ABSTRACT
2-Methoxy-5((3,4,5-trimethosyphenyl)seleninyl) phenol (SQ), a novel synthesized combretastatin A-4 (CA-4) analogue, is identified as a microtubule inhibitor and has been shown to exert anticancer activity in breast cancer cells. Here, we found that SQ reversed epidermal growth factor (EGF)-induced motility and invasion in breast cancer cell lines by the in vitro Wound healing and Transwell assay. Further studies showed that SQ treatment resulted in inhibitory alteration of EGF-stimulated epithelial-to-mesenchymal transition (EMT) and MMP-2 activity. What is more, SQ significantly inhibited the EGF-induced mouse double minute 2- (MDM2) expression and transcription factor Twist1 expression. In addition, compared with the control cells, MDM2 overexpression up-regulated Twist1 expression and dramatically promoted cell migration and invasion, MDM2 under-expression also down-regulated Twist1 expression and suppressed cell motility and invasion. Taken together, our findings suggest that the inhibitory effects of SQ on migration and invasion were related to the suppression of MDM2 and Twist1 signal axis.
Introduction
Breast cancer is a common cause of death in women’s cancers at all ages. The main reason of cancer death results from metastasis instead of the primary tumor itself.Citation1,Citation2 Cancer metastasis includes a series of closely related steps: epithelial-to-mesenchymal transition (EMT), cell motility, local invasion, intravasation, traveling through the blood or lymphatic circulation, extravasation, and transiting in secondary organs.Citation3–Citation5
Breast cancer cells with EMT phenotype lose their epithelial characteristics and escape from the primary tumor.Citation6,Citation7 Multiple pathways including epidermal growth factor (EGF), integrin, Wnt and nuclear factor (NF)-κB pathways have been found to trigger EMT. The main feature of the EMT pathways is that the activated transcription factors (Snail1, Slug and Twist1) initiate EMT program in the epithelial cells, down-regulating E-cadherin and other epithelial cell phenotype-associated genes and activating the mesenchymal cell phenotype-related genes, such as N-cadherin and Vimentin.Citation8 Some studies proved that EGF or the overexpression of transcription factor Twist1 can induce the process of EMT in breast cancer cells.Citation9–Citation13 As reported previously, EGF played a leading role in promoting breast cancer metastasis, probably by activating extracellular signal-regulated kinae 1/2 (ERK 1/2) or phosphoinositide-3 kinase/Akt (PI3K/Akt) signal pathway.Citation11,Citation14,Citation15 Transcription factor Twist1 is involved in the cancer invasiveness and metastasis process.Citation16,Citation17 Twist1 regulates EMT-related proteins, such as down-regulation of E-cadherin and up-regulation of Vimentin and matrix metalloproteinases (MMPs), driving the breast cancer cells toward metastasis Citation12. Furthermore, MMPs including MMP-2 and MMP-9 play an essential role in cancer metastasis by degradation of the extracellular matrix (ECM).Citation18–Citation20
Mouse double minute 2 (MDM2), an oncogene which is usually highly expressed in malignancies including solid tumors, sarcomas and leukemias, regulates the growth in breast cancers.Citation21,Citation22 Recently, some studies have demonstrated that MDM2 overexpression was related to cancer invasiveness and metastasis.Citation23 In recent studies, MDM2 enhances cell mobility and invasion through MDM2-mediated Slug degradation, causes its E3 ubiquitin ligase activity.Citation24,Citation25 In another study, MDM2 promotes migration and invasion through a p53 independent manner instead of depending on its ability as an E3 ubiquitin ligase. However, the exact role of MDM2 in cell metastasis is still unclear.Citation26 Therefore, it is important to find key molecules and molecular mechanisms which regulate breast cancer metastasis and explore new promising therapeutic targets.
Combretastatins are isolated from the South African willow tree Combretum caffrum. Among all combretastatins, Combretastatin A-4 (CA-4) is the most effective with tubulin-binding ability and cytotoxicity ().Citation27 CA-4 is also a vascular-disrupting agent (VDA)Citation28–Citation30 and has an anti-metastatic activity.Citation31 However, CA-4 is unstable and easily isomerized to inactive form. SQ is a novel synthesized CA-4 analogue which hybridizes with organoselenium specifically ().Citation32 In our previous study, SQ has been shown as a promising microtubule destabilizing agent, which can induce apoptosis in human breast cancer cells through p53-independent manner. Besides, SQ directly induces MDM2 degradation through an ubiquitin-proteasome system. Moreover, the apoptosis induced by SQ is associated with the inhibition of MDM2.Citation32,Citation33 In this current study, the effect of SQ on cell migration and invasion and its underlying mechanisms were investigated. We found that SQ suppressed EGF-induced invasion and motility in breast cancer cells by inhibiting MDM2 and down-regulating Twist1 expression.
