miRNAs in all the CAF cell cultures vs dissociated tumor cells (DTs). (B) Treatment of MCF-7/lt E2- cells transfected with dual luciferase reporter vector with different CAF conditioned media. Data represents relative luciferase units (RLU). (C) A volcano plot showing miRNAs that are differentially expressed when MCF-7/lt E2- cells are treated with CAF23BAS vs HMF conditioned media.
Depletion of exosomes in conditioned media from CAF23BAS prevents ER repression
CAFs and other cells of the TME can secrete miRNAs into the media via exosomes/microvesicles.Citation59 It should be noted that due to lack of a sophisticated nomenclature distinguishing exosomes from microvesicles,Citation60 we chose to use the term “exosomes” in our studies. To determine if CAF-secreted exosomes were involved in the ER repression observed in CAF-CM treated MCF-7 cells, CAF-conditioned media was depleted of exosomes by ultracentrifugation. It should be noted that complete removal of exosomes is not readily achievable by conventional means, and though this media was significantly depleted of exosomes, we were nevertheless able to detect trace amounts of miRNAs (data not shown). MCF-7/lt E2- cells were then treated with both concentrated exosomes and exosome-depleted media. Exosome depletion in CAF19LA-CM had no effect on MAPK activation or ER expression of treated MCF-7/ltE2- cells (). Exosome-depleted CAF23BAS-CM no longer repressed ER expression while MAPK activation was still maintained (), suggesting that CAF-secreted exosomes mediate ER repression but not stimulation of MAPK activation. These results further suggest that while MAPK activation within the MCF-7/ltE2- cells would result in ER repression,Citation58 the MAPK activation seen with CM treatment is not the primary driver for ER-repression in MCF-7 cells within the short time frames we observed. These data () along with the repression of ER 3'UTR (), indicate that CAF23BAS CM-mediated ER repression seen within short time frames is attributable to the exosomal miRNAs secreted by the CAFs.
Figure 4. CAF-CM with exosomes containing hMAPK miR221/222 are involved in ER repression.(A) Western blot data showing effect of exosome depleted CAF conditioned media on pERK and ER of MCF/ltE2- cells treated with different CAF conditioned media. GAPDH used as loading control. (B) Cancer associated fibroblast-derived exosomes uptake by MCF-7/lt E2- breast cancer cells. (A) HMF exosomes. (B) CAF19 exosomes. (C) CAF21 exosomes. (D). CAF23 exosomes. (C). A volcano plot showing miRNAs that are differentially expressed in CAF23BAS vs. CAF19LA conditioned media.
Uptake of exosomes secreted by CAFs by breast cancer cells
Nanoparticle tracking analysis and transmission electron microscopy demonstrated that CAFs and HMFs secrete exosomes into conditioned media (Fig. S3A and B). Nanoparticle tracking analysis showed that CAF secreted exosomes were within the exosome size range of 30-100 nm (Fig. S3A).Citation61 Using transmission electron microscopy it can be seen that the exosomes exhibited the characteristic saucer-like morphology (Fig. S3B).Citation61 Additionally, both HMF and CAF secreted exosomes expressed the exosomal marker CD63 (Fig. S3C). MCF-7/ltE2- cells treated with Exo-Red labeled CAF-secreted exosomes display uptake of the exosomes when visualized using confocal microscopy () proving that the exosomes secreted in CAF-CM enter the MCF-7/lt E2- cells.
CAF conditioned media from different breast cancer subtypes contains diverse miRNA profiles
The expression of miRNAs in various CAF-CM, as well as in MCF-7/ltE2- cells following treatment with various CAF CM, was identified using nCounter® Human microRNA Expression Assays. CAF23BAS-CM exhibited overexpression of several miRNAs compared to CAF19LA-CM, including miR221/222 (). These results show that CAF23BAS secreted miR221/222 could be responsible for the ER repression.
Knockdown of miR221/222 in CAF23BAS rescues ER expression
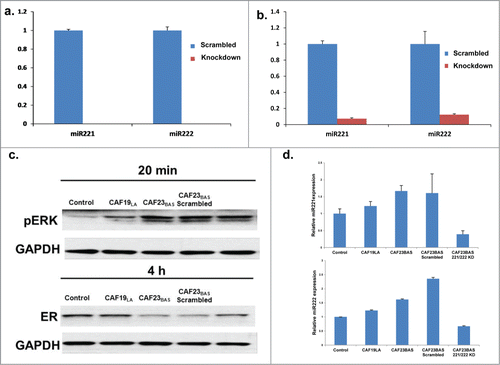
Nanostring miRNA analysis indicated that miR221/222 is secreted in CAF23BAS CM and overexpressed in MCF-7/ltE2- cells treated with CAF23BAS CM, may promote ER repression through miRNA transfer into MCF-7/ltE2- cells. Knockdown of miR221/222 in CAF23BAS was performed using antagomiRs and CM was collected 96hrs after antagomiR transfection. Knockdown of miR221/222 in CAF23BAS cells did not have any impact on cell proliferation during this time-frame (Fig. S4A). miR221/222 knockdown was confirmed by qRT-PCR, and following knockdown miR-221/222 was essentially completely depleted in CAF23BAS cells and significantly reduced in CAF23BAS CM (). Depletion of miR221/222 in CAF23BAS CM attenuates the ability of CAF23BAS CM to repress ER in MCF-7/ltE2- cells () and ER expression correlates with the miR-221/222 expression levels in CAF23BAS CM (). Treatment of MCF-7/ltE2- cells with the miR-221/222 antagomiR for the same amount of time had no effect on ER expression (Fig. S4B). Together, these data indicate that miR221/222 secreted in CAF23BAS CM is a key component involved ER repression caused by CAF23BAS CM.
