ABSTRACT
The long non-coding RNA nuclear paraspeckle assembly transcript 1 (NEAT1) was reported to be upregulated and be involved in oncogenic growth and drug resistance in nasopharyngeal carcinoma (NPC). However, the exact roles of NEAT1 and its underlying mechanisms in the drug resistance of NPC remain largely unclear. In this study, the expressions of NEAT1, let-72-5p and Rsf-1 mRNA were detected by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The effects of NEAT1 and let-72-5p on cell proliferation and cisplatin resistance of NPC cells were investigated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay and 5-ethynyl-20-deoxyuridine (EdU) assay. Western blot analysis was performed to detect the protein levels of Rsf-1, Ras, p-Raf1, Raf1, p-MEK1, MEK1, p-ERK1/2 and ERK1/2. Xenograft tumor assay was done to elucidate the role of NEAT1 involved in NPC tumor growth in vivo. We found that NEAT1 was upregulated and let-7a-5p was downregulated in NPC tissues, as well as NPC cell lines. Inhibition of NEAT1 markedly repressed the cisplatin resistance of NPC cells. NEAT1 was demonstrated to interact with let-7a-5p. Besides, a negative correlation between NEAT1 and let-7a-5p expression was observed in NPC tissues. Rsf-1 was confirmed as a target of let-7a-5p. NEAT1 remarkably reversed the inhibitory effect of let-7q-5p on the cisplatin resistance of NPC cells in vitro. Additionally, NEAT1 knockdown inhibited the Ras-MAPK pathway in NPC cells. NEAT1 knockdown suppressed tumor growth in the presence of cisplatin in vivo. Overall, these findings suggest that NEAT1/let-7a-5p axis regulates the cisplatin resistance in NPC by targeting Rsf-1 and modulating the Ras-MAPK signaling pathway.
Introduction
Nasopharyngeal carcinoma (NPC) is one of the most common cancers the head and neck and is a leading cause of cancer-related death around the world. Despite substantial improvements in treatment in the last decades, the prognosis for advanced NPC remains quite poor with a 5-year survival rate range from 50% to 70%.Citation1,Citation2 Platinum-based chemotherapy is a widely used modality of treatment of NPC. Cisplatin, one of the common platinum-based compounds, has been proven to be effective in treating cancers.Citation3 However, many patients relapse after chemotherapy due to the emergence of drug resistance. Therefore, expanding our knowledge of the molecular mechanisms involved in the emergence of cisplatin resistance is vital for improving the prognosis of patients with NPC.
Generally, non-coding RNAs (ncRNAs) can be divided into two major groups depending on the transcript size: small ncRNAs and long ncRNAs (lncRNAs).Citation4 MicroRNAs (miRNAs) are a class of small ncRNAs with 20∼22 nucleotides in length, while lncRNAs are longer than 200 nucleotides in length and without protein-coding capacity. LncRNAs have been reported to implicated in regulation of gene expression at various levels, including the epigenetic, modification, translational, transcriptional, as well as post-transcriptional levels.Citation5 Previous studies have revealed that lncRNAs may function as miRNA sponges to suppress miRNAs expression, thereby reversing the inhibition of miRNA on the corresponding mRNA expression.Citation6 Dysregulation of lncRNAs has been documented to lead to cancer cell dysfunction and disruption of cell processes, including cell growth, apoptosis and differentiation. Notably, there is a growing evidence to suggest that lncRNAs contribute to the drug resistance in cancers, including NPC.Citation7 The nuclear paraspeckle assembly transcript 1 (NEAT1) is a newly identified nuclear-restricted lncRNA, which has been reported to be highly expressed in diverse cancers, such as prostate cancer,Citation8 colorectal cancer,Citation9 breast cancer,Citation10 and hepatocellular carcinoma.Citation11 In acute promyelocytic leukemia cells, NEAT1 was demonstrated to be downregulated and act as a tumor suppressor by promoting leucocyte differentiation.Citation12 Also, NEAT1 knockdown repressed the cell proliferation, migration and invasion by reversing the epithelial-to-mesenchymal transition phenotype, and increased the sensitivity of renal cell carcinoma to sorafenib in vitro.Citation13 In addition, NEAT1 was upregulated in NPC and reduction of NEAT1 reversed epithelial to mesenchymal transition phenotype and sensitized NPC cells to radiation by modulating miR-204/ZEB1 axis.Citation14 However, the function of NEAT1 and its underlying mechanisms in NPC chemoresistance remain poorly understood.
let-7a-5p, the firstly identified tumor repressing miRNA, can modulate a large number of oncogenes to suppress tumor progression by binding the 3′-untranslated region (3′-UTR) of the target mRNAs, including Ras, MYC and HMGA2.Citation15-17 let-7a-5p was reported to be poorly expressed in colorectal cancer, which could be utilized to predict lymph node metastasis and the disease prognosis.Citation18 In breast cancer, let-7a-5p was downregulated. Moreover, let-7a-5p repressed mammosphere formation capacity and reduced the mammosphere-size via Ras/NF-κB and Ras/MAPK/ERK pathway, respectively.Citation17 let-7a-5p was also underexpressed in NPC and induction of let-7a-5p led to suppression of cell proliferation by downregulating c-Myc expression.Citation19 In addition, let-7a has been proposed to be implicated in the drug resistance.Citation20,Citation21 However, the precise regulatory mechanisms of let-7a-5p in cisplatin resistance in NPC remain unlear.
