ABSTRACT
Lung cancer is the leading cause of cancer deaths in China, and about 60% of the cases are diagnosed with histological adenocarcinoma. The caspase 8 (CASP8) gene is a critical initiator of the extrinsic apoptosis pathway. To explore the relationship between tagSNPs or haplotypes of CASP8 and the efficacy of platinum-based chemotherapy in advanced lung adenocarcinoma patients of China, we recruited 555 advanced adenocarcinoma patients. We extracted the genomic DNA from patients' peripheral blood samples and sequenced tagSNPs of CASP8. We calculated the individual haplotype of CASP8 frequencies using the PHASE 2.0 program. The association between CASP8 tagSNPs and overall survival (OS) was calculated by univariate and multivariate Cox regression analysis. A univariate logistic regression analysis was done to analyze the CASP8 tagSNPs and the toxicity of platinum-based chemotherapy. The same statistical methods were used for exploring haplotypes of CASP8. Rs3769821 and rs1045494 of CASP8 were independent prognosis factors for overall survival (OS) using multivariate Cox's regression models. For the haplotype of the 7 tagSNPs, haplotype AGGAAAGA was correlated with the efficacy of platinum-based chemotherapy. The polymorphisms of CASP8, rs7608692, and haplotype AGAACAG correlated with neutropenia toxicity. The haplotype GGGGAAA was associated with thrombocytopenia toxicity. We conclude that the polymorphisms of CASP8 contribute to the prognosis of advanced lung adenocarcinoma and influence the quality of life and survival.
Abbreviations
| CASP8 | = | Caspase 8 |
| OS | = | overall survival |
| PFS | = | progression-free survival |
| NSCLC | = | non-small-cell lung cancer |
| SCLC | = | small-cell lung cancer |
| ALK | = | anaplastic lymphoma kinase |
| EGFR | = | epidermal growth factor receptor |
| NCCN | = | National comprehensive cancer network |
| DISC | = | death-inducing signaling complex |
| FADD | = | adaptor protein FAS-associated death domain |
| TRADD | = | TNFR-associated death domain |
| SNP | = | Single nucleotide polymorphisms |
| CHB | = | Han Chinese in Beijing |
| CEU | = | Utah residents with Northern and Western European ancestry from the CEPH collection |
| YRI | = | Yoruban in Ibadan |
| TKIs | = | tyrosine kinase inhibitors |
| SmCC | = | small cell carcinoma |
| ECOG PS | = | Eastern Cooperative Oncology Group performance status |
| RECIST | = | Response Evaluation Criteria in Solid Tumors |
| DCR | = | disease control rate |
| CR | = | complete response |
| PR | = | partial response |
| SD | = | stable disease |
| NCI | = | National Cancer Institute |
| CTCAE | = | Common Terminology Criteria for Adverse Events |
Introduction
With the incidence and mortality rates of lung cancer up to 48.3/105 and 39.3/105, respectively, lung cancer remains the most common cancer and leading cause of cancer deaths in both urban and rural areas of China according to the latest report of lung cancer distribution in China.Citation1 The mortality of lung cancer in the Chinese population remained stable from 2000 to 2011.Citation2 Lung cancer is classified as non-small-cell lung cancer (NSCLC) and small-cell lung cancer (SCLC). About 85% of patients are diagnosed with NSCLC, and 60% of them are subdivided into histological adenocarcinoma, one of the histological types of NSCLC.Citation3
Patients are usually in the advanced stage of lung cancer when they are diagnosed, and the surgical option for recovery is usually no longer applicable. For advanced lung cancer patients, chemotherapy presently remains the first-line treatment method.Citation4 According to the National Comprehensive Cancer Network (NCCN) guidelines for NSCLC, Version 6, 20155, targeted therapy has been emphasized as the preferred treatment of advanced lung cancer patients with molecular mutations, and the genetic mutation test is recommended for advanced lung cancer patients, especially for those with anaplastic lymphoma kinase (ALK) gene rearrangements and epidermal growth factor receptor (EGFR) mutations. However, patients with EGFR mutations and ALK rearrangements account for merely 10% and 2%–7%, respectively. A number of patients without typical mutations still rely on chemotherapy, and the platinum-based double chemotherapy regimen is the first-line regimen in advanced lung cancer patients according to the 2015 NCCN guidelines for NSCLC.Citation5
The 5-year overall survival (OS) rate of lung cancer is only about 16.1% in China over the last 40 years.Citation1 A few patients can survive under the same treatment methods because of the individual differences on a genetic level. Patients' survival times are generally dictated by cancer cell's proliferation or apoptosis progression and inevitably involve various genetic variations from a molecular mechanism perspective. Caspase 8 (CASP8), a gene crucial to apoptosis progression, is located on the human chromosome 2q33.1. It is encoded from a spliced message, which is translated initially as a monomer, procaspase-8, and when it is activated by dimerization the extrinsic apoptotic pathway will be initiated.Citation6,7 The extrinsic pathway starts with formation of a death-inducing signaling complex (DISC). Death receptor ligands bind to death receptors, and this process recruits inactivated procaspase-8 and adaptor protein FAS-associated death domain (FADD) or TNFR-associated death domain (TRADD) to form DISC. In turn, DISC converts procaspase-8 to Caspase-8, which then triggers apoptosis via cleavage and activation of executioner caspases −3, −6, and/or −7 or intrinsic pathways to induce cell death.Citation8-10 Various genetic mutations are involved in many diseases. One of the most common genetic mutation is termed single nucleotide polymorphism (SNP). It appears to participate in the progress of tumor formation, metastasis, and drug resistance.Citation11 Kim et al. detected somatic mutations of CASP8 in 180 colon tumors (98 invasive carcinomas and 82 adenomas), and 5 CASP8 mutations were observed in the invasive colon cancer samples rather than in the adenomas.Citation12 Dominant-negative mutations of CASP8 inhibited the progress of the extrinsic apoptotic pathway and were involved in the last stages of tumor carcinogenesis.Citation12, 13 D302H of CASP8 is associated with a risk for breast cancer in the European population.Citation14-17 Many studies have reported the relationship between CASP8 polymorphisms and cancers such as lung, breast, esophageal, and gastric cancers. Qian reported that CASP8 polymorphisms were associated with toxic side effects after platinum-based chemotherapy in NSCLC patients.Citation18 To investigate whether CASP8 tagSNPs have an effect on the efficacy of platinum-based chemotherapy in advanced adenocarcinoma lung cancer, we enrolled 555 advanced adenocarcinoma patients. We collected their clinical characteristics and sequenced the polymorphisms of CASP8, which were chosen from the HapMap SNP database based on 2 criteria: 1) minor allele frequency (MAF) cutoff of 0.05 and 2) a correlation coefficient (r2) threshold of 0.8. The tagSNPs were genotyped following the standard genotyping call rate of > 0.95 and GenCall score > 0.2. The individual haplotype of CASP8 frequencies were calculated using the PHASE 2.0 Program. In this study, we tried to illustrate the relationship between tagSNPs or haplotype of CASP8 and the efficacy of platinum-base chemotherapy.