Figure 5. Knockdown of hMAPK miR221/222 prevents CAF mediated ER repression. (A) qPCR data showing levels of miR 221 and miR 222 in CAF23BAS cells after knockdown. (B) qPCR data showing levels of miR 221 and miR 222 in CAF23BAS conditioned media after knockdown. (C). Western blot data showing effect on pERK and ER of MCF-7lt/E2- cells after treatment with CAF23BAS miR221/222 knockdown conditioned media. GAPDH used as loading control d. qPCR data showing the miR221 and 222 expression in MCF-7/ltE2- cells after treatment with CAF23BAS miR221/222 knockdown conditioned media.
Discussion
It is well appreciated that cancer cells change the behavior of non-cancer stromal cells (i.e immune cells, pericytes, cancer associated fibroblasts, adipocytes, and endothelial cells) to establish a TME that can facilitate tumor progression.Citation1,5-11,60,62-74 Within the TME, CAFs are key regulators of tumor initiation, invasion, EMT, angiogenesis, and metastasis.Citation12,13,Citation5,15-20 Prior studies have demonstrated gene and protein expression heterogeneity between CAFs obtained from different molecular subtypes of breast cancer,Citation75,76 suggesting that aggressive tumor characteristics associated with Basal-like and HER2 breast cancer subtypes may be attributable in part to CAFs. In this study we investigated differences in paracrine interactions between breast cancer cells and CAFs obtained from primary breast cancers from 3 distinct molecular subtypes-Luminal A, HER2-like and Basal (, Fig. S1A, ref.Citation34).
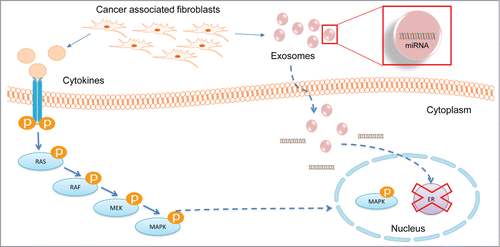
Within the TME, stromal cells communicate with tumor cells and other stromal cells via a multitude of paracrine interactions. Apart from cytokines, hormones, and growth factors, it has recently become appreciated that exosomes also mediate paracrine communication within the TME.Citation45,60 The exosomes that are on the nanometer scale can carry proteins, DNA, mRNA, and miRNA that facilitate communication between different cells within the TME. Exosomal miRNAs and mRNAs can be taken up by tumor cells; this can not only affect the expression of various proteins, but in turn can affect tumor progression and metastasis.Citation45,59, 60,62-66,Citation69,Citation73,74 Additionally, it is known that miRNAs are packaged within exosomes in a non-random fashion.Citation45,60, 64,77-82 The type of cargo the exosome contains depends on the type of cells that it is being secreted from. Different types of cells of the TME secrete different types of exosomal information.Citation60 It has also been shown that exosomes secreted from CAFs can affect breast cancer progression and metastasis via regulation of the Wnt-PCP pathway.Citation59 It is, however, unclear if there is heterogeneity in exosomal miRNAs secreted by CAFs obtained from different molecular subtypes of breast cancer. Using nCounter® Human miRNA Expression Assays analysis, we report that CAFs obtained from luminal exhibit a different profile of secreted miRNA compared to CAFs from basal breast cancer.
Interestingly, we found that a number of miRNAs present in our previously established hMAPK-miRNA signature were secreted in greater amounts by CAF23BAS compared to CAF19LA, including miR221/222, miRNAs known to regulate estrogen receptorCitation38,42,Citation43 (). Because this signature was developed from primary breast tumor miRNA expression data, it is likely that stromal gene expression contributed to this signature, as the presence of non-tumor, stromal cells has been shown to strongly impact gene expression signatures.Citation53-56 Our results reveal a significant association of the hMAPK-miRNA signature with an increased stromal infiltrate score (). CAFs displayed over-expression of hMAPK-miRNAs compared to the dissociated tumor cells (), supporting the notion that CAFs may contribute to the establishment of the hMAPK-miRNA signature.
Activation of the MAPK pathway in tumor cells is one of the mechanisms through which CAFs promote breast cancer progression.Citation15,Citation16,Citation19,21-27 CAFs accomplish this by secretion of cytokines and growth factors, such as EGF (Epidermal Growth Factor), FGF (Fibroblast Growth Factor), IGF (Insulin like growth factor), HGF (Hepatocyte growth factor), and CXCL12,Citation5,15, 19,20 which are potent activators of the MAPK pathway.Citation17,27-33 miRNAs are also able to regulate the tumor-promoting functions of CAFs,Citation45,59 perhaps by disrupting regulation of the MAPK signaling axis.
