ABSTRACT
A fundamental event in the development and progression of malignant melanoma is the de-regulation of cancer-relevant transcription factors. We recently showed that c-Jun is a main regulator of melanoma progression and, thus, is the most important member of the AP-1 transcription factor family in this disease.
Surprisingly, no cancer-related specific c-Jun target genes in melanoma were described in the literature, so far. Therefore, we focused on pre-existing ChIP-Seq data (Encyclopedia of DNA Elements) of 3 different non-melanoma cell lines to screen direct c-Jun target genes. Here, a specific c-Jun antibody to immunoprecipitate the associated promoter DNA was used. Consequently, we identified 44 direct c-Jun targets and a detailed analysis of 6 selected genes confirmed their deregulation in malignant melanoma. The identified genes were differentially regulated comparing 4 melanoma cell lines and normal human melanocytes and we confirmed their c-Jun dependency. Direct interaction between c-Jun and the promoter/enhancer regions of the identified genes was confirmed by us via ChIP experiments. Interestingly, we revealed that the direct regulation of target gene expression via c-Jun can be independent of the existence of the classical AP-1 (5´-TGA(C/G)TCA-3´) consensus sequence allowing for the subsequent down- or up-regulation of the expression of these cancer-relevant genes.
In summary, the results of this study indicate that c-Jun plays a crucial role in the development and progression of malignant melanoma via direct regulation of cancer-relevant target genes and that inhibition of direct c-Jun targets through inhibition of c-Jun is a potential novel therapeutic option for treatment of malignant melanoma.
Introduction
The family of AP-1 (activating protein-1) transcription factors includes the JUN, JDP, FOS/FRA and MAF subfamilies.Citation1 They share a conserved basic DNA-binding domain and a leucine zipper domain (bZIP). The DNA-binding domain determines the spectrum of genes that are controlled by the protein that binds to it, and the transactivation domain (delta domain) is responsible for the regulation of transcriptional activation.Citation2 AP-1 proteins are known to bind to the classical palindromic recognition sequence 5′TGA(C/G)TCA-3′ to regulate target gene expression. In the promoter/enhancer regions, the sequences of the AP-1 regulatory elements often deviate from the classical AP-1 recognition sequence. This variation may contribute to the differential functions of different AP-1 family dimersCitation3; however, a detailed analysis has not yet been performed. AP-1 transcription factor dimers are known to play a crucial role in the development and progression of different cancer types, particularly malignant melanoma.Citation1,4-6 Though, it is still unclear which direct target genes of AP-1 transcription factors cause the functional effects that support the development of melanoma.
Malignant melanoma is the most aggressive skin cancer, and its incidence is growing faster than for any other cancer entity. In the past few decades, many mechanisms of regulation of the development and progression of melanoma and the high migratory and invasive potential of melanoma cells have been identified, but the detailed molecular causes of this disease remain elusive.
Due to the critical influence of the activity of AP-1 transcription factors in malignant melanoma, DNA-protein interactions have been investigated using a variety of biochemical and genomic approaches. In addition to the traditionally used in vitro techniques, such as electrophoretic mobility shift assays (EMSA) and DNase I footprinting assays, chromatin immunoprecipitation (ChIP) has become a very popular technique for identifying DNA-protein interactions in vivo. Apart from identifying interactions between specific proteins and DNA in living cells, the localization of proteins at a specific genomic region can also be determined. ChIP assays can further be combined with sequencing (ChIP-Seq) to allow for a genome-wide analysis of DNA-protein interactions and, thus, the identification of specific DNA-binding sites and direct target genes of individual transcription factors,Citation7,8 such as c-Jun. Recent studies have indicated that the AP-1 family member c-Jun is a main regulator of melanoma progression Citation6,9,10 and that it acts by up-regulating pro-oncogenic genes and down-regulating anti-oncogenic target genes, whose activation promotes the malignant phenotype. We have previously shown that the microRNA miR-125b directly regulates the transcription factor c-Jun and, thus, effects the proliferative and migratory potential of melanoma cells.Citation11 Moreover, we identified an alternative regulatory pathway of c-Jun in melanoma cells that leads to an upregulation of c-Jun activity via the loss of the cell-adhesion molecule E-cadherin.Citation12,13 Although the de-regulation of the transcription factor c-Jun is known to be one of the most important events in the development and progression of malignant melanoma, the specific c-Jun target genes that contribute to the functional effects of c-Jun up-regulation in melanoma cells and their molecular relevance have not been determined.
Consequently, a detailed analysis of directly regulated target genes of the transcription factor c-Jun and their expression level is necessary to determine the role of c-Jun in malignant melanoma. We speculate that targeting transcription factors may be a novel and effective therapeutic approach.