Results
Clinical information and SNP information of CASP8
Clinical characteristics of 555 adenocarcinoma patients
Detailed clinical characteristics of the enrolled 555 advanced lung adenocarcinoma patients are listed in . The median age of the patients was 57, ranging from 26 to 80 y. All the patients received platinum-based chemotherapy, which included DNA-damaging agents, platinum-tubulin-targeting agents, and other combinations. One-hundred 53 patients accepted the platinum-gemcitabine regimen (belong to DNA-damaging agents), 3-hundred 84 patients were treated with platinum-tubulin-targeting drugs, and the other patients were treated with other combinations. showed that age (p = 0.003) and sex (p = 0.009) were associated with the OS of the study patients. However, no significant differences in OS between histology, clinical stages, 3 chemotherapy regimens, and smoking status were observed after using the Kaplan-Meier curve analysis.
Table 1. The clinical characteristics of 555 advanced adenocarcinoma patients treated with platinum-based chemotherapy.
Genotype information in different populations
The genotype information about 7 tagSNPs of CASP8 in different races was shown in . We analyzed the genotype frequencies of the total tagSNPs in the current data or in Han Chinese in Beijing, China (CHB), Utah residents with Northern and Western European ancestry from the CEPH collection (CEU), and Yoruban in Ibadan, Nigeria (YRI). All available data were obtained from the HapMap SNP database (HapMap Genome Browser release #28; phases 1, 2 and 3 – merged genotypes and frequencies). The allele frequencies of the 7 tagSNPs were significantly different among the Chinese, Western, and African population as shown in . The tagSNPs of CASP8 allele frequencies from our data were similar with the CHB data from HapMap SNP database.
Table 2. Genotype frequencies and MAF of CASP8 tag SNPs in current data, or in CHB, CEU and YRI from HapMap SNP database.
Chemotherapy treatment efficacy of CASP8 tagSNPs and haplotypes
The patients with AA genotype of Rs3769821 and rs1045494 of CASP8 have longer OS than patients with non-AA genotype
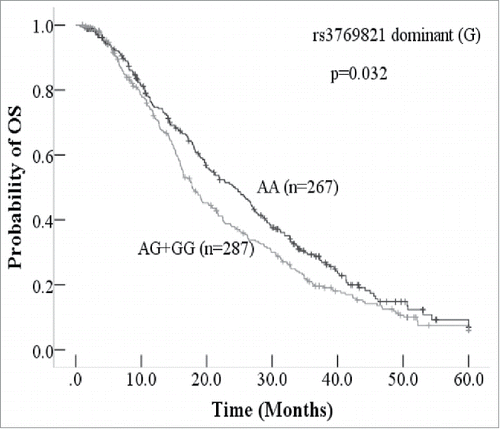
We analyzed the relationship between 7 tagSNPs from CASP8 and OS of 555 advanced adenocarcinoma patients using univariate and multivariate Cox's regression models (multiple testing with FDR). Every tagSNP was classified with genotypic, dominant, recessive, and additive models. The minor allele was considered the disease allele. The result showed that in univariate cox's regression analysis, no significant tagSNPs were associated with patients' OS after multiple testing (). Then we use clinical factors and tagSNPs of CASP8 dominant model and recessive model for multivariate Cox's multivariate regression analysis calculating. However, we found that in multivariate cox's regression analysis, rs3729821 dominant model (p-adjusted = 0.007) and rs1045494 dominant model (p-adjusted = 0.014) were correlated with OS after multiple testing with R language (). It may exist some confounding factor in univariate cox's regression analysis, and we corrected with multivariate cox's regression analysis. Rs3769821 dominant (G) (AA vs. AG+GG)and rs1045494 dominant (G) (AA vs. AG+GG) model were independent factors according to multivariate cox's regression analysis after multiple testing (). The patients with AA genotype of rs3769821 had 6.7 months longer OS than patients with GG+AG genotype (AA = 267, mOS 24.4, 95%CI 20.5–28.2; GG and AG = 287, mOS 17.7, 95%CI 15.4–20.0; HR = 1.52, 95%CI = 1.20–1.94; p-adjusted = 0.007). Patients with AA genotype of rs1045494 also had longer OS than patients with GG+AG genotype (HR = 1.45,95%CI = 1.15–1.82; p-adjusted = 0.014). Sex and age were an significantprognostic clinical factors for 555 advanced adenocarcinoma patients who were treated with platinum-based chemotherapy. We derived a correlation between CASP8 7 tagSNPS and OS using the Kaplan-Meier curves, and the same significant result with rs3769821 was observed (, p = 0.032). The other 6 tagSNPs' Kaplan-Meier OS curves were shown in Fig. S1. No significant associations were observed between the other 6 tagSNPs of CASP8 (rs1045494, rs3769827, rs3754935, rs12990906, rs3769825, and rs7608692) and OS for 555 patients (Fig. S1). Considering progression-free survival (PFS) as an end point, we found that the 7 tagSNPs of CASP8 were not prognostic factors for the patients (Fig. S2), no significant differences between tagSNPs and PFS using Kaplan-Meier curves test was observed.
