ABSTRACT
Background: Colon cancer-associated transcript-1 (CCAT1) has been demonstrated to act as an oncogene and promote chemoresistance in several cancers. However, little is known about the underlying mechanism of CCAT1 in cisplatin (DDP) resistance of non-small-cell lung cancer (NSCLC) cells.
Methods: qRT-PCR was performed to detect the expression levels of CCAT, miR-130a-3p, or sex-determining region Y-box 4 (SOX4) mRNA. Luciferase reporter assay, RNA immunoprecipitation (RIP), and qRT-PCR analysis were carried out to explore the potential targets of CCAT1 or miR-130a-3p. Effect of CCAT1, miR-130a-3p, or SOX4 on IC50 value of DDP and ATP binding cassette subfamily G member 2 (ABCG2) level in NSCLC cells were determined by cell counting kits-8 (CCK-8) assay and western blot, respectively.
Results: CCAT1 and SOX4 were up-regulated, and miR-130a-3p was down-regulated in DDP-resistant NSCLC cells compared with their parental NSCLC cells. CCAT1 directly interacted with miR-130a-3p and negatively regulated miR-130a-3p expression. CCAT1 contributed to DDP resistance of A549/DDP cells by down-regulating miR-130a-3p. miR-130a-3p was found to directly target SOX4 to suppress its expression. SOX4 knockdown reversed miR-130a-3p-inhibition-induced increase of DDP resistance and ABCG2 expression in NSCLC cells. Exogenous expression of SOX4 abrogated CCAT1-knockdown-mediated decrease of DDP resistance and ABCG2 expression in DDP-resistant NSCLC cells.
Conclusion: CCAT1/miR-130a-3p axis enhanced DDP resistance of NSCLC cells by targeting SOX4, providing potential targets to overcome DDP resistance and improve efficacy of chemotherapy for patients with NSCLC.
KEYWORDS:
Introduction
Lung cancer is still the most common prevalent malignancy and remains the primary cause of global cancer-associated morbidity and mortality for both men and women,Citation1 approximately 85% of which is non-small cell lung cancer (NSCLC). The prognosis for patients with advanced NSCLC remains poor, with a five-year survival rate of less than 15%.Citation1 Platinum-based chemotherapy, particularly cisplatin (DDP)-based chemotherapy, has been the first-line adjuvant treatment for NSCLC in the clinic after surgical resection.Citation2 Unfortunately, the clinical therapeutic efficacy of DDP is significantly reduced due to the emergence of resistance to DDP-based therapy.Citation3 Therefore, a better understanding of the mechanisms of DDP resistance in NSCLC would be important to provide useful methods to reverse DDP resistance and improve the efficacy of NSCLC therapeutics.
Long non-coding RNAs (LncRNAs) are defined as a class of non-protein coding transcripts longer than 200 nt. So far, lncRNAs are emerging as crucial regulators of gene expression at different levels through chromatin modification, transcription (or post-transcriptional) and translational regulation. LncRNAs have widely been reported to contribute to malignant physiological or pathological processes such as carcinogenesis, invasion, metastasis, and drug resistance.Citation4
Recently, a novel regulatory circuit, supported by numerous studies, has been proposed that lncRNAs interact with miRNAs, functioning as competing endogenous RNAs (ceRNAs), namely, miRNA sponges or antagomirs, to regulate the expression and activity of miRNAs, thereby leading to the derepression of miRNA targets.Citation5,6 For example, lncRNA tumor suppressor candidate 7 (TUSC7) suppressed epithelial-to-mesenchymal transformation (EMT) through the TUSC7-miR-10a- EphA4 axis in hepatocellular carcinoma (HCC).Citation7 LncRNA nuclear enriched abundant transcript 1 (NEAT1) functioned as a molecular sponge for miR-449b-5p, triggering the upregulation of oncogene c-Met and facilitating the development of glioma.Citation8
Colon cancer-associated transcript-1 (CCAT1), a 2628-bp lncRNA that located at chromosome 8q24.21, was initially discovered to be highly-expressed in colon cancer.Citation9 Several lines of evidence has demonstrated that CCAT1 was aberrantly expressed and exhibited oncogenic roles in various types of tumors, such as gastric cancer,Citation9 HCCCitation10 and lung cancer.Citation11 More importantly, CCAT1 was revealed to act as an oncogene and promote chemoresistance in docetaxel-resistant lung cancer cells.Citation11 However, little is known about the underlying mechanism of CCAT1 in DDP resistance of NSCLC cells.
In this study, we identified the possibility that CCAT1 functioned as a ceRNA for endogenous miRNAs by starBase software. The results showed that CCAT1 effectively acted as a miRNA sponge for miR-130a-3p to enhance sex-determining region Y-box 4 (SOX4) expression. Moreover, CCAT1/miR-130a-3p axis contributed to DDP resistance of NSCLC cells by targeting SOX4.
Materials and methods
Cell lines and culture
Normal human lung bronchial epithelial BEAS-2B cells, parental NSCLC A549 cells, H1299, and DDP-resistant NSCLC A549/DDP cells were purchased from the Cell bank of Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in Dulbecco's modified Eagles medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified air with 5% CO2. To establish DDP-resistant H1299 cell line (H1299/DDP), H1299 cells were initially treated with 0.6 μM of DDP (Sigma, St. Louis, MO, USA), and then were exposed to gradually increasing concentrations of DDP in a stepwise manner, until the final concentration of DDP is 1 µg/ml. Moreover, A549/DDP and H1299/DDP cells was maintained in culture medium additionally containing 1 µg/ml DDP (Sigma) to maintain the drug-resistant phenotype prior to further experiments.