Results
In this study, we used pre-existing ChIP-Seq data of the human cervical adenocarcinoma cell line HeLa S3, the human liver carcinoma cell line HepG2 and the human umbilical vein endothelial cell line HUVEC archived in the Encyclopedia of DNA Elements (ENCODE; http://genome.ucsc.edu/ENCODE/) for in silico analysis to identify genome-wide, general c-Jun target genes. In these data, we screened for enriched DNA sequences (peaks) after ChIP-Seq with a c-Jun antibody (sc-1694). By examining the data in ENCODE, we were able to identify 44 c-Jun binding sites in the promoter/enhancer regions of a variety of target genes in all 3 cell lines (). First, we scanned for known or predicted associations between the 44 identified genes and the transcription factor c-Jun by STRING (Search Tool for the Retrieval of Interacting Genes/Proteins).Citation14 STRING analysis showed known or predicted associations between the transcription factor c-Jun and many of the newly identified c-Jun target genes. The level of confidence of the associations is represented by the thickness of the lines (). Some identified c-Jun targets are not described yet (e.g. SGCE, DKK3 or HPSE), hence STRING analysis showed no relation between those identified c-Jun targets. Then, we analyzed the 44 identified general c-Jun target genes using the Functional Annotation Tool of the Database for Annotation, Visualization and Integrated Discovery (DAVID v6.7).Citation15,16 Functional annotation clustering exemplarily using the category GOTERM_BP_FAT resulted in 7 different significantly enriched annotation clusters (ACs) (Table S1). The analysis of other categories resulted in similar functional ACs. The ACs contained genes that could be summarized according to their molecular function: adhesion (AC 1; enrichment score: 2.76), cell motion (AC 2 and 4; enrichment score: 2.42 and 1.70), phosphorus metabolic process (AC 3; enrichment score: 1.84), cytoskeleton organization (AC 5; enrichment score: 1.62), response to oxidative stress (AC 6; enrichment score: 1.46) and positive regulation of cell proliferation (AC 7; enrichment score: 1.14) ().
Figure 1. All identified potential c-Jun target genes share cancer-relevant molecular functions and contain classical or non-classical AP-1 recognition sequences in their promoter/enhancer regions. (a i) Predicted relation between the 44 identified target genes and the transcription factor c-Jun via STRING. (a ii) Functional annotation clustering using the category GOTERM_BP_FAT resulted in 7 different significantly enriched annotation clusters (ACs). The ACs contained genes that could be summarized according to their molecular function: adhesion (enrichment score (ES): 2.76), cell motion (ES: 2.42), phosphorus metabolic process (ES: 1.84), cytoskeleton organization (ES: 1.62), response to oxidative stress (ES: 1.46) and positive regulation of cell proliferation (ES: 1.14). (b) The promoter/enhancer regions of FosB, WEE1, PVR and LGALS3 contained the classical AP-1 binding sequence with 100% agreement (FosB: 5′-TGA G TCA-3′, WEE1: 5′-TGA(C/G)TCA-3′, PVR: 5′-TGAGTCA-3′, and LGALS3: 5′-TGA G TCA-3′). In the promoter/enhancer regions of NFATC2 and MAP1LC3B, the AP-1 binding motifs each contained a one base pair exchange (NFATC2: 5′-TGA C ACA-3′ and MAP1LC3B: 5′-TGA T TCA-3′). The potential c-Jun binding sites were validated in all 6 detected potential target genes as DNA-motifs, identical or similar to the AP-1 recognition sequence, using the Motiv Discovery program MEME.
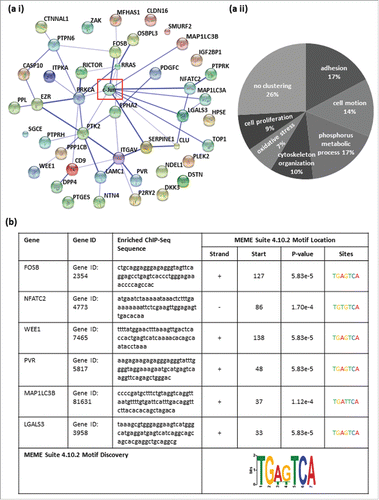
Table 1. General, potential c-Jun target genes identified via ENCODE. The list includes the 44 identified general c-Jun target genes, their gene description and their localization on the human genome (hg19).
For the subsequent analysis, we focused on 6 of the 44 target genes that have previously been described as cancer-relevant genes. The enrichment of DNA fragments displayed as peaks in the promoter/enhancer regions of specific target genes (ENCODE) were localized to the promoter/enhancer regions of FosB, NFATC2, WEE1, PVR, MAP1LC3B and LGALS3Citation17-22 (, ). Furthermore, we confirmed the presence of DNA-motifs identical or similar to the AP-1 recognition site within the enriched ChIP-Seq sequences of the 6 detected potential target genes of c-Jun via the Multiple Em for Motif Elicitation (MEME) Suite 4.10.2 databaseCitation23 (). We revealed that 4 of the 6 identified potential target genes of the transcription factor c-Jun contain a classical AP-1 recognition sequence (5′-TGA(C/G)TCA-3′) within the enriched DNA fragments. In the promoter/enhancer regions of FosB, WEE1, PVR and LGALS3, we observed the classical AP-1 binding sequence with 100% accordance (5′-TGA (C/G) TCA-3′). In the promoter/enhancer regions of NFATC2 and MAP1LC3B, the potential AP-1 binding sequence contained one base pair change compared to the the classical sequence (NFATC2: 5′-TGA C ACA-3′; MAP1LC3B: 5′-TGA T TCA3-´).
Figure 2. Analysis of archived ChIP-Seq data (ENCODE; hg19) showing enrichment of DNA fragments after ChIP with a c-Jun antibody (sc-1694) in the cell lines HUVEC, HepG2 and HEK. The enrichments of DNA fragments are displayed as peaks in the promoter/enhancer regions of specific target genes (labeled in red). The peaks were localized to the promoter/enhancer regions of FosB, NFATC2, WEE1, PVR, LGALS3 and MAP1LC3B.