Materials and methods
Reagents
Compound SQ was synthesized and purified by our group as described previously.Citation33 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT), 4ʹ,6-diamidino-2-phenylindole (DAPI) and EGF were purchased from Sigma-Aldrich (St. Louis, MO, USA). Crystal violet and Lipo6000 tansfection reagent were purchased from Beyotime Biotechnology (Shanghai, China). Antibodies against MDM2, GAPDH, HRP-conjugated goat anti-rabbit IgG and FITC-conjugated goat anti-rabbit IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). E-Cadherin, N-cadherin, Vimentin, FAK, MMP-2, MMP-9, p53, Snail1, Slug and Twist1 antibodies were obtained from Proteintech Group (Chicago, IL, USA).
Cell culture
Human breast cancer cells MCF-7 and MDA-MB-231 were obtained from American Type Culture Collection (Manassas, VA, USA) and grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% streptomycin and 1% penicillin at 37 ºC in humidified incubator under an atmosphere with 5% CO2. MCF-7 and MDA-MB-231 cells were tested monthly for mycoplasma by the GMyc-PCR Mycoplasma Test assay (Yeasen Biotech, Shanghai), and all the cell lines using in our experiment are negative.
Cell viability assay
Cell viability was assessed by the MTT assay. This assay was performed as described previously.Citation33
Small interfering RNA (sirna)
MDM2 siRNA #1 (5ʹ-GCCACAAAUCUGAUAGUAU-3ʹ) and MDM2 siRNA #2 (5ʹ-AGAUCCUGCUGCUUUCGCAUU-3ʹ); scrambled siRNA #1 (5ʹ-GGACGCAUCCUUCUUAAUU-3ʹ) and scrambled siRNA #2 (5ʹ-UUCUUCGAACGUGUCACGUTT-3ʹ) were synthesized by GenePharma (Suzhou, China). Cells were cultured at 50% confluency and transfected with siRNA using Lipo6000 reagent followed the manufacturer’s procedure.
Plasmid construction and transfection
The expression vector for human MDM2 (pGEX4T2-MDM2) was obtained from Addgene (Cambridge, USA). Full-length cDNA of MDM2 was generated by PCR amplification with the primers derived from human MDM2 (5ʹ-CCGGAATTCGCCACCATGTGCAATACCAACAT-3ʹ and 5ʹ-CGCGGATCCCGGGGGAAATAAGTTAGCAC-3ʹ). The PCR products with BamHI/EcoRI were subcloned into eukaryotic pcDNA3.1 expression vector. The resulted MDM2 plasmid was verified by DNA sequencing. For stable transfection, the empty vector or MDM2 plasmid was transfected into MCF-7 cells with Lipo6000 Reagent. After transfected for 24 h, G418 (Life technologies, MA USA) was added to select stable transfected cell lines.
Cell adhesion assay
Cells were pretreated with EGF or the combination with SQ (25 nM and 50 nM) for 24 h and then plated at 2 × 104 per well of collagen I coated 96-well plates (Becton Dickinson, Bedford, USA), followed by incubation for 30 min. Adherent cells were fixed with in 4% paraformaldehyde and stained with 0.2% crystal violet. Then the adherent cells were counted and imaged with an inverted microscope (Olympus IX51).
Wound healing assay
Cells were seeded in 6-well plates and generated confluent cell monolayers. The wound was made across the monolayer by a sterilized pipette tip. Wound closure was measured at 0 and 24 h within scrape line and photographed by an inverted microscope. Quantification of relative wound healing area was calculated using the image J software.
Cell migration and invasion assay
2 × 104 cells suspended in 200 μL serum-free medium were seeded into the upper chamber of Transwell insert (Corning, MA, USA) for the migration assay, while the upper chamber was coated with 50 μl matrix gel for the invasion assay. The plates were incubated for 24 h for migration and 48 h for invasion, the membranes of chamber were then fixed with 4% paraformaldehyde and stained with 0.2% crystal violet. Cells were counted and imaged with an inverted microscope.
Quantitative real-time PCR
MDM2 mRNA level was quantified by real-time PCR as described previously Citation33. The primer sequences were as follows: MDM2, 5ʹ-ATTGGTTGTCTCATACTGG-3ʹ and 5ʹ-CTCAACACAAGCTGAAGAGG-3ʹ; GAPDH, 5ʹ-CAATGACCCCTTCATTGACC-3ʹ and 5ʹ-TGGAAGATGGTGATGGGATT-3ʹ. GAPDH was set as a control.
Immunofluorescence
Cells were cultured on a 6-well plate with cover glass, then treated with EGF or the combination with SQ (25 nM) for 24 h. Immunofluorescence staining was performed as described previously to determine E-cadherin and Vimentin.Citation33
Gelatin zymography
Cells were seeded in 6-well plates and then treated with EGF or EGF combined with SQ for 24 h. The culture supernatants were collected and centrifuged to remove cells and debris. Equal amount of samples were run in 7.5% SDS-polyacrylamide gels containing gelatin. After electrophoresis, the gels were washed and incubated with reaction buffer at 37 ̊C for 48 h. The gels were next stained in 0.5% Coomassie Brilliant Blue solution and destained in 40% methanol and 10% acetic acid solution.