Cell transfection
miR-130a-3p mimic (miR mimic), mimic control (mimic Con), miR-130a-3p inhibitor (miR inhibitor), inhibitor control (inhibitor Con), siRNA against CCAT1 (si-CCAT1), siRNA against SOX4 (si-SOX4) and siRNA control (siRNA Con) were all purchased from GenePharma (Shanghai, China). For overexpressing CCAT1 or SOX4, the sequence of CCAT1 or SOX4 was amplified by PCR and then cloned into eukaryotic expression vector pcDNA3.1 (Invitrogen), forming pcDNA-CCAT1 (CCAT1) or pcDNA-SOX4 (SOX4). All oligonucleotides and plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen). At 48 h posttransfection, transfected cells were harvested for next analysis.
Quantitative real-time PCR (qRT-PCR) assays
Total RNA was isolated using TRIzol reagent (Invitrogen) and quantified by the Nanodrop 2000 (Thermo Scientific, Waltham, MA, USA). The first strand of cDNA was synthesized from approximately 1 μg of extracted total RNA sample with a PrimeScript™ RT-PCR Kit (Takara, Dalian, China). Quantification of miRNA, or lncRNA and mRNA was performed using TaqMan assay kits (Applied Biosystems, Forster City, USA) and SYBR® Premix Ex Taq™ II (Takara) on the ABI 7500 real-time PCR system (Applied Biosystems), respectively. The relative expressions of miR-130a-3p, CCAT1 and SOX4 mRNA were calculated using the 2−ΔΔCT method. U6 small nuclear (snRNA) was used as an internal control for miR-130–3p and GAPDH for CCAT1 and SOX4 mRNA. The specific primer sequences were designed as follows: CCAT1, forward primer, 5′-CATTGGGAAAGGTGCCGAGA-3′, reverse primer, 5′-ACGCTTAGCCATACAGAGCC-3′; miR-130a-3p, forward primer, 5′-TTCACATTGTGCTACTGTCTGC-3′, reverse primer, 5′-GCTCTGACTTTATTGCACTACT-3′; SOX4, forward primer, 5′-GGTCTCTAGTTCTTGCACGCTC-3′, reverse primer, 5′-CGGAATCGGCACTAAGGAG-3′; U6 forward primer, 5′-CTCGCTTCGGCAGCACA-3′, reverse primer, 5′-AACGCTTCACGAATTTGCGT-3′; GAPDH, forward primer, 5′-TATGATGATATCAAGAGGGTAGT-3′, reverse primer, 5′-TGTATCCAAACTCATTGTCATAC-3′.
Western blot
Cells were lysed on ice with cold radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) including protease inhibitors and phosphatase inhibitors (Sigma). Same amounts of protein (50 μg) was subjected to 10% sodium dedecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to PVDF membrane (Millipore, Billerica, MA, USA) for band separation. Subsequently, membranes were blotted with 10% skimmed milk in Tris-buffered saline with Tween-20 (TBS-T), followed by probed with the antibodies against SOX4 (1:2000 dilution; Cell Signaling Technology, Danvers, MA, USA), ABCG2 (1:2000 dilution, Cell Signaling Technology) and β-actin (1:1000 dilution; Cell Signaling Technology) overnight at 4°C. After washing extensively with 0.1% PBS on the following day, membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (1:2000 dilution; Cell Signaling Technology) and visualized by ECL Western Blotting Detection Reagent (BioRad, Hercules, CA, USA).
Luciferase reporter assay
The wild-type CCAT1 sequence, 3′UTR sequences of SOX4 containing the predicted binding sites for miR-130a-3p, or mutated CCAT1 sequence, 3′UTR sequences of SOX4 without miR-130a-3p binding sites were inserted into pGL3 promoter vector (Promega, Madison, WI, USA) and named as pGL3-CCAT1-WT, pGL3-SOX4 3′UTR-WT, pGL3-CCAT1-MUT, and pGL3-SOX4 3′UTR-MUT, respectively. A549/DDP cells were seeded in 96-well plates at 1 × 104 cells per well and then cotransfected with constructed recombinant luciferase vectors and miR mimic or mimic Con using Lipofectamine 2000 (Invitrogen). Luciferase activity was measured at 48 h after transfection with the Dual-Luciferase Reporter Assay System (Promega).
IC50 determination
The transfected cells in the logarithmic phase of growth were seeded in triplicate into 96-well plates at a density of 3000 cells per well. After incubating for 24 h, cells were treated with the indicated concentrations of DDP (2.5, 5, 10, 20, 40, 80 μg/ml) for 48 h. Subsequently, 10 µl of cell counting kits-8 (CCK-8) solution was added to each well, followed by incubation for 1 h at 37°C in 5% CO2. The absorbance at 450 nm was recorded using a microplate reader (BioTek, Winooski, VT, USA). IC50 were determined as the concentration of drug at which DDP produced 50% growth inhibition, with higher IC50 values indicating higher chemoresistance potential.