Conditioned media from macrophages has been shown to repress ER in MCF-7 cells via activation of MAPK pathway.Citation83 In our study, we report that exposure to CAF conditioned media leads to MAPK activation and repression of ER in ER+ breast cancer cells. The MAPK mediated ER-repression effect is more pronounced in conditioned media collected from CAFs isolated from a basal tumor than from CAFs isolated from a luminal tumor. This MAPK mediated ER repression is not seen when MCF-7/ltE2- cells are treated with conditioned media from CAFs isolated from ER-positive tumors (CAF19LA and CAF8LA) (, Fig. S1E). Stimulation of the ER-positive MCF-7/ltE2- cell line with conditioned media from CAF23BAS caused an upregulation of the hMAPK-miRNAs miR221/222 (). Moreover, knockdown of miR221/222 in CAF23BAS cells led to reduced miR221/222 levels in CM. miR221/222 depleted CAF23 CM was unable to repress ER in breast cancer cells (). Collectively, our data indicate that miR221/222 secreted from CAF23BAS are key regulators of MAPK-mediated ER repression observed when ER-positive breast cancer cells were treated with conditioned media from basal CAFs.
Results from our study suggest that CAF populations isolated from different subtypes of primary breast tumor can induce distinct characteristics in breast cancer cell lines. We suggest that there exists a hierarchy and heterogeneity between CAFs obtained from different breast cancer molecular subtypes, specifically in terms of their paracrine interactions with cancer cells. Although the current report focuses on ER-repression as a distinctive phenotype induced by different CAF populations; our findings suggest novel means by which CAFs from different breast cancer subtypes may mediate particular paracrine interactions within the tumor microenvironment, contributing to the breast cancer tumor progression, invasion, EMT, escape from immune surveillance, and metastasis. Furthermore, we report distinct paracrine interactions between cancer cells and CAFs mediated by miRNA depending on whether the cancer cells are surrounded by aggressive (basal) vs indolent (luminal A) CAFs. Understanding these interactions will help to identify biomarkers of CAFs that associate with aggressive breast cancer, potentially identifying specific interactions or miRNAs that could serve as therapeutic targets within the TME. Better characterization of the tumor microenvironment, in addition to characterizing the cancer cells themselves, will foster better classification of tumors, ultimately helping to improve therapy and clinical outcome.
Methods
Stromal, immune, and ESTIMATE score associations with hMAPK-microRNA signature
Stromal, immune, and ESTIMATE scores for breast cancers from the TCGA breast cancer data set were obtained from the ESTIMATE website (http://ibl.mdanderson.org/estimate/disease.html). Breast cancers from the TCGA breast cancer dataset were classified as “high-hMAPK” or “low-hMAPK” according to a published hMAPK-miRNA signature and published method.Citation38 Stromal score, immune score, and ESTIMATE score values were compared between high-hMAPK and low-hMAPK groups using Student's t-test, and were visualized as boxplots.
Tissue culture
All cell lines were grown in a 37°C, 5% CO2 forced air incubator. CAF cells were grown in phenol red containing modified IMEM (Life Technologies) supplemented with 10% fetal bovine serum (FBS, Hyclone). CAFs were passaged and characterized using previously published methods.Citation34 MCF-7/lt E2- cells and (ca)Raf-MCF-7Citation37 cells were maintained in phenol red free modified IMEM (Life Technologies) supplemented with charcoal treated 10% FBS and passaged continuously by detachment using TrypLETM Express (Life Technologies).
Collection of conditioned media
Cultures of CAF cells were grown to 70-80% confluency, and then maintained in serum-free IMEM for 48 hours; this conditioned serum-free IMEM was collected after 48 hours. After conditioned media collection, the media was centrifuged at 1800 rpm for 10 minutes to remove cell debris. The media was then filtered through 0.45 um cellulose acetate syringe filter (Thermo Scientific) and stored at −80°C in 6-9 ml aliquots.
Western blot
The cells were harvested at 70-80% confluency in gold lysis buffer and processed as previously described.Citation84 Protein was transferred on to nitrocellulose membrane, blocked with 5% milk and incubated overnight at 4°C with the following primary antibodies diluted in 0.1 % TBS-T: Anti-ERα (Abcam), anti-pERK1/2 (Cell Signaling), CK19 (Cell Signaling), α-SMA (Santacruz Biotech), Vimentin (Cell Signaling) and anti-GAPDH (Cell Signaling). Membranes were washed with 0.1% TBS-T and incubated with secondary antibody (ECL Rabbit or Mouse IgG, HRP-Linked whole antibody) for 1h at room temperature. Membranes were washed with TBS-T and chemiluminescent detection was performed using ECL western blot substrate or SuperSignal West Pico substrate (Thermo Scientific).
Soft agar colony formation assay
10,000 cells/well were resuspended in 0.3 % agarose IMEM medium [supplemented with 10 % FBS, and penicillin/streptomycin], and were plated in triplicate on top of a 0.9 % agarose IMEM (supplemented with 10 % FBS and penicillin/streptomycin) layer in 6-well plates. All wells were monitored microscopically to assure plating of single-cell suspension. Cultures were maintained at 37°C, 5% CO2 atmosphere for up to 5 weeks, after which cell colonies were stained with crystal violet dye (Sigma) and counted according to established protocol.Citation34
Immunoflorescence staining
Immunostaining of CAFs for presence of Fibroblast Activation Protein (FAP) was performed according to established protocol.Citation34 CAFs were cultured on coverslips and fixed with 10% formalin (VWR). Fixed samples were then blocked with 5% normal goat serum (Rockland) and 0.3% Triton X-100 (Biorad) for 1 hour at room temperature. Samples were then incubated with mouse anti-rabbit antibody against FAP (eBioscience) diluted 1:300 with antibody diluent (Life Technologies). After incubation with primary antibody at room temperature for 2 hours, the samples were incubated with goat anti-mouse Alexa 488 (Life Technologies) at a 1:500 dilution for 1 hr at room temperature. The samples were then mounted by coverslip containing gold-anti-fade DAPI (Life Technologies) and imaged with Nikon Eclipse Ti with 40X oil lens.