Western blot
Western blot was determined as described previously.Citation33 Densitometry analysis was performed using Image J software.
Statistical analysis
Data values were shown as mean ± SD from three independent experiments. Statistical differences were assessed by one-way ANOVA using SPSS software 16.0, as appropriate. P < 0.05 was considered statistically significant.
Results
SQ inhibits EGF-induced EMT
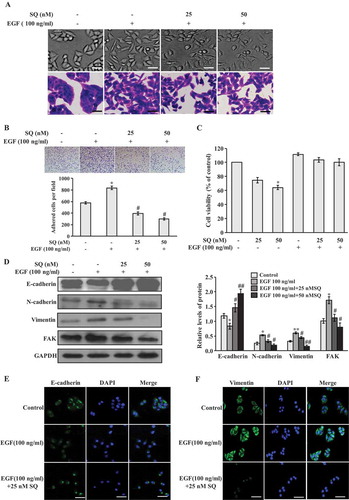
It has been shown that EGF is one of the potent breast cancer motility factors and it can promote metastasis.Citation11,Citation34 In this study, cellular morphological changes of MCF-7 cells under EGF treatment were investigated in the presence or absence of SQ. As shown in , cells formed a typical compact cell-to-cell contact in the control cells. While cells acquire obvious mesenchymal phenotype changes after EGF treatment for 24 h, such as losing cell-to-cell contact, increasing cell scattering and elongating spindle shape. However, SQ (25 nM and 50 nM) treatment mitigated EGF-induced mesenchymal phenotype changes, and the cells exhibited a classical epithelial-like morphology. We also found that EGF increased the adhesion ability, and this increased adhesion ability could be suppressed by SQ (). Moreover, SQ at the concentration of 25 nM and 50 nM has no significant influence on cell proliferation under EGF-treated conditions (). In addition, EGF also caused to cell-to-cell contact decrease and enhanced cell scattering in MDA-MB-231 cells. However, SQ could inhibit cell scattering and mesenchymal phenotype changes at the doses of 5 and 10 nM that do not affect cell proliferation under EGF treatment in MDA-MB-231 cells (supplementary Figure 1a and b). Taken together, these morphological changes indicate the efficacy of SQ in inhibiting EGF-induced EMT. Then, we examined the expressions of EMT-related markers to further clarify whether SQ inhibited EGF-induced EMT. As shown in and supplementary Figure 1c, the expression level of epithelial marker E-cadherin was decreased, the expression levels of mesenchymal marker N-cadherin and Vimentin were increased when cells were treated with EGF in MCF-7 and MDA-MB-231 cells. Moreover, SQ treatment significantly reversed EGF-induced down-regulation of E-cadherin and suppressed EGF-induced the expression level increase of N-cadherin and Vimentin ( and supplementary Figure 1c) in MCF-7 and MDA-MB-231 cells. In addition, Focal adhesion kinase (FAK), a critical mediator of integrin and growth factor receptor signaling pathway, promotes cancer cell adhesion, invasion and metastasis.Citation35 Results from western blot () showed that the treatment with SQ could attenuate EGF-induced FAK expression in MCF-7 cells. Furthermore, the immunofluorescence analysis proved that SQ significantly reversed EGF-downregulated E-cadherin expression increase () and decreased the EGF-induced Vimentin expression increase (). These results demonstrate that EGF induces the occurrence of EMT, and SQ reverses the progress of EGF-induced EMT.
Figure 2. SQ inhibits EGF-induced EMT in MCF-7 cells. (A) Effect of SQ on morphological changes of EGF-induced cells. Cells were treated with EGF (100 ng/ml) or EGF (100 ng/ml) plus SQ (25 or 50 nM) for 24 h. Scale bar = 50 μm. (B) Effect of SQ on EGF-induced adhesion. (C) Effects of SQ on cell viability. Cells were treated with SQ (25 or 50 nM), EGF (100 ng/ml) alone or the combination with SQ at the concentration of 25 or 50 nM for 24 h. (D) EMT-related proteins were determined by western blot. (E, F) Immunofluorescence analysis of E-cadherin and Vimentin. Scale bar = 50 μm. *P < 0.05, ** P < 0.01 vs control; # P < 0.05, ## P < 0.01 vs the cells only treated with 100 ng/ml EGF.(A).