RNA immunoprecipitation
RNA immunoprecipitation (RIP) assays were carried out with EZ-Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Bedford, MA, USA) following the manufacturer's protocol. Briefly, A549/DDP cells at 80–90% confluency were digested, harvested and lyzed in RIP lysis buffer. Then 100 μl of whole cell lysate was co-incubated with RIP buffer containing magnetic beads conjugated with human antibody against Argonaute2 (Ago2) (Abcam, Cambridge, MA, USA) or normal mouse IgG (Millipore) for 6 h at 4°C. The beads were treated with Proteinase K buffer to digest the protein and then target RNA was extracted. Co-precipitated RNAs were finally subjected to qRT-PCR analysis for further study.
Statistical analysis
All statistical analyses were performed using Graph Prism 5.0 software (GraphPad Prism, San Diego, CA). The data were shown as mean ± standard deviation (SD). Student's t test or one-way ANOVA was used to analyze the differences between two or more groups. The results were considered statistically significant when P < 0.05.
Results
Expression levels of CCAT1, miR-130a-3p, and SOX4 in NSCLC parental cells and DDP-resistant cells
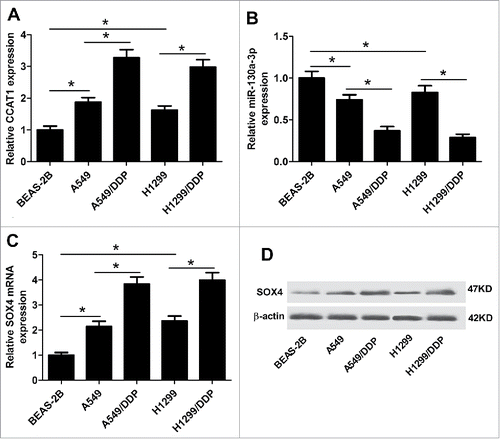
The expression levels of CCAT1, miR-130a-3p, and SOX4 in normal human lung bronchial epithelial cells BEAS-2B, parental NSCLC cells (A549 and H1299) and DDP-resistant NSCLC cells (A549/DDP and H1299/DDP) were evaluated by qRT-PCR. As shown in and , CCAT1 was significantly upregulated, while miR-130a-3p was markedly downregulated in both A549 and H1299 cells compared with BEAS-2B cells. SOX4 expression at mRNA and protein level in A549 and H1299 cells was dramatically higher than BEAS-2B cells ( and ). More interestingly, higher expressions of CCAT1 and SOX4, and lower expression of miR-130a-3p were also observed in A549/DDP and H1299/DDP cells compared with matched A549 and H1299 cells, suggesting that CCAT1, miR-130a-3p, and SOX4 may be implicated in the development of DDP resistance of NSCLC cells.
Figure 1. Expression levels of CCAT1, miR-130a-3p, and SOX4 in NSCLC parental and DDP-resistant cells. qRT-PCR was performed to detect the expression levels of CCAT1 (A), miR-130a-3p (B), and SOX4 mRNA (C) in normal human lung bronchial epithelial BEAS-2B cells, parental NSCLC A549 and H1299 cells, and DDP-resistant A549/DDP and H1299/DDP cells. (D) SOX4 protein expression was measured by western blot in BEAS-2B, A549, H1299, A549/DDP and H1299/DDP cells. *P < 0.05.
CCAT1 enhanced DDP resistance of NSCLC cells
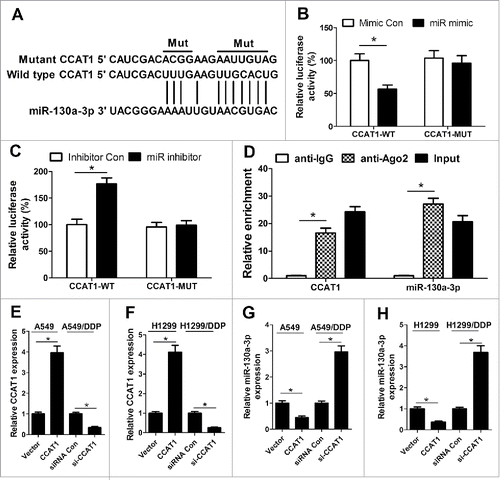
Accumulating evidence has indicated that numerous lncRNAs contain motif with complementary sequence to miRNAs and have an inhibitory effect on the expression and activity of miRNAs. Therefore, we predicted the potential miRNAs which could interact with CCAT1 by the online software starBase v2.0. By searching this database, we discovered that CCAT1 contained potential binding sites complementary to miR-130a-3p, as displayed in . To confirm the direct binding between CCAT1 and miR-130a-3p, luciferase reporter assay was performed. The results demonstrated that cotransfection with pGL3-CCAT1-WT and miR mimic resulted in an obvious suppression of luciferase activity () while cotransfection with pGL3-CCAT1-WT and miR inhibitor led to a marked increase of luciferase activity in A549/DDP cells (), but the luciferase activity in A549/DDP cells cotransfected with pGL3-CCAT1-MUT and miR mimic or miR inhibitor was not significantly changed, suggesting that CCAT1 could interact with miR-130a-3p. Furthermore, miRNAs are known to exert their gene silencing functions through forming RNA-induced silencing complex (RISC) containing Ago2.Citation12 To explore the endogenetic mutual effect between CCAT1 and miR-130a-3p, RIP assay was performed on A549/DDP cell extracts using antibody against Ago2. RNA levels in immunoprecipitates were examined by qRT-PCR. As expected, CCAT1 and miR-130a-3p were both significantly enriched in the Ago2 pellets relative to control IgG immunoprecipitates (), suggesting that CCAT1 and miR-130a-3p might be in the same RISC complex. To investigate the authentic effect of CCAT1 on the expression of miR-130a-3p, qRT-PCR was conducted to determine the expression of miR-130a-3p in A549, H1299, A549/DDP and H1299/DDP cells transfected with pcDAN-CCAT1, si-CCAT1 or respective controls. The transfection efficiency was verified by qRT-PCR and pcDAN-CCAT1 transfection significantly upregulated CCAT1 level in A549 and H1299 cells while si-CCAT1 treatment dramatically repressed CCAT1 expression in A549/DDP and H1299/DDP cells ( and ). As illustrated in and , the expression of miR-130a-3p was strikingly lower in pcDAN-CCAT1-transfected A549 and H1299 cells but remarkably higher in si-CCAT1-transfected A549/DDP and H1299/DDP cells. Collectively, these results revealed that CCAT1 directly targeted miR-130a-3p to suppress its expression.