Table 3. Univariate Cox's regression analysis of 7 tagSNPs of CASP8 with OS in 555 advanced lung adenocarcinoma patients.
Table 4. Multivariate Cox's regression analysis of clinical factors and CASP8 tagSNPs for overall survival in the 555 advanced lung adenocarcinoma patients treated with platinum-based chemotherapy.
Figure 1. Kaplan-Meier curve of overall survival (OS) according to rs3769821 of CASP8 The patients with AA genotype of rs3769821 had longer survival than patients with AG or GG genotype of rs3769821.
Besides OS and PFS, response rate and clinical benefit, both of which belong to treatment efficacy, were considered to analyze. For rs3769821 AA genotype of CASP8, which was the significant variant in OS analysis, no significant association with the response rate and clinical benefit were observed using the univariant logistic regression analysis (for response rate, p = 0.636; for clinical benefit, p = 0.431, Table S2).
The haplotype AGGAAAGA with zero copy patients have longer OS than one or 2 copy patients
Four haplotypes with frequencies over 10% were generated from the 21 haplotypes using PHASE 2.0. (Table S1; Haplotype 1 AGGAAAGA, haplotype 2 AGAACAG, haplotype 3 AAAGAAA, haplotype 4 GGGGAAA). We analyzed the relation between the haplotype of the 7 tagSNPs and OS for the adenocarcinoma patients using log-rank test. The haplotype 1 AGGAAAGA (rs3769827/rs7608692/rs3769825/rs12990906/rs3754935/rs3769821/rs1045494) significantly correlated with OS (, p = 0.015), whereas the other 3 haplotypes were not associated with OS for the patients with adenocarcinoma (). The patients with zero copies of haplotype AGGAAAGA had 6 months longer than patients with one or 2 copies (zero copy, mOS 23.9 months; 1–2 copies, mOS 17.7 months p = 0.016) (). Haplotype AGGAAAGA was a predictive factor for patients with adenocarcinoma treated with platinum-based chemotherapy. However, no significant differences were observed for the other 3 haplotypes. For PFS, the 4 haplotypes of CASP8 were not prognostic factors according to the log-rank test (Table S3, Fig. S3). Haplotype AGGAAAGA did not significantly contribute to the response rate or clinical benefits according to the univariate logistic regression analysis (Table S4).
Figure 2. Kaplan-Meier curve of overall survival according to the haplotypes of CASP8. (A) The patients with zero copy number of haplotype 1 had longer OS than patients with copy number one or 2 of haplotype 1. (B) The Kaplan-Meier curve between haplotype 2 and OS, no significantly difference was observed. (C) The Kaplan-Meier curve between haplotype 3 and OS, no significantly difference was observed. (D) The Kaplan-Meier curve between haplotype 4 and OS, no significantly difference was observed.
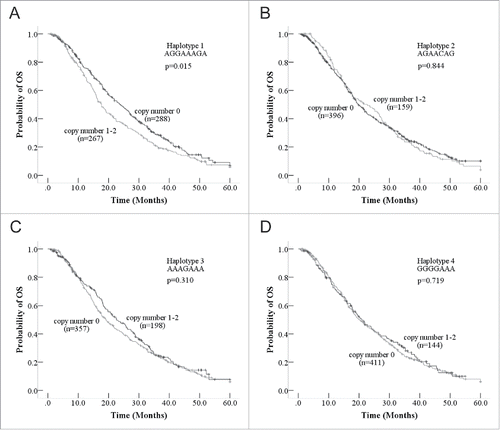
Table 5. Association between CASP8 haplotypes with OS in patients with adenocarcinoma lung cancer.
Patients' toxicity after chemotherapy
Rs7608692 of CASP8 is associated with the toxicity of neutropenia
The toxic effects of first-line platinum-based chemotherapy mainly influenced the OS for the advanced NSCLC patients. A complete set of associations between 7 tagSNPs of CASP8 and each severe toxicity was assessed using multivariate logistic regression analysis, assuming additive model. The only rs7608692 of CASP8 significantly contributed to grades 3 or 4 neutropenia toxicity according to multivariate logistic regression analysis (, additive model, p = 0.009). The homozygous of rs7608692 exhibited a protective effective on neutropenia.
Table 6. Association between CASP8 tagSNPs or haplotypes and toxicity in the 555 patients with adenocarcinoma lung cancer.
Haplotype AGAACAG associated with neutropenia toxicity and GGGGAAA correlated with thrombocytopenia toxicity
The correlation between CASP8 haplotypes and toxic effects was conducted by multivariate logistic regression analysis of the 555 adenocarcinoma patients. Haplotype AGAACAG was associated with grade 3 or 4 neutropenia toxicity, and the incidence rate of severe neutropenia toxicity was significant in patients with 1–2 copies (p = 0.016, incidence rate18.1% vs 10.3%, OR:1.92; 95%CI:1.13–3.25) (). Haplotype GGGGAAA contributed to thrombocytopenia toxicity; patients with zero copies of GGGGAAA haplotype revealed serious thrombocytopenia toxicity after treatment with platinum-based chemotherapy. However, the patients with 1–2 copies of GGGGAAA haplotype showed little thrombocytopenia toxicity after undergoing the regimen (p = 0.042) (). The patients with the other 2 haplotypes (AGAAAGA and AAAGAAA) did not present with any toxic effects after receiving the chemotherapy regimens.