SQ inhibits EGF-induced migration and invasion
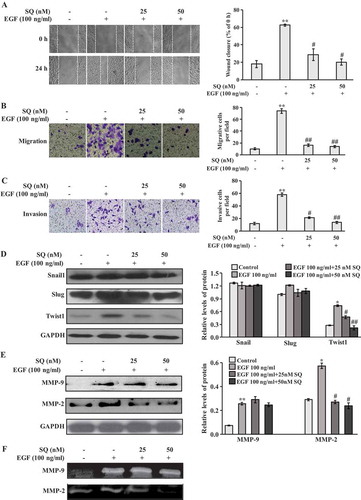
As EMT contributes to tumor cells acquiring migrative and invasive properties,Citation4–Citation6 we determined the effects of SQ on EGF-induced motility and invasion in breast cancer cells. In the wound healing assay, EGF (100 ng/ml) promoted cell motility when compared with the control. Importantly, SQ effectively suppressed EGF-induced motility in MCF-7 and MDA-MB-231 cells ( and supplementary Figure 1d). Moreover, inhibitory effects of SQ on the EGF-induced invasion and migration were detected by the transwell assay. As illustrated in , , supplementary Figure 1e and 1f, under the treatment with EGF, migrative and invasive cells were significantly increased. Notably, SQ reversed the EGF-induced motility and invasion significantly in MCF-7 and MDA-MB-231 cells. Since EMT-related transcription factors such as Snail1, Slug, and Twist1 are usually activated in the process of migration and invasion,Citation8,Citation36 we determined the key EMT-transcriptional inducers to investigate the potential molecular mechanism of SQ on EGF-induced invasion and migration. Our study found that EGF significantly enhanced the expression of Twist1, and Twist1 expression was decreased by the treatment with SQ ( and supplementary Figure 1c). However, the expressions of Snail1 and Slug had almost no change after EGF or EGF plus SQ treatment (). In addition, MMP-2 and MMP-9 are involved in metastasis through the degradation of ECM, which could be regulated by Twist1 signaling.Citation10,Citation12 As a result, western blot and gelatin zymography were performed to assess the protein levels and activities of MMP-2 and MMP-9 in EGF-induced cells. The results showed that SQ inhibited both the protein expression level and activity of EGF-induced MMP-2. However, EGF-induced MMP-9 level was not significantly affected by SQ treatment ( and ).
Figure 3. SQ inhibits EGF-induced migration and invasion in MCF-7 cells. (A) Cell motility was assessed by wound healing assay. Quantitative analyses of the percentage of wound closure at 24 h compared with 0 h were shown on the right. (B, C) Cell migrative and invasive capacities were determined by the transwell assay. Representative images of migrative and invasive cells were shown on the left and the quantitative analyses of migrative and invasive cells per field were shown on the right. (D) Western blots showed the expressions of Snail1, Slug and Twist1. (E) The expression levels of MMP-2 and MMP-9 were detected by western blot. (F) MMP-2 and MMP-9 activities were evaluated by gelatin zymography. * P < 0.05, ** P < 0.01 vs control; # P < 0.05, ## P < 0.01 vs the cells only treated with 100 ng/ml EGF.
SQ inhibits egf-induced MDM2 expression
Recent studies have suggested the linkage between MDM2 expression and the increasing potential of motility, invasion and metastasis.Citation26,Citation37 In our previous studies, SQ inhibited MDM2 expression by a p53-independent manner.Citation32,Citation33 We hypothesized that the regulation of MDM2 was involved in the inhibitory effect of SQ on EGF-induced migration and invasion. As we suspected, the protein and mRNA levels of MDM2 were obviously increased in the present of EGF analyzed by western blot and real-time PCR. However, SQ significantly suppressed EGF-induced MDM2 expression increase ( and ). In addition, EGF also increased the protein and mRNA levels of MDM2 in another breast cancer cell line MDA-MB-231. Furthermore, SQ at the concentration of 5 nM and 10 nM inhibited EGF-induced high expression of MDM2 in MDA-MB-231 cells ( and ). In addition, the total p53 expression was decreased in the presence of EGF, and SQ could not reverse the total p53 expression significantly (supplementary Figure 2).
Figure 4. SQ inhibits EGF-induced expression of MDM2. (A, B) The effects of SQ on EGF-induced MDM2 expression increase were assessed by western blot and real-time PCR in MCF-7 cells. (C, D) The protein and mRNA levels of MDM2 were determined by western blot and real-time PCR in MDA-MB-231 cells. * P < 0.05, ** P < 0.01 vs control; # P < 0.05, ## P < 0.01 vs the cells only treated with 100 ng/ml EGF.