In this study, we determined the expression of NEAT1 and let-7a-5p in NPC tissues and cells. The effect of NEAT1 and let-7a-5p on the course of NPC and the cisplatin resistance and its underlying molecular mechanism were further explored in vitro and in vivo. Our results suggest that NEAT1/let-7a-5p axis may be a potential promising strategy for overcoming cisplatin resistance in the treatment of NPC.
Results
NEAT1 is upregulated and let-7a-5p is downregulated in NPC tissues and NPC cell lines
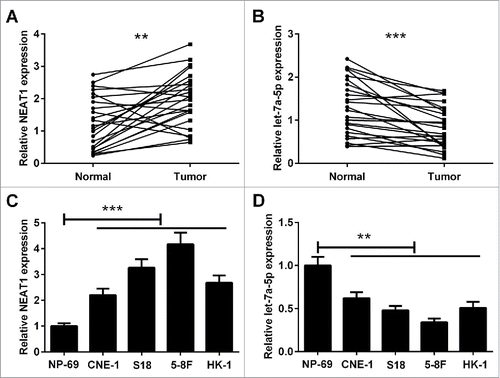
Firstly, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay was conducted to analyze the expression of NEAT1 and let-7a-5p in NPC tissues and paired adjacent normal tissues. As displayed in and , the expression level of NEAT1 was higher in NPC tissues than that in paired adjacent normal tissues. Conversely, let-7a-5p was aberrantly downregulated in NPC tissues in comparison with that in paired adjacent normal tissues. In addition, NEAT1 was obviously upregulated and let-7a-5p was markedly downregulated in NPC cell lines including CNE-1, S18, 5–8F, HK-1 cells compared with NPE cell line NP-69 ( and ). S18 and 5–8F cells were chosen for the following experiment.
Figure 1. The expression levels of NEAT1 and let-7a-5p in NPC tissues and cells. RT-qPCR analysis of expression of NEAT1 (A) and let-7a-5p (B) in 24 paired of NPC tissues and corresponding normal tissues. RT-qPCR analysis of NEAT1 (C) and let-7a-5p (D) expression in NPC cell lines (CNE-1, S18, 5–8F, HK-1) and NPE cell line NP-69. *P < 0.05, **P < 0.01, and ***P < 0.001.
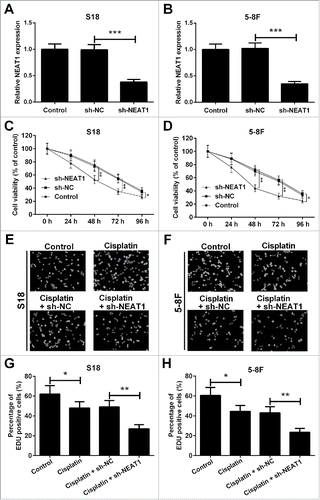
Knockdown of NEAT1 attenuates cisplatin resistance of NPC cells
To investigate the biological effects of NEAT1, we performed loss-of-function studies in S18 and 5–8F cells by transfecting with short-hairpin RNA specific for NEAT (sh-NEAT1) or negative control (sh-NC). As shown in and , the transfection efficiency was identified by RT-qPCR, and the expression level of NEAT1 was dramatically decreased compared with the sh-NC group. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay was used to explore the effect of NEAT1 knockdown on cell viability after administration of 10 μg/ml of cisplatin. Results revealed that NEAT1 knockdown prominently repressed cell viability in the presence of cisplatin in S18 () and 5–8F () cells in comparison with that in sh-NC group. Simultaneously, 5-ethynyl-20-deoxyuridine (EdU) assay was carried out in S18 and 5–8F cells transfected with sh-NEAT1 or sh-NC, following exposure to 10 μg/ml cisplatin for 48 h ( and ). Cell proliferation was reduced by cisplatin treatment in S18 and 5–8F cells, which was dramatically aggravated by NEAT1 knockdown ( and ). Thus, we speculated that knockdown of NEAT1 attenuated cisplatin resistance of NPC cells.
Figure 2. NEAT1 knockdown in hibited cisplatin resistance of NPC cells. The expression level of NEAT1 in S18 (A) and 5–8F (B) cells was obviously downregulated by transfecting with sh-NEAT1. Cell viability was determined by MTT assay in S18 (C) and 5–8F (D) cells after transfection with sh-NEAT1 or sh-NC, following exposing to 10 μg/ml of cisplatin. The representative fluorescent images of EdU assay in S18 (E) and 5–8F (F) cells were shown. EdU assay showed that downregulation of NEAT1 inhibited cell proliferation ability in S18 (G) and 5–8F (H) cells in the presence of 10 μg/ml of cisplatin. *P < 0.05, **P < 0.01, and ***P < 0.001.