Discussion
The caspase families are primarily involved in the progression of cell death, which includes 2 pathways consisting of extrinsic and intrinsic apoptotic pathways. As one of the apoptosis initiating caspases, Caspase 8 was shown to be a critical factor for the progression of extrinsic apoptosis.Citation10 Caspase 8, existing as an inactive procaspase monomer, was activated by dimerization but not by cleavage,Citation19 and then it participated in the apoptosis process with many factors such as death receptors and FADD.Citation20 Once DNA is damaged by drugs or radiation, it can either be repaired or lead to apoptosis/cell death. Caspase 8 is crucial for apoptosis progression; when Caspase 8 was degraded or downregulated, it resulted in immortality of injured cells and induction of various cancers. Many studies reported that knocking out CASP8 in mice resulted in death in the embryonic phase.Citation21, 22 Soung reported that inactivated, mutated CASP8 was related to many cancers, including breast and lung cancers and gastric carcinomas.Citation13 Up- or downregulation of CASP8 was related to drug resistance in cancers, including ovarian and colon cancers.Citation23, 24
Adenocarcinoma is the major histology of NSCLC with approximately 60% of patients presenting with lung adenocarcinoma among NSCLC patients worldwide. A few studies showed that lung adenocarcinoma was related to many genes. Recently, Wu found that the lung adenocarcinoma patients in Xuanwei, China had differentially expressed genes. PAK1, E2F1, CCNE1, EGF, CDC25A, PTTG1, and/or UHRF1 upregulation in lung adenocarcinoma was observed. PIK3R1, RARB, HGF, MAPK11, and SESN1 were downregulated in adenocarcinoma as shown by real-time reverse transcriptase polymerase chain reaction.Citation25 Besides the different gene expressions, various mutations of driver genes were depicted among different racial/ethnic adenocarcinoma patients, and the Asian patients had the highest rate of oncogene drivers in that study (approximately 81%).Citation26 Many genes were related to lung adenocarcinoma according to protein levels or genetic mutations. Gene polymorphisms also correlated with lung adenocarcinoma as reported by many studies. A study reported that XRCC1 Arg194Trp polymorphism was an independent risk factor in female patients with lung adenocarcinoma.Citation27 Our study patients with lung adenocarcinoma were chosen on the basis of the described previously reasons. We found rs3769821 of CASP8 is an independent prognostic factor associated with OS of the lung adenocarcinoma in our study patients. This is the first time that we linked a critical apoptosis genes (CASP8) and lung adenocarcinoma; this connection may provide individualized, targeted therapy in the future.
Targeted therapy for lung cancer patients is being intensely study at present, and many targeted drugs such as EGFR tyrosine kinase inhibitors (TKIs [erlotinib, gefitinib, and afatinib]), are widely used.Citation28 However, the number of patients with driver mutation are limited and consist of only 17% of lung cancer patients with EGFR mutation and 7% of lung adenocarcinoma patients having ALK translocation.Citation29 Most lung cancer patients have no chance for targeted therapy. Platinum-based chemotherapy remains the first-line treatment of most lung cancer patients, and many genes have been reported to correlate with chemotherapy efficacy such as ERCC1 and RRMI genes for platinum and gemcitabine, respectively, and TUBB3 for paclitaxel.Citation30-34 However, the genes' predictive roles for lung cancer patients are still controversial. As biomarkers, these genes have not been tested or verified in other studies or in multicenter clinical trials, possibly because of the instability of the mRNA or protein expression of the biomarkers or unreproducible methods.Citation35, 36 A highly repeatable result could be obtained by detection of the germline or somatic mutations of genes, and this method would be more appropriate for a biomarker in clinical practice. Patients with difference in genotypes of polymorphisms had unequal clinical outcomes, including survival time and toxic effects.Citation18, 37 CASP8 polymorphisms were related to many cancers, including lung cancer and neuroblastoma.Citation38-40 Son et al. found that CASP8 IVS12–19 GG genotype was significantly increased for the risk of small cell carcinoma (SmCC), rather than IVS12–19 AA and IVS12–19 GA genotypes.Citation38 However, this is the first time that rs3769821 and rs1045494 of CASP8 was reported to be associated with lung adenocarcinoma OS in our study, and the patients with AA genotype of rs3769821 had 6.7 months longer OS than patients with GG or AG genotypes (p = 0.007). We analyzed the relation between haplotype of 7 CASP8 tapSNPs and OS in the lung adenocarcinoma. Four haplotypes were acquired using PHASE 2.0, haplotype 1 AGGAAAGA, haplotype 2 AGAACAG, haplotype 3 AAAGAAA, and haplotype 4 GGGGAAA. The haplotype AGGAAAGA was associated with lung adenocarcinoma patients' OS. The patients with zero copies had 6 months longer OS than patients with one or 2 copies of haplotype AGGAAAGA (zero copies, mOS 23.9 months; 1–2 copies, mOS 17.7 months p = 0.016). However, no significant correlation was observed between CASP8 polymorphisms/haplotypes and PFS. Similar results have been seen in treatment studies when the agents used alter immune function A possible reason why CASP8 polymorphisms improved OS but not ORR, DCR, and PFS may be that the role of pharmacogenetics of the treatment drugs altered the metabolism, even the immune function.Citation41 We found that Caspase 8 played crucial role in the immune system. Recently, Phillips reported that Caspase 8 mediated and promoted the bone disease regulating the IL-1β, an important mediator of the inflammatory response.Citation42 Prajwal Gurung found that Caspase 8 activated the Nrpl3 inflammasome priming, which belongs to the innate immune system.Citation43, 44 We searched in the KEGG website (http://www.kegg.jp/) and found that Caspase 8 was involved in the immune system, including Toll-like receptor signaling pathway, NOD-like receptor signaling pathway, and RIG-I-like receptor signaling