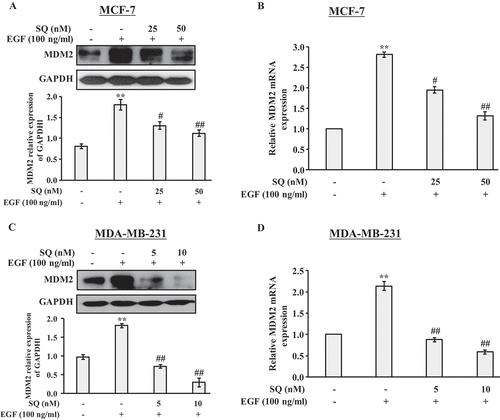
MDM2 overexpression promotes cell migration and invasion
To further confirm our hypothesis, the empty vector and MDM2 plasmid were transfected into MCF-7 cells. The increased protein expression of MDM2 in the stable MDM2-overexpressing cells was shown in . Since we found that Twist1 was participated in the inhibitory effects of SQ on EGF-induced migration and invasion (), we explored the expression of Twsit1 in MDM2-overexpressing cells. The western blot results showed that Twist1 expression was dramatically enhanced followed by MDM2 overexpression (). Additionally, SQ (25 and 50 nM) decreased the expression of MDM2 and Twist1 significantly in MDM2-overexpressing cells. Moreover, wound healing assay showed that MDM2 overexpression promoted cell motility compared with the control. However, SQ (25 and 50 nM) significantly suppressed cell motility in MDM2-overexpressing cells (). Moreover, the effects of MDM2 overexpression on invasion and migration were also determined by the transwell assays. In stable transfected cell lines, MDM2 overexpression promoted migration and invasion ( and ) compared with the control group. However, SQ (25 and 50 nM) significantly reduced migration and invasion ( and ) in MDM2-overexpressing cells.
Figure 5. Up-regulation of MDM2 promotes migration and invasion. (A) Western blot was performed to detect the expression of MDM2 in MCF-7 cells stably transfected with MDM2 plasmid and the empty vector. (B) The MDM2-overexpressing cells were treated with SQ (25 and 50 nM) for 24 h. MDM2 and Twist1 expression levels were detected by western blot. (C) Cell motility was determined by wound healing assay. (D, E) Migrated and invasive cells were analyzed by transwell assay. * P < 0.05, ** P < 0.01 vs the empty vector; # P < 0.05, ## P < 0.01 vs MDM2 plasmid.
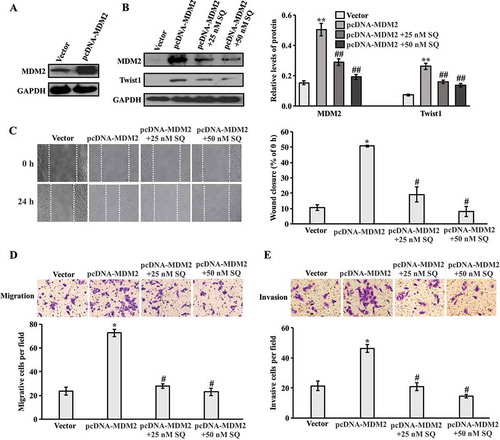
Effect of knockdown MDM2 on migration and invasion
To further explore the role of MDM2 in invasion and migration, MCF-7 cells were transfected with MDM2 siRNA. The decreased expression of MDM2 was proved by western blot ( and supplementary Figure 3a). Furthermore, in the presence of EGF, the expression of MDM2 was down-regulated by siRNA transfection, which resulted in reduction of Twist1 expression ( and supplementary Figure 3b). Down-regulation of MDM2 and Twist1 expression through the treatment with SQ in siRNA tranfected cells were more significant compared with MDM2 siRNA transfected cells in presence of EGF ( and supplementary Figure 3b). For the wound healing assay, the delayed wound closure was detected in cells transfected with MDM2 siRNA compared with scrambled siRNA in the presence of EGF ( and supplementary Figure 3c). The transwell assay was then performed to investigate migration and invasion in siRNA-transfected cell lines. As shown in , , supplementary Figure 3d and 3e, the migrated and invasive cells were decreased in MDM2 siRNA-transfected cells compared with scrambled siRNA-transfected cells in the presence of EGF. Moreover, the addition of SQ significantly inhibited cell motility and invasion in EGF co-existed MDM2 siRNA-transfected cells. Taken together, our data suggests that MDM2 significantly regulated the expression of Twist1, which was involved in the inhibitory effect of SQ on EGF-induced migration and invasion.
Figure 6. Effect of MDM2 knockdown on migration and invasion. (A) Western blot analysis for the expression of MDM2 in MCF-7 cells transfected with MDM2 siRNA and scrambled siRNA. (B) MCF-7 Cells were transfected with MDM2 siRNA for 24 h, and incubated with SQ (25 nM) for another 24 h in the presence of EGF (100 ng/ml). The expression levels of MDM2 and Twist1 were analyzed by western blot. (C) Wound closure area was photographed and measured. (D, E) Migrated and invasive cells were analyzed by transwell assay. * P < 0.05, ** P < 0.01 vs scrambled siRNA.