qRT-PCR
Total RNA was isolated from cells and from serum free conditioned media using the miRNeasy mini or the miRNeasy serum plasma kit RNA isolation kit, respectively, according to the manufacturer's protocol. RNA concentration was measured using Nanodrop 1000 (Thermo Scientific). cDNA synthesis for mRNA or miRNA analysis was performed using High capacity cDNA reverse transcription kit (Applied Biosystems) or miRcury LNATM Universal RT microRNA PCR (Exiqon) kit, respectively. The PCR primer sequences used for quantitative real-time PCR were: ESR1 (Forward primer 5'-CCACCAACCAGTGCACCATT, Reverse primer 5'-GGTCTTTTCGTATCCCACCTTTC) GAPDH (5'-CACCAGGGCTGCTTTTAACT CTGGTA). hsa-miR-221-3p, hsa-miR-222-3p and hsa-miR-16-5p LNATM primers were purchased commercially from Exiqon. TaqMan® Array Human MicroRNA A+B Cards Set v2.0 panel was used to identify differentially expressed miRNAs between CAFs vs DTs using cutoff 1.5 fold change and P value<0.05.
Luciferase reporter assay
The pEZX-MT01 reporter vectors, containing the ESR1 3'UTR or a non-targeting 3'UTR appended to firefly luciferase, were purchased from Genecopoeia. pEZX-MT01 constructs also contain a renilla luciferase enzyme that is constitutively expressed from a CMV promoter element, allowing for dual-luciferase reporter assays. ca-Raf/MCF-7 or MCF-7/lt E2- cells were transfected with the pEZX-MT01 reporter vector using Lipofectamine 2000 (Life Technologies) for 48hours. Dual-luciferase reporter assay was performed using the manufacturer's protocol, and data recorded on a Promega GloMax instrument.
Exosome isolation
Exosomes were isolated from CAF-conditioned media by ultracentifugation. 12ml of frozen serum free conditioned media was ultracentrifuged at 110,000 g for 70 minutes to pellet the small vesicles that correspond to exosomes. The exosome pellet was washed with 12 ml of PBS and ultracentrifuged at 110,000 g for 70 minutes. The exosome pellet was resuspended in 100 µl of PBS.
Transmission electron microscopy
Purified exosomes were fixed with 2% paraformaldehyde. A 20 µl drop of the suspension was loaded onto a formvar coated grid, negatively stained with 2% aqueous uranyl acetate for 2 minutes, and examined under a transmission electron microscope FEI Tecnai G2 Spirit using a digital camera Morada (Olympus Soft Image Solutions).
Nanoparticle tracking analysis
Isolated exosomes were analyzed using the Nanosight NS300 system (Malvern Instruments Company). Exosome preparations were diluted (1:20) in sterile 1X PBS and analyzed by NanoSight NS300 and Nanosight NTA 2.3 Analytical Software. Each individual sample of isolated exosomes was quantified by minimum of 3 replicate analyses.
Exosome labeling, confocal microscopy and live imaging of internalized exosomes
Exosomes were labeled with Exo-Red (System Biosciences), according to the manufacturer's protocol. Briefly, exosome pellets were resuspended in 100µl 1X PBS, mixed with 10μL Exo-Red stain solution, and incubated for 10 minutes at 37°C. The labeling reaction was stopped by adding 100 µl of ExoQuick-TC reagent. Labeled exosomes were incubated 30 minutes at 4°C. Finally exosomes were centrifuged 3 minutes at 14,000 rpm. All experiments were performed on Leica SP5 inverted confocal microscope equipment using excitation wavelengths of 460 nm and a 20x/63x glycerol immersion objectives. For live cell confocal laser scanning microscopy experiments, cells were grown in glass bottom chamber slides, and labeled exosomes were added to subconfluent cells and incubated for 1 hr. Cells were immediately analyzed.
Nanostring analysis
Total RNA from the cells and conditioned media containing exosomes was isolated as mentioned above in qRT-PCR. Prior to Nanostring analysis we analyzed the RNA quality and quantity using Bioanalyzer 2100 Pico (Agilent). Same amount of RNA was used as input for the nCounter® Human microRNA Expression Assays and subsequent analysis.
Knockdown of microRNA 221/222
Locked nucleic acid oligonucleotide microRNA inhibitor (anatgomiR) hsa-miR-221-3p and hsa-miR-222-3p and scrambled control miR (Exiqon) were transfected into ca-Raf/MCF-7 or MCF-7/lt E2- cells using Lipofectamine RNAimax (Life Technologies), at a concentration of 100nm (scrambled) and 50 nM (antagomiR). Knockdown of miR221/222 expression was confirmed by qPCR 48hrs post-transfection.