pathway, which is belongs to the innate immune system. The correlation between Caspase 8 and the immune system may be the reason why rs3769821 is associated with OS, but not for ORR, DCR, PFS in our study. The reason why CASP 8 polymorphisms rs3769821 was associated with OS may can be explained by the protein expression or activation status of Caspase 8 regulated by polymorphisms of CASP8. Rs3769821 was located in promoter of CASP8. Recently, Camp et al. reported that the neutral allele (A) of rs3769821 had higher gene expression than the risk allele as shown by testing luciferase enhancer activity.Citation45 We tried to ultize the public repositories to investigate wheather CASP8 polymorphisms were associated with Caspase 8 expression using eQTL analysis. Using SNPexp, a web tool for calculating correlation between HapMap genotypes and gene expression levels (http://app3.titan.uio.no/biotools/tool.php?app=snpexp), we found that Caspase 8 rs3769821 genotypes (AA/AG/GG) is correlated with Caspase 8 expression in the all populations (p = 0.048, Table S5). Based on the Camp et al. research and the results of public database, we come up with that the reason why patients with AA genotype of rs3769821 had longer OS than the GG or GA genotype may be related with different Caspase 8 protein expression. Many researchers reported rs3769821 of CASP8 was putatively related with development of diseases. For example, Qian reported CASP8 rs3769821 was correlated with the toxicities in advanced NSCLC patients treated with platinum-based chemotherapyCitation18; Qing and coworkers reported that CASP8 rs3769821 was associated with increased risk on (non-Hodgkin lymphoma) NHL overall or its subtypes in 3 populations from the United States of America and AustraliaCitation46
Toxic effects are another crucial clinical factor that influences chemotherapy efficacy. Severe toxic effects are the reason that patients abort the progression of chemotherapy treatment, which affects OS from another perspective. Many researchers reported CASP8 polymorphisms were associated with toxicity.Citation18 In our study, we found results consistent with others; rs7608692 of CASP8 correlated with neutropenia toxicity, and the patients with AA genotype of rs7608692 had minor side effects consisting of neutropenia after platinum-based chemotherapy. In addition, the haplotype AGAACAG of CASP8 was associated with neutropenia toxicity and haplotype GGGGAAA correlated with thrombocytopenia toxicity in our study. Either severe chemotherapy-related neutropenia or thrombocytopenia regularly influences the patients' life quality and even the efficacy of treatment or survival because of serious infection or spontaneous hemorrhage.
Above all, CASP8 is an important apoptosis gene, and a crucial defensive barrier against malignant proliferation and tumorigenesis by triggering the extrinsic apoptosis way. Caspase 8 is a crucial protein for the progression of proliferation and apoptosis. Many researchers reported that CASP8 polymorphisms were a putative biomarker for various cancerssuch as lung and breast cancers.Citation47, 48 Our study first depicted that patients with rs3769821 and rs1045494 AA genotype of CASP8 had longer OS than patients with GG or GA genotype(patients with rs3769821 AA genotype had 6.7 months longer OS), and patients with zero copy haplotype AGGAAAGA had longer OS. The rs7608692 of CASP8 was related to neutropenia toxicity in our study, and the patients with AA genotype had minor neutropenia side effects after platinum-based chemotherapy. However, the toxicity of chemotherapy influences further therapy and even survival time. We concluded that polymorphisms of CASP8 contribute to the prognosis of advanced lung adenocarcinoma patients treated with platinum-based chemotherapy. And we think polymorphisms of CASP8 may be valuable for inducting lung adenocarcinoma patients' treatment in the future. However, this conclusion originated from unselected adenocarcinoma patients separate from EGFR and ALK mutations. It is worth further validation in the future.
Methods
Ethic approval
All procedures performed in our study involving human participants were in accordance with the ethical standards of the Shanghai Changhai Hospital, and Shanghai Pulmonary Hospital, Shanghai Chest Hospital, and Shanghai Zhongshan Hospital research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. We informed all patients who were enrolled in this study and attained their written consent. This article does not contain any studies with animals performed by any of the authors. The experiments in this study were conducted under approved guidelines and regulations.
Study patients
The study cohort enrolled 555 patients who had been diagnosed with advanced adenocarcinoma (stages III or IV) when recruited for this study and had received first-line platinum-based chemotherapy (no prior surgery, radiotherapy, or concurrent chemoradiotherapy) from March 2005 to January 2010. The Eastern Cooperative Oncology Group performance status (ECOG PS) of all the patients were 0–1 points and the chemotherapy side effects on cardiovascular, hepatic, hematologic, and renal functions of the patients were assessed. The association of the CASP8 tagSNPs with the efficacy of platinum-based chemotherapy was reviewed and evaluated. All of the patients had adenocarcinoma as confirmed with by the presence of histological criterion.
Treatment regimen
All 555 patients accepted first-line platinum-based chemotherapy for 2 to 6 cycles. Chemotherapy regimens were classified into 3 main types: 1) DNA-damaging drugs (cisplatin, carboplatin, and gemcitabine); 2) tubulin-targeting agents (cisplatin, carboplatin, navelbine, paclitaxel, and docetaxel); and 3) other combinations (cisplatin and etoposide, bevacizumab). The detailed drug regimens included cisplatin 75 mg/m2 or carboplatin AUC 5 administered on day 1 every 3 weeks in combination with different drugs: 1) gemcitabine 1250 mg/m2 on days 1 and 8 every 3 weeks; 2) tubulin-targeting agents, consisting of navelbine 25 mg/m2 on days 1 and 8 every 3 weeks, paclitaxel 175 mg/m2 on day 1 every 3 weeks, docetaxel 75 mg/m2 on day 1 every 3 weeks; or 3) etoposide or bevacizumab.