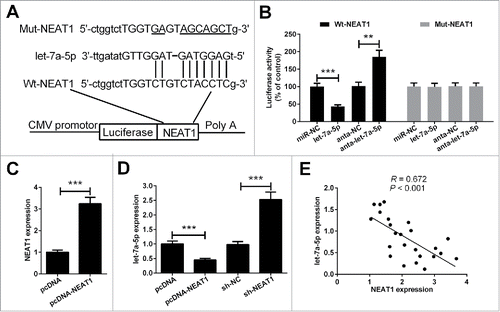
NEAT1 directly interacts with let-7a-5p
Bioinformatics prediction results showed that the NEAT1 sequence harbors a putative let-7a-5p binding region (). To identify whether NEAT1 is a bona fide target of let-7a-5p, the wild type sequence of NEAT1 (Wt-NEAT1) or the mutant (Mut-NEAT1) with a 9-base mutation in the putative let-7a-5p recognition sequence was subcloned into the pMIR luciferase reporter, and then transfected in 5–8F cells together with miR-NC, let-7a-5p, anta-NC or anta-let-7a-5p. The relative luciferase activity of the pMIR-NEAT1-Wt was obviously decreased when let-7a-5p was co-transfected into 5–8F cells. Downregulation of let-7a-5p led to an increase in the luciferase activity of the pMIR-Wt-NEAT1 in 5–8F cells. However, the luciferase activity of the pMIR-NEAT1-Mut was unaffected (). Furthermore, we investigated the correlation between the expression of NEAT1 and let-7a-5p. NEAT1 expression was upregulated by transfection with pcDNA-NEAT1 (). We found that upregulation of NEAT1 decreased let-7a-5p expression, whereas NEAT1 knockdown increased let-7a-5p expression (). Interestingly, a significantly negative correlation was noted between NEAT1 and let-7a-5p expression in NPC tissues (), which further consolidated our findings in vitro.
Figure 3. Verification of NEAT1 as a direct target of let-7a-5p. (A) The sequence of the predicted let-7a-5p targeting site within NEAT1 3′-UTR and the mutated sites were shown. (B) After co-transfection with pMIR-report plasmid and let-71-5p, anta-let-7a-5p or matched controls, the relative luciferase activity of pMIR-report plasmid containing Wt or Mut NEAT1 were evaluated in 5–8F cells. (C) The effect of pcDNA-NEAT1 on NEAT1 expression was validated by RT-qPCR. (D) RT-qPCR analysis of let-7a-5p expression in 5–8F cells transfected with pcDNA-NEAT1, sh-NEAT1 or matched controls. (E) The correlation analysis showed that let-7a-5p was negatively associated with NEAT1 in NPC tissues. **P < 0.01, and ***P < 0.001.
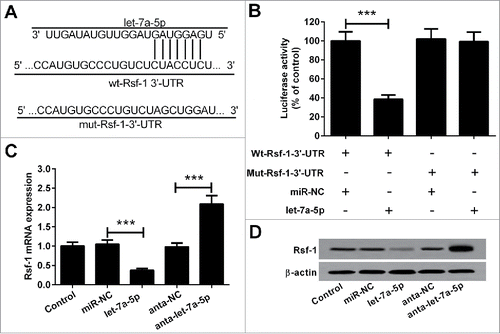
NEAT1 downregulates the expression of Rsf-1 through let-7a-5p (Rsf-1 is a direct let-7a-5p target)
The predicted targeting sites of let-7a-5p and Rsf-1 were shown in . For validation, we fused the wild type sequence of Rsf-1 (Wt-Rsf-1) or the mutant (Mut-Rsf-1) into the pMIR luciferase reporter. Wt-Rsf-1 or Mut-Rsf-1 plasmid and let-7a-5p or negative control (miR-NC) were co-transfected into 5–8F cells. Results of luciferase report assay showed that let-7a-5p markedly reduced the relative luciferase activity of the pMIR-Rsf-1-Wt instead of the pMIR-Rsf-1-Mut (). In addition, we found that transfection with let-7a-5p mimic reduced, but anta-let-7a-5p increased the Rsf-1 mRNA level and protein level in 5–8F cells ( and ). These findings suggested that Rsf-1 was a direct target of let-7a-5p.
Figure 4. Rsf-1 was a direct target of let-7a-5p. (A) The predicted let-7a-5p complementary site within the Rsf-1 3′-UTR and the mutated sites were shown. (B) let-7a-5p obviously repressed the luciferase activity of the pMIR-Rsf-1-Wt but not that of pMIR-Rsf-1-Mut. Upregulation of let-7a-5p decreased, but anta-let-7a-5p increased, the Rsf-1 mRNA level (C) and protein level (D) in 5–8F cells. ***P < 0.001.
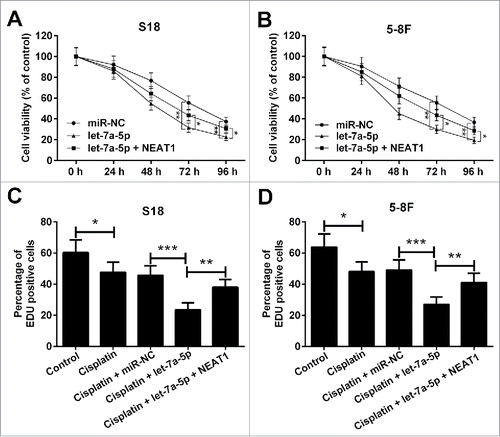
NEAT1 reverses the inhibitory effect of let-7a-5p on the cisplatin resistance of NPC cells
To explore the roles of NEAT1 and let-7a-5p in the development of cisplatin resistance of NPC cells, S18 and 5–8F cells were transfected with let-7a-5p, pcDNA-NEAT1 or matched controls. Upregulation of let-7a-5p reduced cell viability in S18 and 5–8F cells in the presence of 10 μg/ml cisplatin, which was reversed by upregulation of NEAT1 ( and ). In parallel, it was found that cell proliferation was inhibited after administration of cisplatin in S18 and 5–8F cells, which was exaggerated by overexpressing let-7a-5p. However, overexpression of NEAT1 partially reversed the inhibitory effect of let-7a-5p on cell proliferation ( and ). Accordingly, we concluded that NEAT1 reversed the inhibitory effect of let-7a-5p on the cisplatin resistance of NPC cells.