Figure 2. miR-130a-3p is a direct target of CCAT1. (A) The predicted wild-type or mutated binding sites between CCAT1 and miR-130a-3p. (B and C) Luciferase reporter assay was performed to measure luciferase activity in A549/DDP cells cotransfected with pGL3-CCAT1-WT or pGL3-CCAT1-MUT and miR mimic, miR inhibitor, miR Con or inhibitor Con. (D) RIP assay was performed on A549/DDP cell extracts using antibody against Ago2. CCAT1 and miR-130–3p levels were evaluated by qRT-PCR. A549 and H1299 cells were were transfected with CCAT1 or vector, and A549/DDP and H1299/DDP cells were transfected with si-CCAT1 or siRNA Con, then qRT-PCR was carried out to detect the expression levels of CCAT1 (E and F) and miR-130a-3p (G and H). *P < 0.05.
CCAT1 contributed to DDP resistance of NSCLC cells by downregulating miR-130a-3p
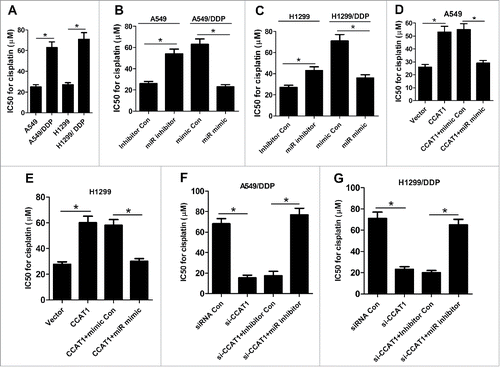
Since a previous study reported that CCAT1 was involved in the development of docetaxel resistance in lung adenocarcinoma cellsCitation11 we further explored the effect of CCAT1 on DDP resistance in NSCLC cells. IC50 values after DDP treatment have always been used to determine DDP resistance.Citation13,14 After A549, H1299, A549/DDP and H1299/DDP cells were treated with the indicated concentrations of DDP (2.5, 5, 10, 20, 40, 80 μg/ml) for 48 h, IC50 value for DDP was calculated with CCK-8 assay. The results showed that IC50 value for DDP was significantly higher in A549/DDP and H1299/DDP cells than A549 and H1299 cells (), respectively. In addition, miR-130a-3p inhibitor notably increased IC50 of DDP in A549 and H1299 cells relative to inhibitor Con group, while ectopic expression of miR-130a-3p greatly decreased IC50 of DDP in A549/DDP and H1299/DDP cells compared with mimic Con group ( and ), as demonstrated by CCK-8 assay. Moreover, CCAT1 overexpression significantly enhanced DDP resistance in A549 () and H1299 () cells, while exogenous expression of miR-130a-3p markedly abolished the improvement of DDP resistance induced by CCAT1 overexpression. Oppositely, CCAT1 knockdown remarkably reduced DDP resistance in A549/DDP () and H1299/DDP () cells, whereas miR-130a-3p inhibition effectively reversed si-CCAT1-induced decrease of DDP resistance. Taken together, these data demonstrated that CCAT1 promoted DDP resistance of NSCLC cells by downregulating miR-130a-3p.
Figure 3. CCAT1 enhanced DDP resistance of NSCLC cells by inhibiting miR-130a-3p. (A) IC50 value for DDP was calculated by CCK-8 assay in A549, H1299, A549/DDP and H1299/DDP cells. (B and C) DDP IC50 value was determined by CCK-8 assay in A549 and H1299 cells transfected with miR inhibitor or inhibitor Con, as well as in A549/DDP and H1299/DDP cells introduced with miR mimic or mimic Con. DDP IC50 value was detected by CCK-8 assay in A549 (D) and H1299 (E) cells transfected with CCAT1, or in combined with miR mimic. DDP IC50 value was measured by CCK-8 assay in A549/DDP (F) and H1299/DDP (G) cells transfected with si-CCAT1, or along with miR inhibitor. *P < 0.05.