Discussion
Cancer metastasis is the most common cause of the death in breast cancers.Citation1,Citation2 Several studies have elucidated that EGF or a combination of other growth factors can induce EMT in breast cancer cells.Citation10,Citation34 The current study found that EGF stimulated motility and invasion, characterized by the regulation of EMT-related markers such as E-cadherin, N-cadherin, Vimentin and increase of the expression levels of MMP-2/9 and FAK in MCF-7 cells. Moreover, EGF significantly enhanced the expression of a EMT-related key transcriptional factor Twist1, but not Snail and Slug. In addition, we also found that EGF promoted cell migration and invasion in MDA-MB-231 cells, accompanied by up-regulation of N-cadherin and Vimentin expressions, and down-regulation of E-cadherin expression. Besides, the transcriptional factor Twist1 was also enhanced by EGF treatment in MDA-MB-231 cells.
MDM2 is an oncogene that was first cloned in a murine double-minute chromosome in a tumorigenic mouse 3T3DM cells.Citation38 MDM2 contains a N-terminal p53-binding domain and a ring finger domain which possesses E3 ligase activity, catalyzing the proteasomal degradation of p53 to promote tumor malignancy.Citation37,Citation39–Citation43 Here, we found that EGF strongly increased the expression of MDM2 at the mRNA and protein levels in human breast MCF-7 and MDA-MB-231 cells. Meanwhile, we demonstrated that the overexpression of MDM2 can significantly increase the capability of cell invasion and migration. Furthermore, the down-regulation of MDM2 after transfection with MDM2 siRNA reduced the movement and invasive ability compared with the control under the treatment with EGF.
Some studies indicate that the expression of transcription factor Twist1 in breast cancers could significantly enhance EMT, cell invasion and metastasis.Citation12,Citation13,Citation44–Citation48 In addition, several studies have shown that Twist1 enhanced MDM2-mediated p53 degradation, which aggravated the malignancy of human sarcoma cells.Citation49,Citation50 Recent study has shown that the expression of MDM2 enhanced migration and invasiveness in cells which were independent of the intact MDM2 ring-finger domain and p53.Citation26 In this study, MDM2 tended to induce the transcription factor Twist1 expression, which was demonstrated by overexpression and silencing of endogenous MDM2, thereby affected cell motility and invasion. However, the relationship between MDM2 and Twist1 requires further study.
Our previous work has demonstrated that SQ, a novel microtubule-destabilizing agent, strongly suppressed tumor proliferation and induced apoptosis, which was associated with MDM2 inhibition.Citation32,Citation33 In this study, we further identified that SQ reversed EGF-induced invasion and migration through the inhibition of MDM2 expression in MCF-7 and MDA-MB-231 cells. In addition, the treatment of SQ significantly inhibited EGF-enhanced key EMT-transcriptional factor Twist1 expression, but had almost no effect on Snail and Slug expression. Taken together, SQ might down-regulate Twist1 through inhibiting MDM2, and ultimately suppress EGF-induced motility and invasion. On the other hand, it was reported that Erk or PI3K signaling may play a role in EGF-induced breast cancer metastasis.Citation11,Citation14,Citation15 As a result, we cannot rule out the involvement of Erk or PI3K signaling in the present study, which will be investigated in the future study. Interestingly, SQ inhibited the high MMP-2 expression induced by EGF, but had little effect on MMP-9 expression increase, which indicates that MMP-2 might be involved in the inhibition of cell migration and invasion induced by SQ treatment in MCF-7 cells.
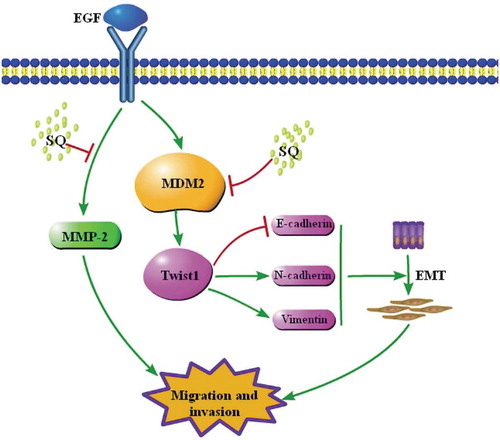
In summary, our study suggests that SQ inhibited EGF-induced migration and invasion by down-regulating the expressions of MDM2 and EMT-related proteins. MDM2-regulated Twist1 signaling pathway might be involved in the inhibitory effects of SQ on migration and invasion, which suggests that SQ is a potential anti-cancer metastasis agent for breast cancer therapy (). These results also provide further evidence for the development of compound SQ as a pharmacological agent. Meanwhile, the axis of MDM2/Twist1 signaling could be a potential drug target for breast cancer treatment.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi:10.3322/caac.21262.
- Weigelt B, Peterse JL, van ‘T Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cance. 2005;5:591–602. doi:10.1038/nrc1670.
- Guan X. Cancer metastases: challenges and opportunities. Acta Pharm Sin B. 2015;5:402–418. doi:10.1016/j.apsb.2015.07.005.