Figure 5. NEAT1 reversed the inhibitory effect of let-7a-5p on the cisplatin resistance of NPC cells. MTT assay was used to assess cell viability in S18 (A) and 5–8F (B) cells stably transfected with let-7a-5p alone or combined with pcDNA-NEAT1, following exposure to 10 μg/ml cisplatin for 48 h. EdU assay was performed to detect the cell proliferation in S18 (C) and 5–8F (D) cells transfected with let-7a-5p or pcDNA-NEAT1, following treatment with 10 μg/ml cisplatin for 48 h. *P < 0.05, **P < 0.01, and ***P < 0.001.
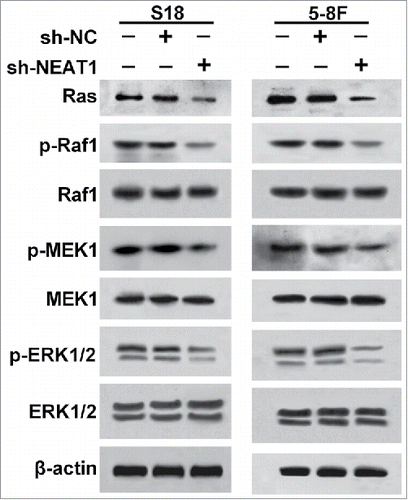
Knockdown of NEAT1 inhibits Ras-MAPK pathway
Previous studies have reported that let-7 is complementary to multiple sequences in the 3′-UTR of human Ras genes, and modulates the Ras-MAPK signaling pathway.Citation22 Therefore, we investigated the impact of NEAT1 knockdown on the Ras-MAPK pathway by measuring protein levels of Ras, p-Raf1, Raf1, p-MEK1, MEK1, p-ERK1/2 and ERK1/2 using western blot. As a result, inhibition of NEAT1 in S18 and 5–8F cells markedly decreased protein levels of Ras, p-Raf1, p-MEK1 and p-ERK1/2 instead of Raf1, MEK1 and ERK1/2 (). These data indicated that inhibition of NEAT1 repressed Ras-MAPK pathway in NPC cells.
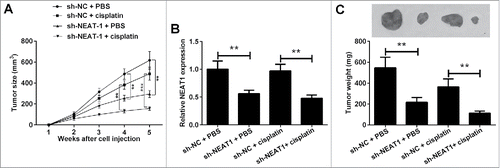
Knockdown of NEAT1 suppresses NPC tumor growth in vivo
Given the function of NEAT1 in vitro, we further characterized whether NEAT1 loss could suppressed the NPC tumor growth in vivo. NPC cells stably expressing sh-NEAT1 or sh-NC were subcutaneously injected into nude mice. Subsequently, the mice were treated with cisplatin or phosphate buffered saline and tumor size was monitored every week using calipers. As a result, xenograft tumors from sh-NEAT1-transfected NPC cells grew obviously slower than the tumors from sh-NC-transfected NPC cells in the presence of cisplatin (). At 5th weeks after injection, the mice were sacrificed and the tumor was resected. The tumor size and weight were strikingly lower in sh-NEAT1-treated group than that in the sh-NC group in the presence of cisplatin (). As except, the expression level of NEAT1 remarkably reduced in xenograft tumors from sh-NEAT1-transfected NPC cells than tumors from sh-NC-transfected NPC cells (). Taken together, these findings suggested that NEAT1 knockdown repressed the cisplatin resistance and tumor growth in vivo.
Figure 7. Knockdown of NEAT1 suppresses NPC tumor growth in vivo. The xenograft tumor model was established by subcutaneously injecting with 5–8F cells stably expressing sh-NEAT1 or sh-NC, following administration with cisplatin or phosphate buffered saline every three days. (A) The tumor size was monitored through measuring the length and width every week. (B) RT-qPCR analysis of NEAT1 expression in xenografted tumors. (C) Mice were sacrificed at 5 weeks after injection and tumors were excised, pictured and weighed. Scale bar: 1 mm. **P < 0.01 and ***P < 0.001.