SOX4 was a functional target of miR-130a-3p
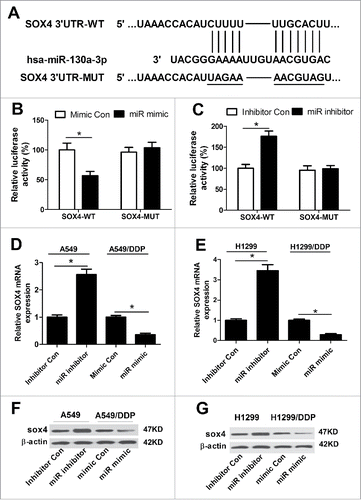
Based on the above results, we intended to explore the potential targets of miR-130a–3p by Targetscan (http://www.targetscan.org) and miRanda (http://www.microrna.org) algorithms. As a result, SOX4, a member of the SOX family of transcription factors, was predicted to be a potential target of miR-130a-3p and the recognition sequence between them were shown in . Next, luciferase reporter assay with luciferase vectors containing the predicted miR-130a-3p binding sites on the 3′UTR of SOX4 was performed to verify the prediction. Luciferase reporter assay showed that enforced expression of miR-130a-3p significantly weakened luciferase activity and miR-130a-3p inhibition conspicuously promoted luciferase activity of pGL3-SOX4 3′UTR-WT in A549/DDP cells ( and C). However, no obvious change was observed in the luciferase activity of pGL3-SOX4 3′UTR-MUT after any transfection ( and ). Then, qRT-PCR and western blot were performed to investigate the effect of miR-130a-3p expression change on mRNA and protein levels of SOX4. As shown in and , miR-130a-3p inhibition dramatically increased the expression of SOX4 at both mRNA and protein levels in A549 ( and ) and H1299 ( and ) cells, while miR-130a-3p overexpression significantly decreased the mRNA and protein levels of SOX4 in A549/DDP ( and ) and H1299/DDP ( and ) cells. Together, we concluded that SOX4 was a direct target of miR-130a-3p.
Figure 4. miR-130a–3p targeted SOX4 to modulate its expression. (A) The predicted miR-130a-3p binding sites and corresponding mutant sites in the 3′UTR of SOX4. (B and C) After A549/DDP cells were cotransfected with pGL3-SOX4 3′UTR-WT or pGL3-SOX4 3′UTR-MUT and miR mimic, miR inhibitor or matched controls, luciferase reporter assay was carried out to determine luciferase activity. Expressions of SOX4 at mRNA and protein levels were examined by qRT-PCR and western blot in A549 (D and F) and H1299 (E and G) cells transfected with miR inhibitor or inhibitor Con, as well as in A549/DDP (D and F) and H1299/DDP (E and G) transfected with miR mimic or mimic Con. *P < 0.05.
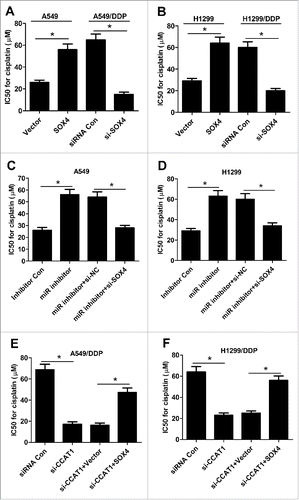
CCAT1/miR-130a-3p axis increased DDP resistance of NSCLC cells by targeting SOX4
The effect of SOX4 on IC50 value of DDP was assessed by transfecting with SOX4 or Vector into A549 and H1299 and introducing with siRNA Con or si-SOX4 into A549/DDP and H1299/DDP cells. As shown in and B, SOX4 overexpression significantly increased DDP resistance in A549 and H1299 cells, while SOX4 knockdown markedly decreased DDP resistance in A549/DDP and H1299/DDP cells, as revealed by CCK-8 assay. To investigate whether CCAT1/miR-130a-3p axis mediated DDP resistance of NSCLC cells by targeting SOX4, A549 and H1299 cells were transfected with miR inhibitor, or miR inhititor + si-SOX4, while A549/DDP and H1299/DDP cells were introduced with si-CCAT1 or si-CCAT1 + SOX4. The results showed that down-regulation of SOX4 notably overturned miR-130a-3p-inhibitor-induced increase in DDP resistance in A549 () and H1299 () cells. Similarly, upregulation of SOX4 remarkably attenuated CCAT1-knockdown-mediated reduction in DDP resistance in A549/DDP () and H1299/DDP () cells. Collectively, these data uncovered that CCAT1/miR-130a-3p axis enhanced DDP resistance of NSCLC cells by regulating SOX4.
Figure 5. CCAT1/miR-130a-3p axis increased DDP resistance of NSCLC cells by targeting SOX4. (A and B) IC50 value of DDP was measured by CCK-8 assay in A549 and H1299 cells transfected with SOX4 or Vector, as well as in A549/DDP and H1299/DDP cells introduced with si-SOX4 or siRNA Con. (C and D) IC50 value for DDP was evaluated by CCK-8 assay in A549 and H1299 cells transfected with miR inhibitor, or along with si-SOX4. (E and F) IC50 value of DDP was determined by CCK-8 assay in A549/DDP and H1299/DDP cells transfected with si-CCAT1, or along with SOX4. *P < 0.05.