Chemotherapy efficacy
The tumor response was based on the Response Evaluation Criteria in Solid Tumors. Short-term responses were estimated after the first 2 cycles of chemotherapy. Overall survival (OS) was an important factor for evaluating the efficacy of chemotherapy treatment, defined from the first day of receiving chemotherapy treatment to the day of death or to the final follow-up. Another important factor reflecting the short-term response of chemotherapy, progression-free survival (PFS), was assessed from the date when the chemotherapy treatment started to objective disease progression or death (whichever occurred first) or the last progression-free follow-up. The disease control rate (DCR) consisted of complete or partial responses (CR and PR, respectively), or stable disease (SD). CR and PR were involved in the objective response rate (ORR).
Chemotherapy toxicity
According to the National Cancer Institute (NCI) Common Terminology Criteria for Adverse Events (CTCAE) version 3.0, chemotherapy toxicities were evaluated twice weekly, and the worst toxic effects records were collected for analysis during the beginning 2 cycles of therapy. Toxicities included vomiting, nausea, neutropenia, thrombocytopenia, and anemia. Severe toxic effects were classified as grades 3 or 4; grade 5 meant death. No grade 5 was observed in this study.
CASP8 tagSNPs and haplotypes information
Seven tagSNPs of CASP8, including rs3769827, rs7608692, rs3769825, rs12990906, rs3754935, rs3769821, and rs1045494 were chosen for study, selected from the CHB data of the phase 2 HapMap SNP database (http://www.hapmap.org/). We extracted genomic DNA from patients' peripheral blood samples using the QIAamp DNA Maxi Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocols for the genotyping purpose. The tagSNPs were genotyped using the iSelect HD Bead-Chip (Illumina, San Diego, CA, USA) with the following quality-control criteria:1) Genotyping call rate > 0.95; 2) MAF > 0.05, and 3) GenCall score > 0.2. The replicates concordance was > 99.9%. The individual haplotype of the 7 tagSNPs of CASP8 frequencies was evaluated using the PHASE 2.0 Program (version 2.0.2) based on the Bayesian algorithm (Table S1).
Statistical analysis
Statistical analysis in our study was done with the SPSS version 20.0 software (SPSS INc. Chicago, IL, USA). The Kaplan-Meier curves were used for illustrating the association between OS or PFS of the adenocarcinoma patients and the tag SNPs of CASP8. We performed univariate and multivariate Cox proportional hazard model analyses to validate the independent significant factors related to the OS or PFS. The R language (Rstudio software) with multiple testing (False discovery rate, FDR) was used for calculating the 7 tagSNPs relationship with patients' OS according to univariate and multivariate cox's rgerssion analysis. The χ2 test was also evaluated for studying the relationship between the significant tagSNPs of CASP8 and the clinical characteristics. The association between tagSNPs/haplotypes of CASP8 and the severe toxicities was calculated with univariate and multivariate logistic analyses. All statistical analyses were defined for 2 sides and the differences of P < 0.05 were considered statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary_Data.zip
Download Zip (4.5 MB)Acknowledgments
This work was funded by National Natural Science Foundation of China (No. 81572269), Science and Technology Commission of Shanghai Municipality (No. 14411966400, No.134119a3400), Foundation of Shanghai Health and Family Planning Commission (No. 201440397, 20134014, XYQ2013115) and Shanghai Jiao Tong University, Med-Engineering Interdisciplinary Research Foundation (No. YG2015MS71).
References
- Zheng R, Zeng H, Zuo T, Zhang S, Qiao Y, Zhou Q, Chen W. Lung cancer incidence and mortality in China, 2011. Thoracic cancer 2016; 7:94-9; PMID:26816543; https://doi.org/10.1111/1759-7714.12286
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66:115-32; PMID:26808342; https://doi.org/10.3322/caac.21338
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin 2014; 64:9-29; PMID:24399786; https://doi.org/10.3322/caac.21208
- Abratt RP, Hart GJ. 10-year update on chemotherapy for non-small cell lung cancer. Ann Oncol 2006; 17 Suppl 5:v33-6; PMID:16807460; https://doi.org/10.1093/annonc/mdj947
- Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR, Cheney RT, Chirieac LR, D'Amico TA, Demmy TL, et al. Non-Small Cell Lung Cancer, Version 6.2015. J Natl Compr Cancer Netw 2015; 13:515-24; PMID:25964637