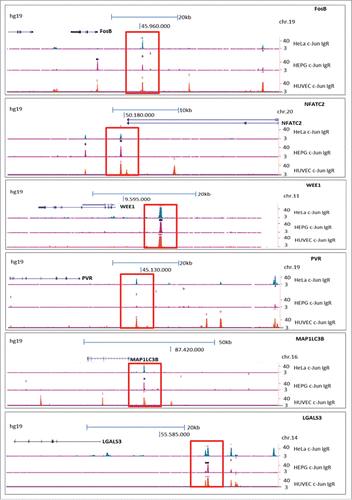
Table 2. Direct c-Jun target genes and described activities. The identified direct c-Jun target genes in malignant melanoma and their recently described cancer-relevant activities.
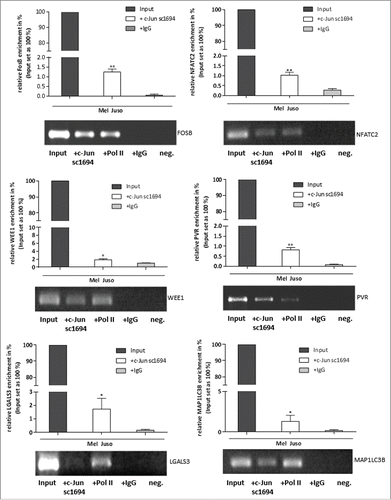
We next validated the results of the in silico analysis of the ENCODE data by performing ChIP in the melanoma cell line Mel Juso, to confirm the direct interaction between c-Jun and the identified promoter/enhancer regions in the new target genes. We performed ChIP experiments with a c-Jun antibody (sc-1694), a RNA polymerase II (Pol II) antibody as a positive control and an IgG antibody as a negative control and analyzed the precipitated DNA fragments by PCR with different primer pairs to amplify specific promoter regions: GAPDH as a positive control for ChIP experiments with the Pol II antibody, negative control primers, ChIP_FosB, ChIP_NFATC2, ChIP_WEE1, ChIP_PVR, ChIP_LGALS3 and ChIP_MAP1LC3B (Table S2). Total DNA of the melanoma cell line Mel Juso served as the Input DNA for the ChIP experiments. Gel electrophoresis of the amplified precipitated DNA fragments after PCR showed a significant enrichment of the promoter/enhancer sequences of the predicted direct c-Jun target genes FosB, NFATC2, WEE1, PVR, LGALS3 and MAP1LC3B (, Fig. S1).
Figure 3. Direct binding of c-Jun to the promoter/enhancer regions of FosB, NFATC2, WEE1, PVR, LGALS3 and MAP1LC3B. ChIP assays demonstrate direct binding of c-Jun to the promoter/enhancer regions of the detected c-Jun target genes. DNA samples from the ChIP reactions (Pol II, IgG, and c-Jun) and input DNA were used for PCR with different primer pairs to amplify specific promoter regions (ChIP_FosB, ChIP_NFATC2, ChIP_WEE1, ChIP_PVR, ChIP_LGALS3 and ChIP_MAP1LC3B). All PCR fragments were detected in the input DNA sample. The bars show the means±s.d. of three independent experiments; measurements were performed in triplicate (**P < 0.01; *P < 0.5).
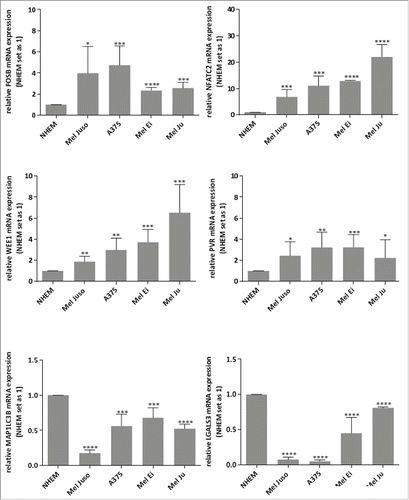
Further, we analyzed whether these newly defined c-Jun target genes (FosB, NFATC2, WEE1, PVR, MAP1LC3B and LGALS3) also are deregulated in malignant melanoma. We performed mRNA expression analysis by qRT-PCR in 4 different melanoma cell lines (Mel Juso, A375, Mel Ei, and Mel Ju) and in primary melanocytes (NHEM) and detected de-regulation of the potential c-Jun target genes in all melanoma cell lines. We observed upregulation of FosB, NFATC2, WEE1 and PVR and down-regulation of MAP1LC3B and LGALS3 in all melanoma cell lines compared to NHEMs ().
Figure 4. Relative FosB, NFATC2, WEE1, PVR, LGALS3 and MAP1LC3B mRNA expressions in melanoma cell lines. qRT–PCR of FosB, NFATC2, WEE1, PVR, LGALS3 and MAP1LC3B in 4 different melanoma cell lines shows up-regulation of FosB, NFATC2, WEE1 and PVR and down-regulation of LGALS3 and MAP1LC3B compared to in NHEMs. The bars indicates the means ± s.d. of three independent experiments; measurements were performed in triplicate (****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.5).