CCAT1/miR-130a-3p axis regulated ABCG2 by targeting SOX4
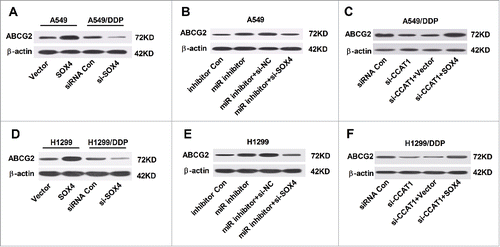
It is well known that chemoresistance of cancer is frequently mediated by overexpression of ATP-binding cassette (ABC) transporter proteins, among which ATP binding cassette subfamily G member 2 (ABCG2) is considered as the main contributing factor to multidrug resistance.15 We further investigated the effect of interaction of SOX4 and CCAT1 or miR-130a-3p on the protein level of ABCG2 in A549, A549/DDP, H1299 and H1299/DDP cells. Western blot analysis showed that the protein level of ABCG2 was exceptionally higher in SOX4-overexpressing A549 cells and H1299 cells, while AGCG2 expression was apparently lowered in si-SOX4-treansfected A549/DDP and H1299/DDP ( and ). Furthermore, inhibition of miR-130a-3p substantially enhanced the protein level of ABCG2 in A549 and H1299 cells, while knockdown of SOX4 effectively abrogated the promotive influence of miR-130a-3p inhibitor on ABCG2 level ( and ). Also, upregulation of SOX4 evidently reversed CCAT1-knockdown-induced decrease in ABCG2 expression in A549/DDP and H1299/DDP ( and ). Taken together, these results demonstrated that CCAT1/miR-130a-3p axis modulated ABCG2 expression by targeting SOX4 in NSCLC cells.
Figure 6. CCAT1/miR-130a-3p axis improved ABCG2 expression by targeting SOX4. (A and D) Protein level of ABCG2 in A549 and H1299 cells transfected with SOX4 or Vector, as well as in A549/DDP and H1299/DDP cells introduced with si-SOX4 or siRNA Con. (B and E) Protein level of ABCG2 in A549 and H1299 cells transfected with miR inhibitor, or in combined with si-SOX4. (C and F) Protein level of ABCG2 in A549/DDP cells transfected with si-CCAT1, or in combined with SOX4.
Discussion
DDP is a commonly used chemotherapy drug against NSCLC, but DDP resistance among NSCLC patients remains a major obstacle to successful chemotherapy.Citation16 Increasing evidence has shown that aberrant expression of lncRNAs participates in chemoresistance of cancers and provides new insights into the biology of tumor progression.Citation17,18 Elucidating the underlying molecular mechanism by which lncRNAs function would contribute to the development of lncRNA-mediated therapeutic approaches against tumors.
It has been proposed that lncRNAs act as ceRNAs or miRNA sponges to abolish the endogenous suppressive effective of these miRNAs on their bona fide targets by interfering with the miRNA pathway. Moreover, these ceRNAs are suggested to be associated with many physiological and pathological processes of tumors and any perturbation of the balance between ceRNAs and miRNAs would affect tumorigenesis.Citation19,20 CCAT1 was identified as an oncogene in various types of tumors, such as HCC, gallbladder cancer and melanoma.Citation21Citation–23 In this study, we focused on the mechanism of CCAT1-mediated DDP chemoresistance in NSCLC cells. The results showed that CCAT1 was upregulated in DDP-resistant NSCLC cells. CCAT1 overexpression increased DDP resistance in NSCLC cells, while CCAT1 knockdown decreased DDP resistance in DDP-resistant NSCLC cells. Consistent with our results, CCAT1 was upregulated in docetaxel-resistant lung adenocarcinoma cells, and forced expression of CCAT1 facilitated the acquisition of chemoresistance by promoting cell proliferation and migration.Citation11 Also, it was reported that upregulation of CCAT1 led to significant enhancement of paclitaxel resistance in nasopharyngeal cancer cells, coincident with our finding.Citation24 Previous studies suggested the involvement of miR-130a in drug resistance in different cancers.Citation25,26 Our study found that miR-130a-3p was down-regulated in DDP-resistant NSCLC cells. Moreover, CCAT1 directly interacted with miR-130a–3p and negatively regulated its expression, functioning as a molecular sponge of miR-130a-3p. Function analysis further revealed that miR-130a-3p inhibition contributed to DDP resistance in NSCLC cells, while exogenetic expression of miR-130a-3p decreased DDP resistance in DDP-resistant NSCLC cells. Subsequent mechanistic analysis demonstrated that CCAT1 increased DDP resistance by downregulating miR-130a–3p. In line with our results, miR-130a was down-regulated in cisplatin-resistant ovarian cancer cell line A2780/DDP, and overexpression of miR-130a sensitized A2780/DDP cells to cisplatin by targeting XIAP.Citation27 Additionally, overexpression of miR-130a was also reported to overcome drug resistance in gefitinib-resistant NSCLC cells and paclitaxel-resistant prostate cancers by hindering cell proliferation and inducing apoptosis.Citation28,29 Conversely, miR-130a level was higher in cisplatin-resistant tumor cells, and overexpression of miR-130a contributed to cisplatin resistance in ovarian cancer and HCC.Citation25,30
SOX4, a member of the SOX family of transcription factors, play instrumental functions in the regulation of transcription during numerous developmental processes.Citation31,32 Convincing evidence has revealed the carcinogenic effect of SOX4 in multiple tumors, such as esophagus cancer, lung cancer and breast cancer.Citation33–35 In our current study, we confirmed the upregulation of SOX4 in parental NSCLC cells, as well as in DDP-resistant NSCLC cells. The present study also discovered that SOX4 was a target of miR-130a-3p and miR-130a-3p could inhibit SOX4 expression. In addition, SOX4 overexpression enhanced DDP resistance and ABCG2 expression in NSCLC cells, while SOX4 knockdown diminished DDP resistance and ABCG2 expression in DDP-resistant NSCLC cells. Similarly, a previous study showed that overexpression of SOX4 contributed to resistance to chemotherapeutic drug via upregulation of ABCG2 in cervical cancer.Citation36 The resistance to DDP has previously been demonstrated to be attributable to overexpression of ABCG2 in various cancers including lung cancer.Citation37 Besides, SOX4 knockdown substantially abated miR-130a-3p inhibition-triggered increase of DDP resistance and ABCG2 expression in NSCLC cells. Meanwhile, exogenous expression of SOX4 significantly abrogated CCAT1-knockdown-mediated decrease of DDP resistance and ABCG2 expression in DDP-resistant NSCLC cells. These results indicated that CCAT1/miR-130a-3p axis contributed DDP resistance of NSCLC cells by targeting SOX4.