Discussion
Chemoresistance is a main obstacle to the effective treatment of NPC in clinical settings, which is associated with poor prognosis and high mortality in patients with advanced NPC. Therefore, clarifying the exact mechanism of cisplatin resistance in NPC is an urgent problem. Recently, a growing body of evidence shows that lncRNAs play a critical role in regulating drug resistance of NPC. For instance, upregulation of colon cancer associated transcript-1 (CCAT1) resulted in repression of paclitaxel sensitivity in NPC cells due to its regulatory effect on CPEB2 expression by targeting miR-181a.Citation23 Additionally, n375709, a newly identified lncRNA, was documented to be highly expressed in NPC cells and implicated in the regulation of NPC paclitaxel resistance.Citation24 However, the roles of NEAT1 and its underlying mechanisms in cisplatin resistance remain unclear. In our study, we confirmed the upregulation of NEAT1 in NPC tissues and cells. As expected, functional analyses proved that inhibition of NEAT1 enhanced the sensitivity of NPC cells to cisplatin. Consistently, in vivo experiments further confirmed that NEAT1 knockdown suppressed tumor growth in the presence of cisplatin.
Rsf-1, located on chromosome 11p13.5–14, was highly expressed in many cancers, such as prostate cancer, breast cancer and colon cancer.Citation25-27 Previously, there were several literatures elucidating the involvement of Rsf-1 in the emergence of drug resistance in cancers. For example, upregulation of Rsf-1 obviously repressed paclitaxel sensitivity in ovarian cancer cells.Citation28,Citation29 Besides, elevated expression of Rsf-1 promoted tumor progression, aggressiveness and radioresistance of NPC, which predicted poor therapeutic response and worse survival.Citation30 Furthermore, Rsf-1 inhibition potentiated NPC cells to be sensitive to paclitaxel by modulating the NF-κB signaling pathway.Citation31 In the present study, we found that let-7a-5p was downregulated in NPC tissues, as well as NPC cells. Besides, a negative correlation between NEAT1 and let-7a-5p expression was observed in NPC tissues. In addition, Rsf-1 was confirmed as a direct target of let-7a-5p. Thus, we speculated that NEAT1 might downregulated the expression of Rsf-1 through direct interaction with let-7a-5p.
Ras–mitogen activated protein kinase (MAPK) pathway is a vital intracellular signaling pathway that modulates multiple cellular progresses including proliferation, apoptosis, transcription, differentiation and cell cycle arrest. Ras, the most frequented mutated oncogene, belongs to the superfamily of small GTPases and is the first intracellular effector of the Ras-MAPK signaling pathway. Activation of Ras is triggered by various extracellular stimuli, which alter the status of Ras from a GDP-bound inactivated form to a GTP-bound activated form, and recruit the Raf-1 kinase to the plasma membrane.Citation32 Activated Raf-1 induces phosphorylation of MEK 1 and MEK 2 (p-MEK 1 and p-MEK 2), which in turn phosphorylates and activates ERK 1/2. p-ERK is then translocated into the nucleus and participates in cell cycle progression or other cellular processes.Citation33 Deregulation of the Ras–MAPK pathway is an early event in diverse cancers and functions as an important contributor to chemoresistance.Citation34 In basal cell carcinoma, activation of the RAS/MAPK signaling pathway repressed Shh signaling and reduced the sensitivity of basal cell carcinoma cells to smoothened inhibitor.Citation35 In the present study, NEAT1 was demonstrated to interact with let-7a-5p. NEAT1 knockdown inhibited the Ras-MAPK signaling in both S18 and 5–8F NPC cells. Moreover, NEAT1 reversed the inhibitory effect of let-7q-5p on the cisplatin resistance of NPC cells in vitro.
In general, our study suggested that NEAT1 was upregulated and let-7a-5p was downregulated in NPC tissues, as well as NPC cells. Functional and mechanistic analyses suggested that NEAT1 knockdown enhanced sensitivity of NPC cells to cisplatin by modulating Rsf-1 expression and the Ras-MAPK pathway through inhibiting let-7a-5p. Thus, targeting NEAT1/let-7a-5p axis might have important therapeutic implications for overcoming cisplatin resistance in NPC.
Material and methods
Cell culture and transfection
Four NPC cell lines HK-1 (from the China Center for Type Culture Collection, Wuhan, China), CNE-1, CNE-2 subclone S18 (from the Chinese Academy of Sciences, Shanghai, China) and SUNE1 subclone 5–8F (from the Cancer Research Institute of Sun Yatsen University, Guangzhou, China) were cultured in RPMI-1640 medium (Invitrogen, Grand Island, NY, USA) with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA). A nasopharyngeal epithelial (NPE) cell line NP-69 (from the Chinese Academy of Medical Sciences, Beijing, China) was cultured in keratinocyte/serum-free medium (Invitrogen) supplemented with bovine pituitary extract at 37°C in a humidified incubator with 5% CO2.
To overexpress NEAT1, full NEAT1 sequence was amplified and cloned into pcDNA-3.1 vector. Then pcDNA- NEAT1 plasmid was transfected into NPC cells. To knockdown NEAT1, shRNA oligonucleotide of NEAT1 and its corresponding control sequence were fused into the pGPU6/GFP/Neo vectors (GenePharma, Shanghai, China), respectively.
To overexpress or silence let-7a-5p, let-7a-5p mimic, anta-let-7a-5p, miR-NC and antagomir control (anta-NC) were designed and synthesized by Sangon Biotech (Shanghai, China), and transfected into NPC cells. Cell transfection was performed using Lipofectamine 2000 reagent (Invitrogen). At 48 h after transfection, cells were collected for further study.