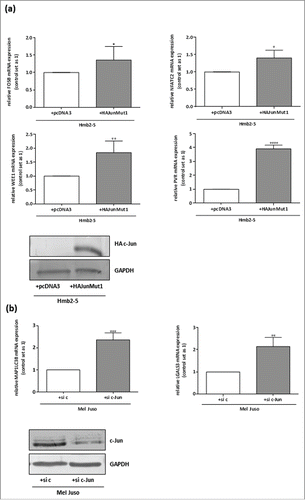
To further ensure that the predicted target genes are regulated by the transcription factor c-Jun in malignant melanoma, we transfected a melanocyte-resembling cell clone lacking c-Jun expression (HMB2-5Citation24) with a c-Jun expression construct (HA-JunMut1Citation11) and confirmed an increase in c-Jun accumulation by Western blotting with a transfection efficiency of about 80% (). qRT-PCR of mRNA from the transfected cells revealed that the increase in c-Jun expression was accompanied by an increase in expression of FosB, NFATC2, WEE1 and PVR compared to the pcDNA3 (negative control)-transfected Hmb2-5 cells (). Vice versa, we analyzed the c-Jun dependent regulation of MAP1LC3B and LGALS3 in Mel Juso after knockdown of c-Jun via siRNA. The successful knockdown with a transfection efficiency of about 85% of c-Jun was confirmed by Western blot analysis (). We observed up-regulation of MAP1LC3B and LGALS3 mRNA expression in the si c-Jun cells compared to the control transfected cells (si c) ().
Figure 5. FosB, NFATC2, WEE1, PVR, LGALS3 and MAP1LC3B are regulated by the transcription factor c-Jun. (a) qRT-PCR after c-JUN re-expression with a HA-tagged-c-Jun expression construct (HA-Jun Mut1) Citation11 in the melanocyte-resembling cell clone Hmb2-5 lacking c-Jun expression Citation24 showed upregulation of FosB, NFATC2, WEE1 and PVR. pcDNA3 served as a negative control. Transfection efficiency after transfection with the HA-tagged-c-Jun expression construct was verified by Western Blot analysis. (b) qRT-PCR after transfection of siRNA against c-Jun in the melanoma cell line Mel Juso revealed downregulation of MAP1LC3B and LGALS3. Transfection experiments with a control siRNA served as the negative control. Transfection efficiency after transfection with the siRNA against c-Jun was verified by Western Blot analysis. The bars show the means ± s.d. of three independent experiments; measurements were performed in triplicate (****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0 .5).
Discussion
Malignant melanoma is an aggressive cancer derived from melanocytes and is resistant to most current therapeutic approaches. In recent studies with melanoma cells, we demonstrated that the transcription factor c-Jun plays a crucial role in the development and progression of this cancer type.Citation5,11,12 However, the detailed molecular mechanism of c-Jun's influence on melanoma progression and development remains elusive, as only a subset of target genes is known. Previous studies have shown that cancer-relevant genes, such as cyclin D1, p53, and INK4ACitation25-27 are regulated by c-Jun. In the present study, we identified 40 4 general c-Jun target genes in non-melanoma cell lines by in silico analysis of pre-existing ChIP-Seq data (ENCODE). The identified c-Jun target genes within this study did not include the genes cyclin D1, p53, and INK4A described as c-Jun regulated genes in previous studies, indicating that these genes are potentially indirectly regulated by the transcription factor c-Jun, but not by direct binding in the promotor-/enhancer region of these genes. However, we observed that these via ENCODE identified c-Jun target genes also share known cancer-relevant molecular functions, such as adhesion, cell motion and positive regulation of cell proliferation (GO terms), indicating that c-Jun supports cancer development. Moreover, for the first time, we confirmed the direct regulation of 6 selected target genes by the transcription factor c-Jun in melanoma cells. Based on the results of this study, the c-Jun target genes FosB, WEE1, PVR, LGALS3 contain the classical AP-1 consensus sequence, whereas MAP1LC3B and NFATC2 contain variations of the classical AP-1 DNA binding site. Thus, interactions between c-Jun and DNA are not strictly dependent on the classical AP-1 consensus sequence and, consequently, we suppose an interaction between c-Jun and 2 novel DNA binding sites most similar to the consensus sequence. Moreover, our research showed that the newly identified c-Jun target genes are de-regulated in melanoma cell lines compared to melanocytes: FosB, NFATC2, WEE1, PVR are up-regulated, whereas MAP1LC3B and LGALS3 are down-regulated. It is possible that the detected up- and downregulation of c-Jun target genes depends on the observed differences in the c-Jun binding sites in the different target genes. Additionally, we suggest a mechanism resulting in the up- or downregulation of the potential c-Jun target genes. One possible mechanism is that the binding site of c-Jun overlaps a binding site of an activator and prevents its binding. Such a mechanism was previously suggested for the crosstalk between c-Jun and the glucocorticoid receptor in the regulation of some genes.Citation28