In conclusion, our study demonstrated the important role of CCAT1-miR-130a-3p-SOX4 regulatory network in DDP resistance of NSCLC cells, providing potential targets to overcome DDP resistance and enhance efficacy of chemotherapy during the treatment of NSCLC.
Authors' Contribution
Baoli Hu, Haifeng Zhang and Li Li designed the experiment. Baoli Hu, Haifeng Zhang and Zuopei Wang performed the experiment and analyzed the data. Baoli Hu, Haifeng Zhang and Feng Zhang prepared the manuscript. Haitao Wei and Li Li supervised the study and reviewed the manuscript
Acknowledgments
This work was supported by Science and Technology Research Key Project in Henan Province Department of Education (16A320034).
Disclosure Statement
No potential conflicts of interest were disclosed.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi:10.3322/caac.20138.
- Ardizzoni A, Boni L, Tiseo M, Fossella FV, Schiller JH, Paesmans M, Radosavljevic D, Paccagnella A, Zatloukal P, Mazzanti P, et al. Cisplatin- versus carboplatin-based chemotherapy in first-line treatment of advanced non-small-cell lung cancer: an individual patient data meta-analysis. J Natl Cancer Inst. 2007;99:847–57. doi:10.1093/jnci/djk196.
- Barabas K, Milner R, Lurie D, Adin C. Cisplatin: a review of toxicities and therapeutic applications. Vet Comp Oncol. 2008;6:1–18. doi:10.1111/j.1476-5829.2007.00142.x.
- Ren K, Li Y, Lu H, Li Z, Li Z, Wu K, Li Z, Han X. Long Noncoding RNA HOTAIR Controls Cell Cycle by Functioning as a Competing Endogenous RNA in Esophageal Squamous Cell Carcinoma. Transl Oncol. 2016;9:489–97. doi:10.1016/j.tranon.2016.09.005.
- Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PloS one. 2013;8:e53823. doi:10.1371/journal.pone.0053823.
- Juan L, Wang G, Radovich M, Schneider BP, Clare SE, Wang Y, Liu Y. Potential roles of microRNAs in regulating long intergenic noncoding RNAs. BMC Med Genomics. 2013;6:S7. doi:10.1186/1755-8794-6-S1-S7.
- Wang Y, Liu Z, Yao B, Dou C, Xu M, Xue Y, Ding L, Jia Y, Zhang H, Li Q, et al. Long non-coding RNATUSC7acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumor Biol. 2016;37:11429–41. doi:10.1007/s13277-016-4892-6.
- Zhen L, Yun-Hui L, Hong-Yu D, Jun M, Yi-Long Y. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumor Biol. 2016;37:673–83. doi:10.1007/s13277-015-3843-y.
- Yang F, Xue X, Bi J. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–45. doi:10.1007/s00432-012-1324-x.
- Liang D, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi:10.1186/s13046-015-0136-7.
- Chen J, Zhang K, Song H, Wang R, Chu X, Chen L. Long noncoding RNA CCAT1 acts as an oncogene and promotes chemoresistance in docetaxel-resistant lung adenocarcinoma cells. Oncotarget. 2016;7:62474–89. doi:10.18632/oncotarget.11518.
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC Couples MicroRNA Biogenesis and Posttranscriptional Gene Silencing. Cell. 2005;123:631–40. doi:10.1016/j.cell.2005.10.022.
- Peng F, Zhang H, Du Y, Tan P. miR-23a promotes cisplatin chemoresistance and protects against cisplatin-induced apoptosis in tongue squamous cell carcinoma cells through Twist. Oncol Rep. 2014;33:942–50. doi:10.3892/or.2014.3664.
- Yashiro M, Nishii T, Hasegawa T, Matsuzaki T, Morisaki T, Fukuoka T, Hirakawa K. A c-Met inhibitor increases the chemosensitivity of cancer stem cells to the irinotecan in gastric carcinoma. Br J Cancer. 2013;109:2619–28. doi:10.1038/bjc.2013.638.