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, et al. A unified model for apical caspase activation. Mol Cell 2003; 11:529-41; PMID:12620239; https://doi.org/10.1016/S1097-2765(03)00051-0
- Fiandalo MV, Schwarze SR, Kyprianou N. Proteasomal regulation of caspase-8 in cancer cell apoptosis. Apoptosis 2013; 18:766-76; PMID:23456622; https://doi.org/10.1007/s10495-013-0821-y
- Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity 2000; 12:611-20; PMID:10894161; https://doi.org/10.1016/S1074-7613(00)80212-5
- Bhattacharya S, Ghosh MK. Cell death and deubiquitinases: perspectives in cancer. Bio Med Res Int 2014; 2014:435197. PMID:25121098; https://doi.org/10.1155/2014/435197
- McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol 2013; 5:a008656. PMID:23545416; https://doi.org/10.1101/cshperspect.a008656
- Syvanen AC. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat Rev Genet 2001; 2:930-42; PMID:11733746; https://doi.org/10.1038/35103535
- Kim HS, Lee JW, Soung YH, Park WS, Kim SY, Lee JH, Park JY, Cho YG, Kim CJ, Jeong SW, et al. Inactivating mutations of caspase-8 gene in colorectal carcinomas. Gastroenterology 2003; 125:708-15; PMID:12949717; https://doi.org/10.1016/S0016-5085(03)01059-X
- Soung YH, Lee JW, Kim SY, Jang J, Park YG, Park WS, Nam SW, Lee JY, Yoo NJ, Lee SH. CASPASE-8 gene is inactivated by somatic mutations in gastric carcinomas. Cancer Res 2005; 65:815-21; PMID:15705878
- Sun T, Gao Y, Tan W, Ma S, Shi Y, Yao J, Guo Y, Yang M, Zhang X, Zhang Q, et al. A 6-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet 2007; 39:605-13; PMID:17450141; https://doi.org/10.1038/ng2030
- Pittman AM, Broderick P, Sullivan K, Fielding S, Webb E, Penegar S, Tomlinson I, Houlston RS. CASP8 variants D302H and -652 6N ins/del do not influence the risk of colorectal cancer in the United Kingdom population. Br J Cancer 2008; 98:1434-6; PMID:18362937; https://doi.org/10.1038/sj.bjc.6604314
- MacPherson G, Healey CS, Teare MD, Balasubramanian SP, Reed MW, Pharoah PD, Ponder BA, Meuth M, Bhattacharyya NP, Cox A. Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J Natl Cancer Institute 2004; 96:1866-9; PMID:15601643; https://doi.org/10.1093/jnci/dji001
- Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, Scollen S, Baynes C, Ponder BA, Chanock S, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet 2007; 39:352-8; PMID:17293864; https://doi.org/10.1038/ng1981
- Qian J, Qu HQ, Yang L, Yin M, Wang Q, Gu S, Wu Q, Zhao X, Wu W, Wu J, et al. Association between CASP8 and CASP10 polymorphisms and toxicity outcomes with platinum-based chemotherapy in Chinese patients with non-small cell lung cancer. Oncologist 2012; 17:1551-61; PMID:22843554; https://doi.org/10.1634/theoncologist.2011-0419
- Chang DW, Xing Z, Capacio VL, Peter ME, Yang X. Interdimer processing mechanism of procaspase-8 activation. EMBO J 2003; 22:4132-42; PMID:12912912; https://doi.org/10.1093/emboj/cdg414
- Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat Immunol 2009; 10:348-55; PMID:19295631; https://doi.org/10.1038/ni.1714
- Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature 2011; 471:373-6; PMID:21368761; https://doi.org/10.1038/nature09878
- Oberst A, Green DR. It cuts both ways: reconciling the dual roles of caspase 8 in cell death and survival. Nat Rev Mol Cell Biol 2011; 12:757-63; PMID:22016059; https://doi.org/10.1038/nrm3214
- Duiker EW, Meijer A, van der Bilt AR, Meersma GJ, Kooi N, van der Zee AG, de Vries EG, de Jong S. Drug-induced caspase 8 upregulation sensitises cisplatin-resistant ovarian carcinoma cells to rhTRAIL-induced apoptosis. Br J Cancer 2011; 104:1278-87; PMID:21487429; https://doi.org/10.1038/bjc.2011.84
- Van Geelen CM, Pennarun B, Ek WB, Le PT, Spierings DC, De Vries EG, De Jong S. Downregulation of active caspase 8 as a mechanism of acquired TRAIL resistance in mismatch repair-proficient colon carcinoma cell lines. Int J Oncol 2010; 37:1031-41; PMID:20811726; https://doi.org/10.3892/ijo_00000755
- Wu H, Meng S, Xu Q, Wang X, Wang J, Gong R, Song Y, Duan Y, Zhang Y. Gene expression profiling of lung adenocarcinoma in Xuanwei, China. Eur J Cancer Prev 2015; 25(6):508-17; PMID:26695082; https://doi.org/10.1097/CEJ.0000000000000214
- Steuer CE, Behera M, Berry L, Kim S, Rossi M, Sica G, Owonikoko TK, Johnson BE, Kris MG, Bunn PA, et al. Role of race in oncogenic driver prevalence and outcomes in lung adenocarcinoma: Results from the Lung Cancer Mutation Consortium. Cancer 2015; 122:766-72; PMID:26695526; https://doi.org/10.1002/cncr.29812.