Clinical tissues and RNA isolation
Twenty-four NPC tissues and paired corresponding normal tissues were obtained from NPC patients who did not receive radiotherapy and chemotherapy Henan Provincial People's Hospital from 2013 to 2016. All tissues were confirmed by pathological evaluation. NPC tissues and paired corresponding normal tissues were snap-frozen in liquid nitrogen and stored at −80°C for further experiments. The project protocols were approved by the Institutional Review Board of Henan Provincial People's Hospital and patients were given informed consent before participating in this project.
RT-qPCR
Total RNA from tissues and cells were isolated using Trizol reagent (Invitrogen) and reverse transcribed to generate cDNA with the use of PrimeScript RT reagent kit (Takara Bio, Shiga, Japan), according to the manufacturer's instruction. qPCR analysis was performed using SYBR Green PCR Kit (Takara) on the ABI prism 7900 sequence detection system (Life Technologies, Carlsbad, CA, USA). All reactions were performed in triplicate. β-actin was used as a reference for NEAT1 and Rsf-1 mRNA. U6 was employed as an endogenous control for let-7a-5p. Relative gene expression levels were quantified using the 2−ΔΔCt method. Gene-specific RT-qPCR primers were as follows: NEAT1 forward, 5′-GTA CGC GGG CAG ACT AAC AC-3′ and reverse, 5′-TGC GTC TAG ACA CCA CAA CC-3′; Rsf-1 forward, 5′-TCC CAG ATG GAG AAT GGT TC-3′ and reverse, 5′-AGT CTG GCT CTT GTG GAG GA-3′; β-actin forward, 5′-TGA GCG CGG CTA CAG CTT-3′ and reverse, 5′-TCC TTA ATG TCA CGC ACG ATT T-3′. let-7a-5p forward, 5′-CGA TTC AGT GAG GTA GTA GGT TGT-3′ and reverse, 5′-TAT GGT TGT TCT GCT CTC TGT CTC-3′; U6 forward, 5′-CTC GCT TCG GCA GCA CA-3′ and reverse, 5′-AAC GCT TCA CGA ATT TGC GT-3′.
Luciferase report assay
The 3′-UTR of Rsf-1 or let-7a-5p was amplified and subcloned into the pMIR luciferase reporter vector. Similarly, the fragment of mutant Rsf-1 3′-UTR or let-7a-5p was inserted into the pMIR luciferase reporter vector. The luciferase report vector (carrying NEAT1-Wt or NEAT1-Mut) was transfected into 5–8F cells together with let-7a-5p, anta-let-7a-5p or matched controls. The luciferase report vector (carrying Rsf-1-Wt or Rsf-1-Mut) was co-transfected into 5–8F cells with let-7a-5p or miR-NC. The relative luciferase activity of NEAT1 or Rsf-1 was determined at 48 h post-transfection using a luciferase assay kit (Pharmingen, San Diego, CA, USA).
Western blot analysis
Cells were lysed with ice-cold RIPA buffer containing protease inhibitor (Roche, Mannheim, Germany). The protein concentration was measured with a BCA Protein Assay Kit (Pierce, Rockford, IL, USA). A total of 30 μg protein lysate was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and then transferred onto polyvinylidene fluoride membranes (Millipore Corporation, Billerica, MA, USA). Following blocking with a 5% skim milk solution, membranes were incubated at 4°C overnight with specific antibodies against Rsf-1 (Abcam, Cambridge, MA, USA), Ras (Cell Signaling Technology, Inc., Beverly, MA, USA), p-Raf1 (Abcam), Raf1 (Abcam), p-MEK1 (Cell Signaling Technology), MEK1 (Cell Signaling Technology), p-ERK1/2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and ERK1/2 (Santa Cruz). Subsequently, membranes were incubated with the horseradish peroxidase (HRP)-conjugated antibody (Santa Cruz) for 1 h at room temperature. Signal detection was performed using a chemiluminescence detection kit (Pierce, Rockford, IL, USA).
Detection of cell viability and proliferative capacity
Cell viability was measured with a MTT kit (Sigma-Aldrich). The transfected cells were seeded in 96-well plates and treated with indicated concentrations 10 μg/ml of cisplatin (Sigma). Following 48 h of incubation, 20 μl of MTT (2 mg/ml) was added to each well and cultured for 4 h in a humidified atmosphere containing 5% CO2. MTT solution was replaced with DMSO. At 5 min after incubation, plates were analyzed using a microplate reader at 570 nm wavelength.
EdU assay were performed to evaluate cell proliferation. For the EdU assay, the transfected cells were incubated in EdU solution (50 nmol/l) for 2 h and then visualized. Pictures were captured using a fluorescence microscope and analyzed by Image J software.
Tumor xenograft model in nude mice
Male BALB/c nude mice (5-weeks old; 18–20 g) were purchased from the Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and acclimated for 1 week under standard conditions of temperature, relative humidity and light. 1 × 106 5–8F cells stably transfected with sh-NEAT1 or sh-NC were injected subcutaneously into the posterior fossa of nude mice (n = 8 per group). After 5 d, when average tumor volume was about 50 mm3, mice were intraperitoneally injected with cisplatin (3.5 mg/kg body weight) or phosphate buffered saline every three days. The tumor size was measured every week by using calipers [(length × widthCitation2)/2]. Mice were sacrificed at 5th week after injection and tumors were removed, weighed and analyzed by RT-qPCR. All the experimental procedures and protocols used in this study were reviewed and approved by the Institutional Animal Ethics Committee of Henan Provincial People's Hospital.