Cell proliferation assay
Following 48 hrs of transfection with scrambled control siRNA or miR-221/222, 10,000 CAF23BAS cells were seeded in 24-wells in triplicates in IMEM+10%FBS media. Counts of viable cells were determined by trypan blue exclusion at 48 hr intervals, and enumerated using hand counts on a hemocytometer
Statistical analysis
The nSolver software v 1.0 (nCounter) was used to evaluate the results of the Nanostring analysis. 95% confidence interval from the negative control was selected as a threshold for signal. All the microRNAs with intensity less than the threshold were discarded from the analysis. For all samples, microRNAs with % coefficient of variance less than 15% and intensity greater than signal threshold were used as normalization codesets. Student t-test was performed to identify statistically differentially expressed microRNAs (P<0.05)
Reagents and Supplies
Tissue Culture
IMEM with phenol red (Life Technologies Cat # A10489-01), IMEM phenol red free (Life Technologies Cat # A10488-01), FBS (Thermo Scientific Cat # SH30071.03), charcoal treated FBS (Thermo Scientific Cat # SH30068.03), Tryple Express (Life Technologies Cat # 12604.013)Antibodies: ERα (Abcam Cat # ab16660), pERK1/2 (Cell signaling Cat # 9101L), GAPDH (Cell signaling Cat # 3683S), FAP (eBioscience Cat # BMS168), Vimentin (Cell Signaling Cat # 50741S) CK19 (Cell signaling Cat # 4558S), α-SMA (Santacruz Biotech Cat # sc32251), Rabbit IgG (GE Healthcare Cat # LNA934V) Mouse IgG (GE Healthcare Cat # LNXA931), Alexa 488 (Life technologies Cat # A11029)
Kits
miRNeasy mini/serum plasma kit (Qiagen Cat # 217004/217184) miRNA Primers and AntagomiRs: Primers Exiqon (hsa-miR-221-3p Cat # 204532, hsa-miR-222-3p Cat # 204551, hsa-miR-16-5p Cat # 205702) and AntagomiRs Exiqon (hsa-miR-221-3p Cat # 4103821-001) and hsa-miR-222-3p Cat # 4101984-001)
Miscellaneous
10% Formalin (VWR Cat # 89370-094), Triton x-100 (Biorad Cat # 161-0407), 5 % normal goat serum (Rockland Cat # B304), Antibody diluent (Dako Cat # K8006), gold-antifade DAPI (Life Technologies Cat # P36931), Lipofectamine 2000/RNAiMAX (Life Technologies Cat # 11668-019/13778-075), Exored (System Biosciences Cat # EXOR100A-1), Syringe filter (Thermo Scientific Cat # 190-2545) and ECL western blot substrate/SuperSignal West Pico substrate (Thermo Scientific Cat # 32106/34077)
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Supplemental Figures
Download Zip (863 KB)Acknowledgments
The authors would like to thank Dr. Marc E. Lippman and members of Dr. El-Ashry and Dr. Lippman laboratory group, Drs. Bal Lokeshwar, Kerry Burnstein, Ram Datar and Sharon Elliot for thoughtful discussions. Sanket H. Shah would like to thank partial support and assistance from the Sheila and David Fuente Graduate Program in Cancer Biology, University of Miami. Marta Garcia-Contreras would like to thank partial support and assistance from School of Medicine and Dentistry, Catholic University of Valencia, Valencia, Spain and Ri.Med Foundation, Palermo, Italy. We thank Sylvester Comprehensive Cancer Center development funds for partial support to DEA. We would like to thank Sion Williams at Oncogenomics Core Facility of University of Miami for their technical support with the Nanostring analysis. We would like to acknowledge Mario Soriano-Navarro for his technical support at the Electron Microscopy Service of Centro de Investigación Príncipe Felipe, Valencia, Spain. Also we thank the Microvesicles and Exosomes core at Diabetes Research Institute, University of Miami for the nanoparticle tracking analysis technical support. And Maria Boulina for technical support at the Analytical Imaging Core Facility core of University of Miami.
References
- Tlsty TD. Stromal cells can contribute oncogenic signals. Semin Cancer Biol 2001; 11:97-104; PMID:11322829; http://dx.doi.org/10.1006/scbi.2000.0361
- Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer cell 2004; 6:17-32.
- Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 2009; 9:274-U65; PMID:19308067; http://dx.doi.org/10.1038/nrc2622
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/10.1016/j.cell.2011.02.013
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer 2006; 6:392-401; PMID:16572188; http://dx.doi.org/10.1038/nrc1877
- Folkman J. Tumor angiogenesis: therapeutic implications. N Eng J Med 1971; 285:1182-6; PMID:4938153; http://dx.doi.org/10.1056/NEJM197108122850711
- Tlsty TD, Hein PW. Know thy neighbor: stromal cells can contribute oncogenic signals. Curr Opin Genet Dev 2001; 11:54-9; PMID:11163151; http://dx.doi.org/10.1016/S0959-437X(00)00156-8
- Ronnov-Jessen L, Petersen OW, Bissell MJ. Cellular changes involved in conversion of normal to malignant breast: importance of the stromal reaction. Physiol Rev 1996; 76:69-125; PMID:8592733
- Schedin P, Elias A. Multistep tumorigenesis and the microenvironment. Breast Cancer Res 2004; 6:93-101; PMID:14979914; http://dx.doi.org/10.1186/bcr772
- Shekhar MP, Pauley R, Heppner G. Host microenvironment in breast cancer development: extracellular matrix-stromal cell contribution to neoplastic phenotype of epithelial cells in the breast. Breast Cancer Res 2003; 5:130-5; PMID:12793893; http://dx.doi.org/10.1186/bcr580