Previous studies suggest that the newly identified direct c-Jun target genes FosB, WEE1, PVR, MAP1LC3B and LGALS3 have crucial roles in tumorigenesis in several cancer types. Moreover, according to the The Human Protein Atlas database (http://www.proteinatlas.org/)Citation29 the immunohistochemistry stainings with antibodies against the identified potential c-Jun target genes of malignant melanoma tissue samples and normal melanocytes show a similar expression pattern in melanoma patients as suggested by our data. FOS proteins have been implicated as regulators of cell proliferation, differentiation and transformation. Kim and colleaguesCitation30 detected under-expression of FosB in pancreatic cancer tissues with lymph node metastasis compared to pancreatic cancer tissues without lymph node metastasis and showed that decreased expression of FosB is associated with reduced survival. In contrast, other studies showed a bipolar FosB expression pattern in breast cancer,Citation31 whereas our study indicated high FosB expression in melanoma cell lines compared to NHEMs. Moreover, the gene product of NFATC2 was found to be functionally important in malignant melanoma, as it inhibits apoptosis.Citation18 Braeuer and colleaguesCitation32 showed a decrease in tumor growth and the number of experimental lung metastasis after silencing NFATC2 in A375SM melanoma cells, which indicates a crucial role in melanoma growth and metastasis. Furthermore, several studies have confirmed the functional relevance of WEE1 overexpression in multiple cancer types, such as breast cancer,Citation33 ovarian carcinoma,Citation34 medulloblastomaCitation35 and melanoma.Citation36 Other studies have shown overexpression of PVR in lung adenocarcinoma and cutaneous melanoma, which influences invasive activity and, thus, the development of a malignant phenotype.Citation37,38 In contrast to our melanoma data, Wu and colleagues described MAP1LC3B overexpression in hepatocellular carcinoma and its correlation with malignant progression and poor prognosis.Citation39 Interestingly, our study showed down-regulation of MAP1LC3B in melanoma cells, which was previously and concordantly with our findings observed in hypopharyngeal squamous cell carcinoma (HSCC) correlating with poor prognosis.Citation40 Therefore, we speculate that the status and role of MAP1LC3 expression vary across cancer types. Some other studies confirmed our expression data in case of LGALS3, which is known to be associated with melanomaCitation41, 42 and has been reported to play an essential role in the acquisition of vasculogenic mimicry and angiogenic properties associated with melanoma progression. Thus, c-Jun target genes seem to have both pro-oncogenic and anti-oncogenic functions, which possibly depend on cancer type and/or variations in DNA binding sites recognized by c-Jun. The identification of melanoma-specific, direct c-Jun target genes by ChIP-Seq would offer detailed insight into the molecular role of c-Jun in the development of the malignant phenotype. Moreover, inhibiting direct c-Jun target genes whose expressions and/or activities support the development and progression of malignant melanoma, by inhibiting the transcription factor c-Jun itself is a potential novel therapeutic option.
In summary, the results of this study indicate that the transcription factor c-Jun plays a crucial role in the development and progression of malignant melanoma through direct regulation of cancer-relevant target genes. Moreover, the expression of c-Jun can lead to upregulation and downregulation of specific target genes; thus, we hypothesize that c-Jun enhances the expression of pro-oncogenic target genes and inhibits the expression of anti-oncogenic target genes. A further detailed analysis of direct c-Jun target genes and their possible functions in the development and progression of malignant melanoma is warranted to determine whether c-Jun could serve as a therapeutic target for malignant melanoma.
Material and methods
In silico analysis
ChIP-Seq data archived in the Encyclopedia of DNA Elements (ENCODE; http://genome.ucsc.edu/ENCODE/) were used for in silico analysis. The data were screened for c-Jun binding regions within the human genome (hg19) of HUVEC, HepG2 and HeLa-S3.
STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database was used for the identification of known and predicted protein associations, including indirect (functional) associations.
To discover novel, ungapped c-Jun binding motifs in nucleotide sequences the database MEME Suite 4.10.2 (Multiple Em for Motif Elicitation) was utilized.
Cell culture
Melanoma cell lines Mel Juso, Mel Ju, A375, Mel Ei and normal human epidermal melanocytes (NHEMs) were described previously.Citation43 The cell lines Mel Juso, Mel Ei and A375 were derived from primary cutaneous melanomas; Mel Ju was derived from metastasis of melanoma. Cells were maintained in DMEM supplemented with penicillin (400 units/ml), streptomycin (50mg/ml), L-glutamine (300 mg/ml), 10% FCS (Sigma-Aldrich, Steinheim, Germany) and split at a 1:5 ratio every 3 d. NHEMs (PromoCell, Heidelberg, Germany) were derived from neonatal foreskin and were used between passages 2 and 6. HMB2-5 is a cell clone in our laboratory resembling melanocytes.Citation24 Once a week this cell line is treated with G418 (2 mg/ml) to ensure clone selection.
Analysis of gene expression by quantitative PCR
cDNAs of total RNA fractions were generated using SuperScript II Reverse Transcriptase Kit (Invitrogen, Groningen, The Netherlands). qRT-PCR was performed on a Lightcycler (Roche, Mannheim, Germany). cDNA template (500 ng), 0.5 µl (20 µM) each of forward and reverse primers and 10 µl of SybrGreen LightCycler Mix in a total of 20 µl were applied to the PCR program as described previously.Citation11 Annealing and melting temperatures were optimized for each primer set (Table S2). The PCR reaction was evaluated by melting curve analysis and determining the PCR products on agarose gels. β-Actin was used for normalization.