Statistical analysis
SPSS 20.0 software (IBM, SPSS, Chicago, IL, USA) was applied for the general statistical. Measurement data are presented as mean ± SEM from at least three independent experiments and analyzed for statistical significance by the Student's t test or One-Way Analysis of Variance followed by Newman-Keuls post hoc test. A value of P < 0.05 was considered significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Pan F, Ruan Z, Li J, Pang X, Zhang Y, Zou L, Liang H. Radiotherapy combined docetaxel and oxaliplatin chemotherapy is effective in patients with locally advanced nasopharyngeal carcinoma. Med Oncol. 2015;32(11):252. doi:10.1007/s12032-015-0698-4. PMID:26471179.
- Colaco RJ, Betts G, Donne A, Swindell R, Yap BK, Sykes AJ, Slevin NJ, Homer JJ, Lee LW. Nasopharyngeal carcinoma: a retrospective review of demographics, treatment and patient outcome in a single centre. Clin Oncol (R Coll Radiol). 2013;25(3):171–7. doi:10.1016/j.clon.2012.10.006. PMID:23337060.
- Bergamini A, Pisano C, Di Napoli M, Arenare L, Della Pepa C, Tambaro R, Facchini G, Gargiulo P, Rossetti S, Mangili G, et al. Cisplatin can be safely administered to ovarian cancer patients with hypersensitivity to carboplatin. Gynecol Oncol. 2017;144(1):72–76. doi:10.1016/j.ygyno.2016.10.023. PMID:28094039.
- Johnsson P, Morris KV. Expanding the functional role of long noncoding RNAs. Cell Res. 2014;24(11):1284–5. doi:10.1038/cr.2014.104. PMID:25104732.
- Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol. 2014;26:10–18. doi:10.1016/j.ceb.2013.08.005. PMID:24529241.
- Hu X, Jiang H, Jiang X. Downregulation of lncRNA ANRIL inhibits proliferation, induces apoptosis, and enhances radiosensitivity in nasopharyngeal carcinoma cells through regulating miR-125a. Cancer Biol Ther. 2017;18(5):331–8. doi:10.1080/15384047.2017.1310348. PMID:28402230.
- Deng H, Zhang J, Shi J, Guo Z, He C, Ding L, Tang JH, Hou Y. Role of long non-coding RNA in tumor drug resistance. Tumour Biol. 2016;37(9):11623–31. doi:10.1007/s13277-016-5125-8. PMID:27380056.
- Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat commun. 2014;5:5383. doi:10.1038/ncomms6383. PMID:25415230.
- Li Y, Li Y, Chen W, He F, Tan Z, Zheng J, Wang W, Zhao Q, Li J. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6(29):27641–50. doi:10.18632/oncotarget.4737. PMID:26314847.
- Almnaseer ZA, Mourtada-Maarabouni M. NEAT1, a long non-coding RNA, controls cell survival and is up-regulated in breast cancer. Eur J Cancer. 2016;61:S75–6. doi:10.1016/S0959-8049(16)61263-7..
- Fang L, Sun J, Pan Z, Song Y, Zhong L, Zhang Y, Liu Y, Zheng X, Huang P. Long non-coding RNA NEAT1 promotes hepatocellular carcinoma cell proliferation through the regulation of miR-129-5p-VCP-IkappaB. Am J Physiol Gastrointest Liver Physiol. 2017;313(2):G150–g156. doi:10.1152/ajpgi.00426.2016. PMID:28526689.
- Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L, Chen S, Li Y. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693. doi:10.1186/1471-2407-14-693. PMID:25245097.
- Liu F, Chen N, Gong Y, Xiao R, Wang W, Pan Z. The long non-coding RNA NEAT1 enhances epithelial-to-mesenchymal transition and chemoresistance via the miR-34a/c-Met axis in renal cell carcinoma. Oncotarget. 2017;8(38):62927–38. doi:10.18632/oncotarget.17757. PMID:28968960.
- Lu Y, Li T, Wei G, Liu L, Chen Q, Xu L, Zhang K, Zeng D, Liao R. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. 2016;37(9):11733–41. doi:10.1007/s13277-015-4773-4. PMID:27020592.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi:10.1016/j.cell.2009.01.002. PMID:19167326.
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67(20):9762–70. doi:10.1158/0008-5472.CAN-07-2462. PMID:17942906.
- Xu C, Sun X, Qin S, Wang H, Zheng Z, Xu S, Luo G, Liu P, Liu J, Du N, et al. Let-7a regulates mammosphere formation capacity through Ras/NF-kappaB and Ras/MAPK/ERK pathway in breast cancer stem cells. Cell Cycle. 2015;14(11):1686–97. doi:10.1080/15384101.2015.1030547. PMID:25955298.
- Liu TP, Huang CC, Yeh KT, Ke TW, Wei PL, Yang JR, Cheng YW. Down-regulation of let-7a-5p predicts lymph node metastasis and prognosis in colorectal cancer: implications for chemotherapy. Surg Oncol. 2016;25(4):429–34. doi:10.1016/j.suronc.2016.05.016. PMID:27262492.