- Kang Y, Pantel K. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer Cell. 2013;23:573–581. doi:10.1016/j.ccr.2013.04.017.
- Welch DR, Steeg PS, Rinker-Schaeffer CW. Molecular biology of breast cancer metastasis. Genetic regulation of human breast carcinoma metastasis. Breast Cancer Res. 2000;2:408–416.
- Radisky DC. Epithelial-mesenchymal transition. J Cell Sci. 2005;118:4325–4326. doi:10.1242/jcs.02552.
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi:10.1038/nrm1835.
- Bartis D, Mise N, Mahida RY, Eickelberg O, Thickett DR. Epithelial-mesenchymal transition in lung development and disease: does it exist and is it important? Thorax. 2014;69:760–765. doi:10.1136/thoraxjnl-2013-204608.
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi:10.1158/0008-5472.CAN-07-0575.
- Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei Y, Abbruzzese JL, Hortobagyi GN, Hung M-C. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–9076. doi:10.1158/0008-5472.CAN-07-0575.
- Olsen DA, Bechmann T, Ostergaard B, Wamberg PA, Jakobsen EH, Brandslund I. Increased concentrations of growth factors and activation of the EGFR system in breast cancer. Clin Chem Lab Med. 2012;50:1809–1818. doi:10.1515/cclm-2011-0823.
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi:10.1016/j.cell.2004.06.006.
- Fu J, Qin L, He T, Qin J, Hong J, Wong J, Liao L, Xu J. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011;21:275–289. doi:10.1038/cr.2010.118.
- Ellerbroek SM, Halbleib JM, Benavidez M, Warmka JK, Wattenberg EV, Stack MS, Hudson LG. Phosphatidylinositol 3-kinase activity in epidermal growth factor-stimulated matrix metalloproteinase-9 production and cell surface association. Cancer Res. 2001;61:1855–1861. PMID: 11280738.
- Grunert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol. 2003;4:657–665. doi:10.1038/nrm1175.
- Sun T, Zhao N, Zhao XL, Gu Q, Zhang SW, Che N, Wang X-H, Du J, Liu Y-X, Sun B-C. Expression and functional significance of Twist1 in hepatocellular carcinoma: its role in vasculogenic mimicry. Hepatology (Baltimore, Md). 2010;51:545–556. doi:10.1002/hep.23311.
- Eckert MA, Lwin TM, Chang AT, Kim J, Danis E, Ohno-Machado L, Yang J. Twist1-induced invadopodia formation promotes tumor metastasis. Cancer Cell. 2011;19:372–386. doi:10.1016/j.ccr.2011.01.036.
- Zheng H, Takahashi H, Murai Y, Cui Z, Nomoto K, Niwa H, Tsuneyama K, Takano Y. Expressions of MMP-2, MMP-9 and VEGF are closely linked to growth, invasion, metastasis and angiogenesis of gastric carcinoma. Anticancer Res. 2006;26:3579–3583. PMID: 17094486.
- Chandler S, Miller KM, Clements JM, Lury J, Corkill D, Anthony DC, Adams SE, Gearing AJ. Matrix metalloproteinases, tumor necrosis factor and multiple sclerosis: an overview. J Neuroimmunol. 1997;72:155–161. PMID: 9042108.
- Overall CM, Lopez-Otin C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat Rev Cance. 2002;2:657–672. doi:10.1038/nrc884.
- Turbin DA, Cheang MC, Bajdik CD, Gelmon KA, Yorida E, De Luca A, Nielsen TO,Huntsman DG, Gilks CB. MDM2 protein expression is a negative prognostic marker in breast carcinoma. Modern pathology: an official journal of the United States and Canadian. Acad Pathology. 2006;19:69–74. doi:10.1038/modpathol.3800484.
- Rayburn E, Zhang R, He J, Wang H. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets. 2005;5:27–41. PMID: 15720187.
- Rayburn ER, Ezell SJ, Zhang R. Recent advances in validating MDM2 as a cancer target. Anticancer Agents Med Chem. 2009;9:882–903. PMID: 19538162.
- Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, Yuan A, Lin C-W, Yang S-C, Chan W-K, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11:694–704. doi:10.1038/ncb1875.
- Lin TY, Hsu HY. Ling Zhi-8 reduces lung cancer mobility and metastasis through disruption of focal adhesion and induction of MDM2-mediated Slug degradation. Cancer Lett. 2016;375:340–348. doi:10.1016/j.canlet.2016.03.018.
- Polanski R, Warburton HE, Ray-Sinha A, Devling T, Pakula H, Rubbi CP, Vlatković N, Boyd MT. MDM2 promotes cell motility and invasiveness through a RING-finger independent mechanism. FEBS Lett. 2010;584:4695–4702. doi:10.1016/j.febslet.2010.10.049.
- Pettit GR, Singh SB, Hamel E, Lin CM, Alberts DS, Garcia-Kendall D. Isolation and structure of the strong cell growth and tubulin inhibitor combretastatin A-4. Experientia. 1989;45:209–211. PMID: 2920809.