- Hou Z, Xu C, Xie H, Xu H, Zhan P, Yu L, Fang X. Long Noncoding RNAs Expression Patterns Associated with Chemo Response to Cisplatin Based Chemotherapy in Lung Squamous Cell Carcinoma Patients. Plos One. 2014;9:e108133. doi:10.1371/journal.pone.0108133.
- Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H, Wang YX, Zeng J, Song Y, Gao W, et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med. 2015;13:84. doi:10.1186/s12967-015-0442-z.
- Liu J, Wan L, Lu K, Sun M, Pan X, Zhang P, Lu B, Liu G, Wang Z. The Long Noncoding RNA MEG3 Contributes to Cisplatin Resistance of Human Lung Adenocarcinoma. PloS one. 2015;10:e0114586. doi:10.1371/journal.pone.0114586.
- Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Dan G, Morris KV. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20:440–6. doi:10.1038/nsmb.2516.
- Yokoyama Y, Wan X, Shinohara A, Takahashi S, Takahashi Y, Niwa K, Tamaya T. Expression of PTEN and PTEN pseudogene in endometrial carcinoma. Int J Mol Med. 2000;6:47–50.
- Zhu H, Zhou X, Chang H, Li H, Liu F, Ma C, Lu J. CCAT1 promotes hepatocellular carcinoma cell proliferation and invasion. Int J Clin Exp Pathol. 2015;8:5427–34.
- Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi:10.1038/cddis.2014.541.
- Lv L, Jia JQ, Chen J. LncRNA CCAT1 Upregulates Proliferation and Invasion in Melanoma Cells Via Suppressing miR-33a. Oncol Res. 2017; doi:10.3727/096504017X14920318811749.
- Wang Q, Zhang W, Hao S. LncRNA CCAT1 modulates the sensitivity of paclitaxel in nasopharynx cancers cells via miR-181a/CPEB2 axis. Cell Cycle. 16:795–801. doi:10.1080/15384101.2017.1301334.
- Li N, Yang L, Wang H, Yi T, Jia X, Chen C, Xu P. MiR-130a and MiR-374a Function as Novel Regulators of Cisplatin Resistance in Human Ovarian Cancer A2780 Cells. Plos One. 2015;10:e0128886. doi:10.1371/journal.pone.0128886.
- Yang L, Li N, Wang H, Jia X, Wang X, Luo J. Altered microRNA expression in cisplatin-resistant ovarian cancer cells and upregulation of miR-130a associated with MDR1/P-glycoprotein-mediated drug resistance. Oncol Rep. 2012;28:592–600. doi:10.3892/or.2012.1823.
- Zhang XA, Huang L, Zhao Y, Tan W. Downregulation of miR-130a contributes to cisplatin resistance in ovarian cancer cells by targeting X-linked inhibitor of apoptosis (XIAP) directly. Acta Biochim Biophys Sin (Shanghai). 2013;45:995–1001. doi:10.1093/abbs/gmt113.
- Zhou YM, Liu J, Sun W. MiR-130a Overcomes Gefitinib Resistance by Targeting Met in Non-Small Cell Lung Cancer Cell Lines. Asian Pac J Cancer Prev. 2014;15:1391–6.
- Fujita Y, Kojima T, Kawakami K, Mizutani K, Kato T, Deguchi T, Ito M. miR‐130a activates apoptotic signaling through activation of caspase‐8 in taxane‐resistant prostate cancer cells. Prostate. 2015;75:1568–78. doi:10.1002/pros.23031.
- Xu N, Shen C, Luo Y, Xia L, Xue F, Xia Q, Zhang J. Upregulated miR-130a increases drug resistance by regulating RUNX3 and Wnt signaling in cisplatin-treated HCC cell. Biochem Biophys Res Commun. 2012;425:468–72. doi:10.1016/j.bbrc.2012.07.127.
- Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–70.
- Penzo-Méndez AI. Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol. 2010;42:425–8. doi:10.1016/j.biocel.2009.07.018.
- Kang M, Li Y, Liu W, Wang R, Tang A, Hao H, Liu Z, Ou H. miR-129-2 suppresses proliferation and migration of esophageal carcinoma cells through downregulation of SOX4 expression. Int J Mole Med. 2013;32:51–8. doi:10.3892/ijmm.2013.1384.
- Li Y, Zu L, Wang Y, Wang M, Chen P, Zhou Q. miR-132 inhibits lung cancer cell migration and invasion by targeting SOX4. J Thorac Dis. 2015;7:1563–9. doi:10.3978/j.issn.2072-1439.2015.09.06.
- Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X, Feng J, Zhang Y, Gao H, Liu DX, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72:4597–608. doi:10.1158/0008-5472.CAN-12-1045.
- Sun R, Jiang B, Qi H, Zhang X, Yang J, Duan J, Li Y, Li G. SOX4 contributes to the progression of cervical cancer and the resistance to the chemotherapeutic drug through ABCG2. Cell Death Dis. 2015;6:e1990. doi:10.1038/cddis.2015.290.
- Ke SZ, Ni XY, Zhang YH, Wang YN, Wu B, Gao FG. Camptothecin and cisplatin upregulate ABCG2 and MRP2 expression by activating the ATM/NF-κB pathway in lung cancer cells. Int J Oncol. 2013;42:1289–96. doi:10.3892/ijo.2013.1805.