Chromatin Immunoprecipitation
ChIP assays were performed following the manufacturer's instructions (ChIP-IT Express; Active Motif, Carlsbad, CA, USA) as described previously.Citation44 Mel Juso cells grown to 70–80 % confluence on 3 T-175 flasks were used for one chromatin isolation (˜20 million cells/ ChIP). Samples were immunoprecipitated using a specific c-Jun antibody (3 µg of anti-c-Jun (sc-1694; Santa Cruz Biotechnology, Heidelberg, Germany). A RNA polymerase II antibody was used as a positive, and an IgG antibody as a negative control, following the protocol provided with the control kit (ChIP-IT control Kit-human; Active Motif). DNA samples from the ChIP experiments were used for analysis by PCR utilizing the real-time PCR LightCycler system (Roche) as described previously.Citation12,45 PCR was performed on 4 DNA templates: the input DNA (1: 10), DNA isolated through RNA polymerase II ChIP (Pol II), DNA isolated through the negative control IgG ChIP (IgG), and DNA isolated through the c-Jun ChIP (c-Jun). A control reaction (H2O) with no DNA template was also performed. Sets of primer pairs amplifying a specific promotor region of a control gene and of each predicted target gene were used: the positive control (glyceraldehyde-3-phosphate dehydrogenase; GAPDH) and the negative control (Active Motif; amplifies a 78 base pair fragment from a gene desert on human chromosome 12) - primer pairs provided by the kit, and primer pairs spanning the c-Jun-binding site within the promoter regions of the predicted c-Jun target genes (ChIP Primers; Table S2). DNA fragments precipitated with the c-Jun and Pol II antibodies were analyzed on a 1.5 % agarose gel compared to the Input DNA and the fragments precipitated with the IgG antibody.
Transfection experiments
Two times 105 cells were seeded each well in 6-well plates and transfected with 0.5 mg of HA-tagged c-Jun (HAJunMut) or pcDNA3 constructs using Lipofectamine Plus (transfection efficiency of 80%) (Invitrogen). The HA-tagged c-Jun expression vector was generated in our laboratory and results in a complete loss of repression by miR-125b. Exogenous expression of HA-tagged wt c-Jun (HAJun wt) in the HMB2-5 cell clone (expressing miR-125b) did not lead to c-Jun protein expression, whereas the mutated version (HAJun Mut miR-125b) led to strong induction of protein expression in this cell type.Citation11 Twenty-four h after transfection, the cells were used for further analysis.
siRNA transfection experiments
siRNA transfection of Mel Juso cells was performed using the reverse transfection protocol of the Lipofectamine RNAiMAX reagent (transfection efficiency of 85%) (Invitrogen) according to the manufacturer´s instructions. Eight times 104 cells were transfected with 10 nM of c-Jun (JUNVHS40918; Invitrogen) or negative control siRNA (Qiagen, Hilden, Germany) for 16 h, respectively. Each experiment was repeated at least 3 times.
Western blotting
The transfected cells were resuspended in 200 ml RIPA buffer (Roche) and lysed for 15 min at 4°C. Insoluble fragments were removed by centrifugation at 13.000 r.p.m. for 10 min and the supernatant was stored at −20°C. Twenty µg of RIPA complete cell lysates was loaded per lane and separated on SDS–PAGE gels (Invitrogen) and subsequently blotted onto a PVDF membrane. After blocking for 1 h blocking with 5 % MP/TBST the membrane was incubated for 16h with one of the following antibodies: anti-c-Jun (1 in 1000 dilution; Cell Signaling, Frankfurt am Main, Germany), anti-GAPDH (1 in 3000 dilution; Cell Signaling) and anti-HA-tag (1 in 1000 dilution; Cell Signaling). After three washing steps with TBS-T, the membrane was incubated for 1 h with an alkaline phosphate-coupled secondary anti-mouse (1 in 3000 dilution in TBS-T) or anti-rabbit (1 in 3000 dilution in TBS-T) IgG antibody (Chemicon, Hofheim, Germany) and then washed again for 3 times in TBS-T. Finally, immunoreactions were visualized by NBT/BCIP (Sigma-Aldrich) staining.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Data.zip
Download Zip (66.9 KB)Acknowledgment
We thank Susanne Wallner for excellent technical assistance. This work was supported by grants from the German cancer Aid and the Melanoma Research Network to A.K.B and S.K.
References
- Shaulian E. AP-1–The Jun proteins: Oncogenes or tumor suppressors in disguise? Cellular signalling 2010; 22(6):894-9; PMID:20060892; http://dx.doi.org/10.1016/j.cellsig.2009.12.008
- Vogt PK. Jun, the oncoprotein. Oncogene 2001; 20(19):2365-77; PMID:11402333; http://dx.doi.org/10.1038/sj.onc.1204443
- Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 2001; 20(19):2438-52; PMID:11402339; http://dx.doi.org/10.1038/sj.onc.1204385
- Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene 2001; 20(19):2390-400; PMID:11402335; http://dx.doi.org/10.1038/sj.onc.1204383