- Wong TS, Man OY, Tsang CM, Tsao SW, Tsang RK, Chan JY, Ho WK, Wei WI, To VS. MicroRNA let-7 suppresses nasopharyngeal carcinoma cells proliferation through downregulating c-Myc expression. J Cancer Res Clin Oncol. 2011;137(3):415–22. doi:10.1007/s00432-010-0898-4. PMID:20440510.
- Yu CC, Chen YW, Chiou GY, Tsai LL, Huang PI, Chang CY, Tseng LM, Chiou SH, Yen SH, Chou MY, et al. MicroRNA let-7a represses chemoresistance and tumourigenicity in head and neck cancer via stem-like properties ablation. Oral Oncol. 2011;47(3):202–10. doi:10.1016/j.oraloncology.2010.12.001. PMID:21292542.
- Chen Y, Jacamo R, Konopleva M, Garzon R, Croce C, Andreeff M. CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. J Clin Invest. 2013;123(6):2395–407. doi:10.1172/JCI66553. PMID:23676502.
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–47. doi:10.1016/j.cell.2005.01.014. PMID:15766527.
- Wang Q, Zhang W, Hao S. LncRNA CCAT1 modulates the sensitivity of paclitaxel in nasopharynx cancers cells via miR-181a/CPEB2 axis. Cell Cycle. 2017;16(8):795–801. doi:10.1080/15384101.2017.1301334. PMID:28358263.
- Ren S, Li G, Liu C, Cai T, Su Z, Wei M, She L, Tian Y, Qiu Y, Zhang X, et al. Next generation deep sequencing identified a novel lncRNA n375709 associated with paclitaxel resistance in nasopharyngeal carcinoma. Oncol Rep. 2016;36(4):1861–7. doi:10.3892/or.2016.4981. PMID:27498905.
- Li H, Zhang Y, Zhang Y, Bai X, Peng Y, He P. Rsf-1 overexpression in human prostate cancer, implication as a prognostic marker. Tumour Biol. 2014;35(6):5771–6. doi:10.1007/s13277-014-1766-7. PMID:24584698.
- Ren J, Chen QC, Jin F, Wu HZ, He M, Zhao L, Yu ZJ, Yao WF, Mi XY, Wang EH, et al. Overexpression of Rsf-1 correlates with pathological type, p53 status and survival in primary breast cancer. Int J Clin Exp Pathol. 2014;7(9):5595–608. PMID:25337201.
- Liu S, Dong Q, Wang E. Rsf-1 overexpression correlates with poor prognosis and cell proliferation in colon cancer. Tumour Biol. 2012;33(5):1485–91. doi:10.1007/s13277-012-0399-y. PMID:22528946.
- Choi JH, Sheu JJ, Guan B, Jinawath N, Markowski P, Wang TL, Shih IeM. Functional analysis of 11q13.5 amplicon identifies Rsf-1 (HBXAP) as a gene involved in paclitaxel resistance in ovarian cancer. Cancer Res. 2009;69(4):1407–15. doi:10.1158/0008-5472.CAN-08-3602. PMID:19190325.
- Yang YI, Ahn JH, Lee KT, Shih Ie M, Choi JH. RSF1 is a positive regulator of NF-kappaB-induced gene expression required for ovarian cancer chemoresistance. Cancer Res. 2014;74(8):2258–69. doi:10.1158/0008-5472.CAN-13-2459. PMID:24566868.
- Tai HC, Huang HY, Lee SW, Lin CY, Sheu MJ, Chang SL, Wu LC, Shiue YL, Wu WR, Lin CM, et al. Associations of Rsf-1 overexpression with poor therapeutic response and worse survival in patients with nasopharyngeal carcinoma. J Clin Pathol. 2012;65(3):248–53. doi:10.1136/jclinpath-2011-200413. PMID:22081787.
- Liu Y, Li G, Liu C, Tang Y, Zhang S. RSF1 regulates the proliferation and paclitaxel resistance via modulating NF-kappaB signaling pathway in nasopharyngeal carcinoma. J Cancer. 2017;8(3):354–62. doi:10.7150/jca.16720. PMID:28261335.
- Dhawan NS, Scopton AP, Dar AC. Small molecule stabilization of the KSR inactive state antagonizes oncogenic Ras signalling. Nature. 2016;537(7618):112–6. doi:10.1038/nature19327. PMID:27556948.
- Delire B, Starkel P. The Ras/MAPK pathway and hepatocarcinoma: pathogenesis and therapeutic implications. Eur J Clin Invest. 2015;45(6):609–23. doi:10.1111/eci.12441. PMID:25832714.
- Hrustanovic G, Bivona TG. RAS-MAPK signaling influences the efficacy of ALK-targeting agents in lung cancer. Mol Cell Oncol. 2016;3(2):e1091061. doi:10.1080/23723556.2015.1091061. PMID:27308613.
- Zhao X, Ponomaryov T, Ornell KJ, Zhou P, Dabral SK, Pak E, Li W, Atwood SX, Whitson RJ, Chang AL, et al. RAS/MAPK Activation Drives Resistance to Smo Inhibition, Metastasis, and Tumor Evolution in Shh Pathway-Dependent Tumors. Cancer Res. 2015;75(17):3623–35. doi:10.1158/0008-5472.CAN-14-2999-T. PMID:26130651.