- van Kempen LC, Ruiter DJ, van Muijen GN, Coussens LM. The tumor microenvironment: a critical determinant of neoplastic evolution. Eur J Cell Biol 2003; 82:539-48; PMID:14703010; http://dx.doi.org/10.1078/0171-9335-00346
- Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle 2006; 5:1597-601; PMID:16880743; http://dx.doi.org/10.4161/cc.5.15.3112
- Sappino AP, Skalli O, Jackson B, Schurch W, Gabbiani G. Smooth-muscle differentiation in stromal cells of malignant and non-malignant breast tissues. Int J Cancer 1988; 41:707-12; PMID:2835323; http://dx.doi.org/10.1002/ijc.2910410512
- Augsten M. Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front Oncol 2014; 4:62; PMID:24734219; http://dx.doi.org/10.3389/fonc.2014.00062
- Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012; 487:500-4; PMID:22763439; http://dx.doi.org/10.1038/nature11183
- Shekhar MP, Santner S, Carolin KA, Tait L. Direct involvement of breast tumor fibroblasts in the modulation of tamoxifen sensitivity. Am J Pathol 2007; 170:1546-60; PMID:17456761; http://dx.doi.org/10.2353/ajpath.2007.061004
- Gao MQ, Kim BG, Kang S, Choi YP, Park H, Kang KS, Cho NH. Stromal fibroblasts from the interface zone of human breast carcinomas induce an epithelial-mesenchymal transition-like state in breast cancer cells in vitro. J Cell Sci 2010; 123:3507-14; PMID:20841377; http://dx.doi.org/10.1242/jcs.072900
- Mueller MM, Fusenig NE. Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer 2004; 4:839-49; PMID:15516957; http://dx.doi.org/10.1038/nrc1477
- Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005; 121:335-48; PMID:15882617; http://dx.doi.org/10.1016/j.cell.2005.02.034
- Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature 2004; 432:332-7; PMID:15549095; http://dx.doi.org/10.1038/nature03096
- Giulianelli S, Cerliani JP, Lamb CA, Fabris VT, Bottino MC, Gorostiaga MA, Novaro V, Góngora A, Baldi A, Molinolo A, et al. Carcinoma-associated fibroblasts activate progesterone receptors and induce hormone independent mammary tumor growth: A role for the FGF-2/FGFR-2 axis. Int J Cancer 2008; 123:2518-31; PMID:18767044; http://dx.doi.org/10.1002/ijc.23802
- Giannoni E, Bianchini F, Masieri L, Serni S, Torre E, Calorini L, Chiarugi P. Reciprocal activation of prostate cancer cells and cancer-associated fibroblasts stimulates epithelial-mesenchymal transition and cancer stemness. Cancer Res 2010; 70:6945-56; PMID:20699369; http://dx.doi.org/10.1158/0008-5472.CAN-10-0785
- Grugan KD, Miller CG, Yao Y, Michaylira CZ, Ohashi S, Klein-Szanto AJ, Diehl JA, Herlyn M, Han M, Nakagawa H, et al. Fibroblast-secreted hepatocyte growth factor plays a functional role in esophageal squamous cell carcinoma invasion. Proc Natl Acad Sci U S A 2010; 107:11026-31; PMID:20534479; http://dx.doi.org/10.1073/pnas.0914295107
- Tyan SW, Kuo WH, Huang CK, Pan CC, Shew JY, Chang KJ, Lee EY, Lee WH. Breast cancer cells induce cancer-associated fibroblasts to secrete hepatocyte growth factor to enhance breast tumorigenesis. PloS One 2011; 6:e15313; PMID:21249190; http://dx.doi.org/10.1371/journal.pone.0015313
- Stetler-Stevenson WG, Aznavoorian S, Liotta LA. Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu Rev Cell Biol 1993; 9:541-73; PMID:8280471; http://dx.doi.org/10.1146/annurev.cb.09.110193.002545
- Sternlicht MD, Lochter A, Sympson CJ, Huey B, Rougier JP, Gray JW, Pinkel D, Bissell MJ, Werb Z. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell 1999; 98:137-46; PMID:10428026; http://dx.doi.org/10.1016/S0092-8674(00)81009-0
- Studebaker AW, Storci G, Werbeck JL, Sansone P, Sasser AK, Tavolari S, Huang T, Chan MW, Marini FC, Rosol TJ, et al. Fibroblasts isolated from common sites of breast cancer metastasis enhance cancer cell growth rates and invasiveness in an interleukin-6-dependent manner. Cancer Res 2008; 68:9087-95; PMID:18974155; http://dx.doi.org/10.1158/0008-5472.CAN-08-0400
- Bogoyevitch MA, Glennon PE, Andersson MB, Clerk A, Lazou A, Marshall CJ, Parker PJ, Sugden PH. Endothelin-1 and fibroblast growth factors stimulate the mitogen-activated protein kinase signaling cascade in cardiac myocytes. The potential role of the cascade in the integration of two signaling pathways leading to myocyte hypertrophy. J Biol Chem 1994; 269:1110-9; PMID:7507104
- Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res 2010; 16:2927-31; PMID:20484021; http://dx.doi.org/10.1158/1078-0432.CCR-09-2329
- Javelaud D, Mauviel A. Crosstalk mechanisms between the mitogen-activated protein kinase pathways and Smad signaling downstream of TGF-β: implications for carcinogenesis. Oncogene 2005; 24:5742-50; PMID:16123807; http://dx.doi.org/10.1038/sj.onc.1208928
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 2003; 425:577-84; PMID:14534577; http://dx.doi.org/10.1038/nature02006
- Qiu Q, Yang M, Tsang BK, Gruslin A. EGF-induced trophoblast secretion of MMP-9 and TIMP-1 involves activation of both PI3K and MAPK signalling pathways. Reproduction 2004; 128:355-63; PMID:15333786; http://dx.doi.org/10.1530/rep.1.00234
- Ogata A, Chauhan D, Teoh G, Treon SP, Urashima M, Schlossman RL, Anderson KC. IL-6 triggers cell growth via the Ras-dependent mitogen-activated protein kinase cascade. J Immunol 1997; 159:2212-21.