- Kappelmann M, Bosserhoff A, Kuphal S. AP-1/c-Jun transcription factors: Regulation and function in malignant melanoma. Eur J Cell Biol 2014; 93(1-2): p. 76-81. -81 PMID:24315690
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nature reviews Cancer 2003; 3(11):859-68; PMID:14668816; http://dx.doi.org/10.1038/nrc1209
- Mundade R, Ozer HG, Wei H, Prabhu L, Lu T. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell cycle (Georgetown, Tex) 2014; 13(18):2847-52; PMID:25486472; http://dx.doi.org/10.4161/15384101.2014.949201
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nature protocols 2006; 1(2):729-48; PMID:17406303; http://dx.doi.org/10.1038/nprot.2006.98
- Jochum W, Passegue E, Wagner EF. AP-1 in mouse development and tumorigenesis. Oncogene 2001; 20(19):2401-12; PMID:11402336; http://dx.doi.org/10.1038/sj.onc.1204389
- Weiss C, Bohmann D. Deregulated repression of c-Jun provides a potential link to its role in tumorigenesis. Cell cycle (Georgetown, Tex) 2004; 3(2):111-3; PMID:14712066; http://dx.doi.org/10.4161/cc.3.2.648; 20 and 23
- Kappelmann M, Kuphal S, Meister G, Vardimon L, Bosserhoff AK. MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 2013; 32(24):2984-91; PMID:22797068; http://dx.doi.org/10.1038/onc.2012.307
- Spangler B, Kappelmann M, Schittek B, Meierjohann S, Vardimon L, Bosserhoff AK, Kuphal S. ETS-1/RhoC signaling regulates the transcription factor c-Jun in melanoma. Int J Cancer Journal international du cancer 2012; 130(12):2801-11; PMID:21732343; http://dx.doi.org/10.1002/ijc.26277
- Spangler B, Vardimon L, Bosserhoff AK, Kuphal S. Post-transcriptional regulation controlled by E-cadherin is important for c-Jun activity in melanoma. Pigment Cell Melanoma Res 2011; 24(1):148-64; PMID:20977688; http://dx.doi.org/10.1111/j.1755-148X.2010.00787.x
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43(Database issue):D447-52; PMID:25352553; http://dx.doi.org/10.1093/nar/gku1003
- Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 2007; 8(9):R183; PMID:17784955; http://dx.doi.org/10.1186/gb-2007-8-9-r183
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 2007; 35(Web Server issue):W169-75; PMID:17576678; http://dx.doi.org/10.1093/nar/gkm415
- Yang S, McNulty S, Meyskens FL, Jr. During human melanoma progression AP-1 binding pairs are altered with loss of c-Jun in vitro. Pigment Cell Res 2004; 17(1):74-83; PMID:14717848; http://dx.doi.org/10.1046/j.1600-0749.2003.00114.x
- Perotti V, Baldassari P, Bersani I, Molla A, Vegetti C, Tassi E, Dal Col J, Dolcetti R, Anichini A, Mortarini R. NFATc2 is a potential therapeutic target in human melanoma. J Investigative Dermatol 2012; 132(11):2652-60; PMID:22718120; http://dx.doi.org/10.1038/jid.2012.179
- Bhattacharya A, Schmitz U, Wolkenhauer O, Schönherr M, Raatz Y, Kunz M. Regulation of cell cycle checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene 2013; 32(26):3175-83; PMID:22847610; http://dx.doi.org/10.1038/onc.2012.324
- Sloan KE, Eustace BK, Stewart JK, Zehetmeier C, Torella C, Simeone M, Roy JE, Unger C, Louis DN, Ilag LL, et al. CD155/PVR plays a key role in cell motility during tumor cell invasion and migration. BMC Cancer 2004; 4:73; PMID:15471548
- Liu H, He Z, Simon HU. Autophagy suppresses melanoma tumorigenesis by inducing senescence. Autophagy 2014; 10(2):372-3; PMID:24300435; http://dx.doi.org/10.4161/auto.27163
- Schoof N, Iles MM, Bishop DT, Newton-Bishop JA, Barrett JH, Genomel Consortium. Pathway-based analysis of a melanoma genome-wide association study: analysis of genes related to tumour-immunosuppression. PloS One 2011; 6(12):e29451; PMID:22216283; http://dx.doi.org/10.1371/journal.pone.0029451
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 1994; 2:28-36; PMID:7584402
- Poser I, Tatzel J, Kuphal S, Bosserhoff AK. Functional role of MIA in melanocytes and early development of melanoma. Oncogene 2004; 23(36):6115-24; PMID:15208686; http://dx.doi.org/10.1038/sj.onc.1207797
- Bakiri L, Lallemand D, Bossy-Wetzel E, Yaniv M. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO JJ 2000; 19(9):2056-68; PMID:10790372; http://dx.doi.org/10.1093/emboj/19.9.2056
- Schreiber M, Kolbus A, Piu F, Szabowski A, Möhle-Steinlein U, Tian J, Karin M, Angel P, Wagner EF. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev 1999; 13(5):607-19; PMID:10072388; http://dx.doi.org/10.1101/gad.13.5.607
- Passegue E, Wagner EF. JunB suppresses cell proliferation by transcriptional activation of p16(INK4a) expression. EMBO J 2000; 19(12):2969-79; PMID:10856241; http://dx.doi.org/10.1093/emboj/19.12.2969
- Yang-Yen HF, Chambard JC, Sun YL, Smeal T, Schmidt TJ, Drouin J, Karin M. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell 1990; 62(6):1205-15; PMID:2169352; http://dx.doi.org/10.1016/0092-8674(90)90396-V