- Catana A, Pop M, Hincu BD, Pop IV, Petrisor FM, Porojan MD, Popp RA. The XRCC1 Arg194Trp polymorphism is significantly associated with lung adenocarcinoma: a case-control study in an Eastern European Caucasian group. OncoTargets Ther 2015; 8:3533-8; PMID:26664136; https://doi.org/10.2147/OTT.S92361
- Somasundaram A, Socinski MA, Burns TF. Personalized treatment of EGFR mutant and ALK-positive patients in NSCLC. Exp Opin Pharmacother 2014; 15:2693-708; PMID:25381900; https://doi.org/10.1517/14656566.2014.971013
- Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson SL, Su PF, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. Jama 2014; 311:1998-2006; PMID:24846037; https://doi.org/10.1001/jama.2014.3741
- Lord RV, Brabender J, Gandara D, Alberola V, Camps C, Domine M, Cardenal F, Sanchez JM, Gumerlock PH, Taron M, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res 2002; 8:2286-91; PMID:12114432
- Tantraworasin A, Saeteng S, Lertprasertsuke N, Arayawudhikul N, Kasemsarn C, Patumanond J. The prognostic value of ERCC1 and RRM1 gene expression in completely resected non-small cell lung cancer: tumor recurrence and overall survival. Cancer Manag Res 2013; 5:327-36; PMID:24124391; https://doi.org/10.2147/CMAR.S52073
- Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med 2007; 356:800-8; PMID:17314339; https://doi.org/10.1056/NEJMoa065411
- Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper HH, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006; 355:983-91; PMID:16957145; https://doi.org/10.1056/NEJMoa060570
- Zhang HL, Ruan L, Zheng LM, Whyte D, Tzeng CM, Zhou XW. Association between class III beta-tubulin expression and response to paclitaxel/vinorebine-based chemotherapy for non-small cell lung cancer: a meta-analysis. Lung cancer (Amsterdam, Netherlands) 2012; 77:9-15; PMID:22306125; https://doi.org/10.1016/j.lungcan.2012.01.005
- Yamashita F, Azuma K, Yoshida T, Yamada K, Kawahara A, Hattori S, Takeoka H, Zaizen Y, Kawayama T, Kage M, et al. Prognostic value of EGFR mutation and ERCC1 in patients with non-small cell lung cancer undergoing platinum-based chemotherapy. PloS one 2013; 8:e71356. PMID:23940741; https://doi.org/10.1371/journal.pone.0071356
- Bepler G, Williams C, Schell MJ, Chen W, Zheng Z, Simon G, Gadgeel S, Zhao X, Schreiber F, Brahmer J, et al. Randomized international phase III trial of ERCC1 and RRM1 expression-based chemotherapy versus gemcitabine/carboplatin in advanced non-small-cell lung cancer. J Clin Oncol 2013; 31:2404-12; PMID:23690416; https://doi.org/10.1200/JCO.2012.46.9783
- Liu D, Wu C, Jiao Y, Hou L, Lu D, Zheng H, Chen C, Qian J, Fei K, Su B. WEE1 kinase polymorphism as a predictive biomarker for efficacy of platinum-gemcitabine doublet chemotherapy in advanced non-small cell lung cancer patients. Scientific Rep 2015; 5:11114. PMID:26057002; https://doi.org/10.1038/srep11114
- Son JW, Kang HK, Chae MH, Choi JE, Park JM, Lee WK, Kim CH, Kim DS, Kam S, Kang YM, et al. Polymorphisms in the caspase-8 gene and the risk of lung cancer. Cancer Genet cytogenetics 2006; 169:121-7; PMID:16938569; https://doi.org/10.1016/j.cancergencyto.2006.04.001
- Hart K, Landvik NE, Lind H, Skaug V, Haugen A, Zienolddiny S. A combination of functional polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk of non-small cell lung cancer. Lung cancer (Amsterdam, Netherlands) 2011; 71:123-9; PMID:20471133; https://doi.org/10.1016/j.lungcan.2010.04.016
- Rihani A, De Wilde B, Zeka F, Laureys G, Francotte N, Tonini GP, Coco S, Versteeg R, Noguera R, Schulte JH, et al. CASP8 SNP D302H (rs1045485) is associated with worse survival in MYCN-amplified neuroblastoma patients. PloS one 2014; 9:e114696. PMID:25502557; https://doi.org/10.1371/journal.pone.0114696
- Hattinger CM, Serra M. Role of pharmacogenetics of drug-metabolizing enzymes in treating osteosarcoma. Exp Opin Drug Metab Toxicol 2015; 11:1449-63; PMID:26095223; https://doi.org/10.1517/17425255.2015.1060220
- Phillips FC, Gurung P, Kanneganti TD. Microbiota and caspase-1/caspase-8 regulate IL-1beta-mediated bone disease. Gut Microbes 2016; 7:1-8; PMID:27148834; https://doi.org/10.1080/19490976.2016.1182289
- Gurung P, Anand PK, Malireddi RK, Vande Walle L, Van Opdenbosch N, Dillon CP, Weinlich R, Green DR, Lamkanfi M, Kanneganti TD. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J Immunol 2014; 192:1835-46; PMID:24453255; https://doi.org/10.4049/jimmunol.1302839
- Gurung P, Li B, Subbarao Malireddi RK, Lamkanfi M, Geiger TL, Kanneganti TD. Chronic TLR Stimulation Controls NLRP3 Inflammasome Activation through IL-10 Mediated Regulation of NLRP3 Expression and Caspase-8 Activation. Sci Rep 2015; 5:14488. PMID:26412089; https://doi.org/10.1038/srep14488
- Camp NJ, Lin WY, Bigelow A, Burghel GJ, Mosbruger TL, Parry MA, Waller RG, Rigas SH, Tai PY, Berrett K, et al. Discordant Haplotype Sequencing Identifies Functional Variants at the 2q33 Breast Cancer Risk Locus. Cancer Res 2016; 76:1916-25; PMID:26795348; https://doi.org/10.1158/0008-5472.CAN-15-1629
- Lan Q, Morton LM, Armstrong B, Hartge P, Menashe I, Zheng T, Purdue MP, Cerhan JR, Zhang Y, Grulich A, et al. Genetic variation in caspase genes and risk of non-Hodgkin lymphoma: a pooled analysis of 3 population-based case-control studies. Blood 2009; 114:264-7; PMID:19414860; https://doi.org/10.1182/blood-2009-01-198697
- Zhang YJ, Zhong XP, Chen Y, Liu SR, Wu G, Liu YF. Association between CASP-8 gene polymorphisms and cancer risk in some Asian population based on a HuGE review and meta-analysis. Genet Mol Res 2013; 12:6466-76; PMID:23479148; https://doi.org/10.4238/2013.February.28.3
- George GP, Mandal RK, Kesarwani P, Sankhwar SN, Mandhani A, Mittal RD. Polymorphisms and haplotypes in caspases 8 and 9 genes and risk for prostate cancer: a case-control study in cohort of North India. Urologic Oncol 2012; 30:781-9; PMID:21396853; https://doi.org/10.1016/j.urolonc.2010.08.027