- Drews-Elger K, Brinkman JA, Miller P, Shah SH, Harrell JC, da Silva TG, Ao Z, Schlater A, Azzam DJ, Diehl K, et al. Primary breast tumor-derived cellular models: characterization of tumorigenic, metastatic, and cancer-associated fibroblasts in dissociated tumor (DT) cultures. Breast Cancer Res Treat 2014; 144:503-17; PMID:24567196; http://dx.doi.org/10.1007/s10549-014-2887-9
- Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D. Activation of mitogen-activated protein kinase in estrogen receptor α-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor α-negative human breast tumors. Cancer Res 2006; 66:3903-11; PMID:16585219; http://dx.doi.org/10.1158/0008-5472.CAN-05-4363
- Bayliss J, Hilger A, Vishnu P, Diehl K, El-Ashry D. Reversal of the estrogen receptor negative phenotype in breast cancer and restoration of antiestrogen response. Clin Cancer Res 2007; 13:7029-36; PMID:18056179; http://dx.doi.org/10.1158/1078-0432.CCR-07-0587
- Oh AS, Lorant LA, Holloway JN, Miller DL, Kern FG, El-Ashry D. Hyperactivation of MAPK induces loss of ERalpha expression in breast cancer cells. Mol Endocrinol 2001; 15:1344-59; PMID:11463858
- Miller P, Clarke J, Koru-Sengul T, Brinkman J, El-Ashry D. A novel MAPK-microRNA signature is predictive of hormone-therapy resistance and poor outcome in ER-positive breast cancer. Clin Cancer Res 2015 Jan 15;21(2):373-85.
- Pandey DP, Picard D. miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor α mRNA. Mol Cell Biol 2009; 29:3783-90; PMID:19414598; http://dx.doi.org/10.1128/MCB.01875-08
- Xiong J, Yu D, Wei N, Fu H, Cai T, Huang Y, Wu C, Zheng X, Du Q, Lin D, et al. An estrogen receptor α suppressor, microRNA-22, is downregulated in estrogen receptor α-positive human breast cancer cell lines and clinical samples. FEBS J 2010; 277:1684-94; PMID:20180843; http://dx.doi.org/10.1111/j.1742-4658.2010.07594.x
- Cochrane DR, Cittelly DM, Howe EN, Spoelstra NS, McKinsey EL, LaPara K, Elias A, Yee D, Richer JK. MicroRNAs link estrogen receptor α status and Dicer levels in breast cancer. Horm Cancer 2010; 1:306-19; PMID:21761362; http://dx.doi.org/10.1007/s12672-010-0043-5
- Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor α and is associated with tamoxifen resistance in breast cancer. J Biol Chem 2008; 283:31079-86; PMID:18790736; http://dx.doi.org/10.1074/jbc.M806041200
- Wei Y, Lai X, Yu S, Chen S, Ma Y, Zhang Y, Li H, Zhu X, Yao L, Zhang J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res Treat 2014; 147:423-31; PMID:25007959; http://dx.doi.org/10.1007/s10549-014-3037-0
- Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 2008; 27:5959-74; PMID:18836476; http://dx.doi.org/10.1038/onc.2008.274
- Kohlhapp FJ, Mitra AK, Lengyel E, Peter ME. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene 2015; ePub ahead of print; PMID:25867073
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A 2004; 101:11755-60; PMID:15284443; http://dx.doi.org/10.1073/pnas.0404432101
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435:834-8; PMID:15944708; http://dx.doi.org/10.1038/nature03702
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65:7065-70; PMID:16103053; http://dx.doi.org/10.1158/0008-5472.CAN-05-1783
- Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res 2009; 11:R27; PMID:19432961; http://dx.doi.org/10.1186/bcr2257
- Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer 2006; 5:24; PMID:16784538; http://dx.doi.org/10.1186/1476-4598-5-24
- Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG, Rønneberg JA, Johnsen H, Navon R, Rødland E, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PloS One 2011; 6:e16915; PMID:21364938; http://dx.doi.org/10.1371/journal.pone.0016915
- Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol 2007; 8:R214; PMID:17922911; http://dx.doi.org/10.1186/gb-2007-8-10-r214
- de Ridder D, van der Linden CE, Schonewille T, Dik WA, Reinders MJ, van Dongen JJ, Staal FJ. Purity for clarity: the need for purification of tumor cells in DNA microarray studies. Leukemia 2005; 19:618-27; PMID:15744349
- Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013; 4:2612; PMID:24113773; http://dx.doi.org/10.1038/ncomms3612
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012; 30:413-21; PMID:22544022; http://dx.doi.org/10.1038/nbt.2203
- Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W, Weigman VJ, Marynen P, Zetterberg A, Naume B, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A 2010; 107:16910-5; PMID:20837533; http://dx.doi.org/10.1073/pnas.1009843107
- An J, Enomoto A, Weng L, Kato T, Iwakoshi A, Ushida K, Maeda K, Ishida-Takagishi M, Ishii G, Ming S, et al. Significance of cancer-associated fibroblasts in the regulation of gene expression in the leading cells of invasive lung cancer. J Cancer Res Clin Oncol 2013; 139:379-88; PMID:23108890; http://dx.doi.org/10.1007/s00432-012-1328-6
- El-Ashry D, Miller DL, Kharbanda S, Lippman ME, Kern FG. Constitutive Raf-1 kinase activity in breast cancer cells induces both estrogen-independent growth and apoptosis. Oncogene 1997; 15:423-35; PMID:9242379; http://dx.doi.org/10.1038/sj.onc.1201198
- Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, Wrana JL. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 2012; 151:1542-56; PMID:23260141; http://dx.doi.org/10.1016/j.cell.2012.11.024
- Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol 2012; 22:125-32; PMID:22260888; http://dx.doi.org/10.1016/j.tcb.2011.12.001
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002; 2:569-79; PMID:12154376
- Boon RA, Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol 2013; 33:186-92; PMID:23325475; http://dx.doi.org/10.1161/ATVBAHA.112.300139
- Lotvall J, Valadi H. Cell to cell signalling via exosomes through esRNA. Cell Adh Migr 2007; 1:156-8; PMID:19262134; http://dx.doi.org/10.4161/cam.1.3.5114
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9:654-9; PMID:17486113; http://dx.doi.org/10.1038/ncb1596
- Umezu T, Ohyashiki K, Kuroda M, Ohyashiki JH. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene 2013; 32:2747-55; PMID:22797057; http://dx.doi.org/10.1038/onc.2012.295
- Bobrie A, Colombo M, Raposo G, Thery C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 2011; 12:1659-68; PMID:21645191; http://dx.doi.org/10.1111/j.1600-0854.2011.01225.x
- Pegtel DM, van de Garde MD, Middeldorp JM. Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion. Biochim Biophys Acta 2011; 1809:715-21; PMID:21855666; http://dx.doi.org/10.1016/j.bbagrm.2011.08.002
- Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer 2011; 10:117; PMID:21939504; http://dx.doi.org/10.1186/1476-4598-10-117
- Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 2010; 39:133-44; PMID:20603081; http://dx.doi.org/10.1016/j.molcel.2010.06.010
- Collino F, Deregibus MC, Bruno S, Sterpone L, Aghemo G, Viltono L, Tetta C, Camussi G. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PloS One 2010; 5:e11803; PMID:20668554; http://dx.doi.org/10.1371/journal.pone.0011803
- Ogawa R, Tanaka C, Sato M, Nagasaki H, Sugimura K, Okumura K, Nakagawa Y, Aoki N. Adipocyte-derived microvesicles contain RNA that is transported into macrophages and might be secreted into blood circulation. Biochem Biophys Res Commun 2010; 398:723-9; PMID:20621060; http://dx.doi.org/10.1016/j.bbrc.2010.07.008
- Muller G, Schneider M, Biemer-Daub G, Wied S. Microvesicles released from rat adipocytes and harboring glycosylphosphatidylinositol-anchored proteins transfer RNA stimulating lipid synthesis. Cell Signal 2011; 23:1207-23; PMID:21435393; http://dx.doi.org/10.1016/j.cellsig.2011.03.013
- Muralidharan-Chari V, Clancy JW, Sedgwick A, D'Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci 2010; 123:1603-11; PMID:20445011; http://dx.doi.org/10.1242/jcs.064386
- Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med 2013; 91:431-7; PMID:23519402; http://dx.doi.org/10.1007/s00109-013-1020-6
- Tchou J, Kossenkov AV, Chang L, Satija C, Herlyn M, Showe LC, Puré E. Human breast cancer associated fibroblasts exhibit subtype specific gene expression profiles. BMC Med Genom 2012; 5:39; PMID:22954256; http://dx.doi.org/10.1186/1755-8794-5-39
- Park SY, Kim HM, Koo JS. Differential expression of cancer-associated fibroblast-related proteins according to molecular subtype and stromal histology in breast cancer. Breast Cancer Res Treat 2015; 149:727-41; PMID:25667103; http://dx.doi.org/10.1007/s10549-015-3291-9
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008; 10:1470-6; PMID:19011622; http://dx.doi.org/10.1038/ncb1800
- Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 2009; 11:1143-9; PMID:19684575; http://dx.doi.org/10.1038/ncb1929
- Guduric-Fuchs J, O'Connor A, Camp B, O'Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom 2012; 13:357; PMID:22849433; http://dx.doi.org/10.1186/1471-2164-13-357
- Nolte-'t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, t Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res 2012; 40:9272-85; PMID:22821563; http://dx.doi.org/10.1093/nar/gks658
- Simons M, Raposo G. Exosomes–vesicular carriers for intercellular communication. Curr Opin Cell Biol 2009; 21:575-81; PMID:19442504; http://dx.doi.org/10.1016/j.ceb.2009.03.007
- Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, Ibberson M, De Palma M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep 2014; 8:1432-46; PMID:25159140; http://dx.doi.org/10.1016/j.celrep.2014.07.035
- Stossi F, Madak-Erdogan Z, Katzenellenbogen BS. Macrophage-elicited loss of estrogen receptor-α in breast cancer cells via involvement of MAPK and c-Jun at the ESR1 genomic locus. Oncogene 2012; 31:1825-34; PMID:21860415; http://dx.doi.org/10.1038/onc.2011.370
- Bayraktar UD, Kim TK, Drews-Elger K, Benjamin C, El-Ashry D, Wieder E, Komanduri KV. Simultaneous measurement of ERalpha, HER2, and phosphoERK1/2 in breast cancer cell lines by flow cytometry. Breast Cancer Res Treat 2011; 129:623-8; PMID:21607585; http://dx.doi.org/10.1007/s10549-011-1586-z