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Proteomics. Tissue-based map of the human proteome. Science (New York, NY) 2015;347(6220):1260419; PMID:25613900; http://dx.doi.org/10.1126/science.1260419
- Kim JH, Lee JY, Lee KT, Lee JK, Lee KH, Jang KT, Heo JS, Choi SH, Rhee JC. RGS16 and FosB underexpressed in pancreatic cancer with lymph node metastasis promote tumor progression. Tumour Biol 2010; 31(5):541-8; PMID:20571966; http://dx.doi.org/10.1007/s13277-010-0067-z
- Bamberger AM, Methner C, Lisboa BW, Städtler C, Schulte HM, Löning T, Milde-Langosch K. Expression pattern of the AP-1 family in breast cancer: association of fosB expression with a well-differentiated, receptor-positive tumor phenotype. Int J Cancer Journal international du cancer 1999; 84(5):533-8; PMID:10502734; http://dx.doi.org/10.1002/(SICI)1097-0215(19991022)84:5%3c533::AID-IJC16%3e3.0.CO;2-J
- Braeuer RR, Zigler M, Kamiya T, Dobroff AS, Huang L, Choi W, McConkey DJ, Shoshan E, Mobley AK, Song R, et al. Galectin-3 contributes to melanoma growth and metastasis via regulation of NFAT1 and autotaxin. Cancer Res 2012; 72(22):5757-66; PMID:22986745; http://dx.doi.org/10.1158/0008-5472.CAN-12-2424
- Ghiasi N, Habibagahi M, Rosli R, Ghaderi A, Yusoff K, Hosseini A, Abdullah S, Jaberipour M. Tumour suppressive effects of WEE1 gene silencing in breast cancer cells. Asian Pac J Cancer Prev 2014; 14(11):6605-11; PMID:24377575; http://dx.doi.org/10.7314/APJCP.2013.14.11.6605
- Slipicevic A, Holth A, Hellesylt E, Tropé CG, Davidson B, Flørenes VA. Wee1 is a novel independent prognostic marker of poor survival in post-chemotherapy ovarian carcinoma effusions. Gynecol Oncol 2014; 135(1):118-24; PMID:25093290; http://dx.doi.org/10.1016/j.ygyno.2014.07.102
- Harris PS, Venkataraman S, Alimova I, Birks DK, Balakrishnan I, Cristiano B, Donson AM, Dubuc AM, Taylor MD, Foreman NK, et al. Integrated genomic analysis identifies the mitotic checkpoint kinase WEE1 as a novel therapeutic target in medulloblastoma. Mol Cancer 2014; 13:72; PMID:24661910; http://dx.doi.org/10.1186/1476-4598-13-72.
- Haarberg HE, Paraiso KH, Wood E, Rebecca VW, Sondak VK, Koomen JM, Smalley KS. Inhibition of Wee1, AKT, and CDK4 underlies the efficacy of the HSP90 inhibitor XL888 in an in vivo model of NRAS-mutant melanoma. Mol Cancer Ther 2013; 12(6):901-12; PMID:23538902; http://dx.doi.org/10.1158/1535-7163.MCT-12-1003
- Tane S, Maniwa Y, Hokka D, Tauchi S, Nishio W, Okita Y, Yoshimura M. The role of Necl-5 in the invasive activity of lung adenocarcinoma. Exp Mol Pathol 2013; 94(2):330-5; PMID:23276719; http://dx.doi.org/10.1016/j.yexmp.2012.12.003
- Bevelacqua V, Bevelacqua Y, Candido S, Skarmoutsou E, Amoroso A, Guarneri C, Strazzanti A, Gangemi P, Mazzarino MC, D'Amico F, et al. Nectin like-5 overexpression correlates with the malignant phenotype in cutaneous melanoma. Oncotarget 2012; 3(8):882-92; PMID:22929570; http://dx.doi.org/10.18632/oncotarget.594
- Wu DH, Jia CC, Chen J, Lin ZX, Ruan DY, Li X, Lin Q, Min-Dong, Ma XK, Wan XB, et al. Autophagic LC3B overexpression correlates with malignant progression and predicts a poor prognosis in hepatocellular carcinoma. Tumour Biol 2014; 35(12):12225-33; PMID:25256671; http://dx.doi.org/10.1007/s13277-014-2531-7
- Wang J, Pan XL, Ding LJ, Liu DY, Da-Peng Lei, Jin T. Aberrant expression of Beclin-1 and LC3 correlates with poor prognosis of human hypopharyngeal squamous cell carcinoma. PloS One 2013; 8(7):e69038; PMID:23935917; http://dx.doi.org/10.1371/journal.pone.0069038
- Olbryt M, Habryka A, Tyszkiewicz T, Rusin A, Cichoń T, Jarząb M, Krawczyk Z. Melanoma-associated genes, MXI1, FN1, and NME1, are hypoxia responsive in murine and human melanoma cells. Melanoma Res 2011; 21(5):417-25; PMID:21912348; http://dx.doi.org/10.1097/CMR.0b013e328348db2f
- Mourad-Zeidan AA, Melnikova VO, Wang H, Raz A, Bar-Eli M. Expression profiling of Galectin-3-depleted melanoma cells reveals its major role in melanoma cell plasticity and vasculogenic mimicry. Am J Pathol 2008; 173(6):1839-52; PMID:18988806; http://dx.doi.org/10.2353/ajpath.2008.080380
- Braig S, Bosserhoff AK. Death inducer-obliterator 1 (Dido1) is a BMP target gene and promotes BMP-induced melanoma progression. Oncogene 2013; 32(7):837-48; PMID:22469980; http://dx.doi.org/10.1038/onc.2012.115
- Wenke AK, Niebler S, Grassel S, Bosserhoff AK. The transcription factor AP-2varepsilon regulates CXCL1 during cartilage development and in osteoarthritis. Osteoarthritis Cartilage 2011; 19(2):206-12; PMID:21134476; http://dx.doi.org/10.1016/j.joca.2010.11.011
- Kappelmann M, Bosserhoff A, Kuphal S. AP-1/c-Jun transcription factors: regulation and function in malignant melanoma. Eur J Cell Biol 2014; 93(1-2):76-81; PMID:24315690; http://dx.doi.org/10.1016/j.ejcb.2013.10.003