- Kanthou C, Tozer GM. Tumour targeting by microtubule-depolymerizing vascular disrupting agents. Expert Opin Ther Targets. 2007;11:1443–1457. doi:10.1517/14728222.11.11.1443.
- Tozer GM, Kanthou C, Baguley BC. Disrupting tumour blood vessels. Nat Rev Cance. 2005;5:423–435. doi:10.1038/nrc1628.
- Kanthou C, Tozer GM. Microtubule depolymerizing vascular disrupting agents: novel therapeutic agents for oncology and other pathologies. Int J Exp Pathol. 2009;90:284–294. doi:10.1111/j.1365-2613.2009.00651.x.
- Mahal K, Biersack B, Caysa H, Schobert R, Mueller T. Combretastatin A-4 derived imidazoles show cytotoxic, antivascular, and antimetastatic effects based on cytoskeletal reorganisation. Invest New Drugs. 2015;33:541–554. doi:10.1007/s10637-015-0215-9.
- Xu J, Zuo D, Qi H, Shen Q, Bai Z, Han M, Li Z, Zhang W, Wu Y. 2-Methoxy-5((3,4,5-trimethosyphenyl)seleninyl) phenol (SQ0814061), a novel microtubule inhibitor, evokes G2/M cell cycle arrest and apoptosis in human breast cancer cells. Biomed Pharmacother. 2016;78:308–321. doi:10.1016/j.biopha.2016.01.040.
- Xu J, Han M, Shen J, Guan Q, Bai Z, Lang B, Zhang H, Li Z, Zuo D, Zhang W, et al. 2-Methoxy-5((3,4,5-trimethosyphenyl)seleninyl) phenol inhibits MDM2 and induces apoptosis in breast cancer cells through a p53-independent pathway. Cancer Lett. 2016;383:9–17. doi:10.1016/j.canlet.2016.09.011.
- Hardy KM, Booth BW, Hendrix MJ, Salomon DS, Strizzi L. ErbB/EGF signaling and EMT in mammary development and breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:191–199. doi:10.1007/s10911-010-9172-2.
- Lee BY, Timpson P, Horvath LG, Daly RJ. FAK signaling in human cancer as a target for therapeutics. Pharmacol Ther. 2015;146:132–149. doi:10.1016/j.pharmthera.2014.10.001.
- Sun L, Fang J. Epigenetic regulation of epithelial-mesenchymal transition. Cell Mol Life Sci. 2016;73:4493–4515. doi:10.1007/s00018-016-2303-1.
- Thut CJ, Goodrich JA, Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–1986. PMID: 9271120. doi:10.1101/gad.11.15.1974.
- Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569–573. doi:10.1038/35023507.
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi:10.1038/387296a0.
- Uhrinova S, Uhrin D, Powers H, Watt K, Zheleva D, Fischer P, McInnes C, Barlow PN. Structure of free MDM2 N-terminal domain reveals conformational adjustments that accompany p53-binding. J Mol Biol. 2005;350:587–598. doi:10.1016/j.jmb.2005.05.010.
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science (New York, NY). 2004;303:844–848. PMID: 18051440. doi:10.1126/science.1092472.
- Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res. 2003;1:993–1000.
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. PMID: 9450543.
- Soini Y, Tuhkanen H, Sironen R, Virtanen I, Kataja V, Auvinen P, Mannermaa A, Kosma V-M. Transcription factors zeb1, twist and snai1 in breast carcinoma. BMC Cancer. 2011;11:73. doi:10.1186/1471-2407-11-73.
- Qin L, Liu Z, Chen H, Xu J. The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Res. 2009;69:3819–3827. doi:10.1158/0008-5472.CAN-08-4389.
- Vernon AE, LaBonne C. Tumor metastasis: a new twist on epithelial-mesenchymal transitions. Curr Biol. 2004;14:R719–21. doi:10.1016/j.cub.2004.08.048.
- Karreth F, Tuveson DA. Twist induces an epithelial-mesenchymal transition to facilitate tumor metastasis. Cancer Biol Ther. 2004;3:1058–1059. PMID: 15640618. doi:10.4161/cbt.3.11.1302.
- Yang J, Mani SA, Weinberg RA. Exploring a new twist on tumor metastasis. Cancer Res. 2006;66:4549–4552. doi:10.1158/0008-5472.CAN-05-3850.
- Piccinin S, Tonin E, Sessa S, Demontis S, Rossi S, Pecciarini L, Zanatta L, Pivetta F, Grizzo A, Sonego M, et al. A “twist box” code of p53 inactivation: twist box: p53 interaction promotes p53 degradation. Cancer Cell. 2012;22:404–415. doi:10.1016/j.ccr.2012.08.003.
- Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 2012;22:90–106. doi:10.1038/cr.2011.144.