ABSTRACT
Background: Leptomeningeal metastases (LM), associated with poor prognosis, are frequent complications of advanced non-small cell lung cancer (NSCLC) patients, especially in patients with epidermal growth factor receptor (EGFR) mutations. Due to limited access to leptomeningeal lesions, the mutational landscape of LM has not been comprehensively investigated in large cohorts and the underlining biology of LM remains elusive. Some studies have explored the potential of cerebrospinal fluid (CSF) in reflecting the molecular profile of LM but with limited number of patients enrolled.
Methods: In this study, we performed capture-based targeted sequencing using a panel consisting of 168 lung cancer-related genes on matched CSF and plasma samples from 72 advanced NSCLC patients with confirmed LM to interrogate the potential of CSF as a source of liquid biopsy.
Results: We revealed a rate of detection of 81.5% and 62.5% for CSF and plasma, respectively (p = 0.008). The maximum allelic fraction (MaxAF) was also significantly higher in CSF (43.6% vs. 4.6%) (p < 0.001). CSF, harboring a unique genomic profile by having a significant number of CSF-specific mutations, primarily copy number variations, is superior to plasma in reflecting the mutational profile of LM. Further pathway enrichment analysis revealed that most of CSF-specific mutations participated in pathways relevant to the tumorigenesis and the development of metastases. Moreover, our data also revealed that TP53 loss of heterozygosity (LOH) predominantly existed in CSF (p < 0.001).
Conclusions: Collectively, we demonstrated that CSF provides a more comprehensive profile of LM than plasma in a large cohort, thus can be used as an alternative source of liquid biopsy for LM patients.
Introduction
Leptomeningeal metastases (LM), a form of central nervous system (CNS) metastases, associated with high mortality, occurring in 3–5% of advanced non-small cell lung cancer (NSCLC) patients and 9–10% of epidermal growth factor receptor (EGFR)-mutant patients.Citation1,Citation2 LM often represents a terminal event of NSCLC, with a historical (pre-approval of contemporary systemic treatment) median survival of 1–3 months to 3–11 months with radiotherapy or chemotherapy.Citation3–Citation5 EGFR-tyrosine kinase inhibitors (TKIs) such as erlotinib, osimertinib and AZD3759,Citation6–Citation8 has been used as a targeted therapy of LM from EGFR mutation-positive NSCLC, and markedly prolong survival.Citation9 However, the aggressive nature and increasing prevalence highlight the need to understand the underlying mechanisms of LM, which remains a devastating complication due to the difficulties in accessing leptomeningeal lesions.Citation10–Citation12
Multiple alternative sampling sources have been investigated to represent the genomic profile of LM. Numerous studies have shown circulating tumor DNA (ctDNA) derived from plasma can be used as a surrogate for reflecting the genomic profile of solid tumors as well as for diagnosis, disease monitoring and identification of resistant mechanisms.Citation13 However, only low amounts of ctDNA originating from brain tumors are present in the plasma due to the blood-brain barrier (BBB).Citation14 Recently multiple studies have demonstrated the ability of cerebrospinal fluid (CSF), circulating through the central nervous system (CNS), to recapitulate the genomic profile of brain lesions.Citation15,Citation16 Moreover, cfDNA in CSF better represents the genomic landscape of brain tumors than plasma, thus may serve as a liquid biopsy to monitor brain tumor evolution.Citation16–Citation18 However, only a very limited number of patients with NSCLC were included in these studies, and little is known about the genomic aberrations during LM development in NSCLC patients and the signaling pathways implicated in this metastasis process.Citation12,Citation16,Citation18
In this study, we performed capture-based targeted sequencing on matched CSF and plasma samples from 72 patients with confirmed LM to evaluate the ability of both liquid biopsy media in repopulating the genomic profile of LM. Moreover, we performed pathway analysis to further analyze singling pathways participated in this metastatic process.
Materials and methods
Patients and sample collection
Ninety-two NSCLC adenocarcinoma patients diagnosed with LM at any of the participating hospitals: Taizhou First People’s Hospital, Taizhou University Hospital, Nantong University Affiliated Hospital or Second Affiliated Hospital of Fujian Medical University between February 2016 and April 2018 were enrolled in this study. The diagnosis criteria of LM included 1) a positive brain MRI imaging, characterized by linear or micronodularpial enhancement 2) a positive CSF cytology test. Ten milliliter CSF was obtained from 92 patients by lumbar puncture and 10 mL of plasma was collected from 72 of them. All patients provided informed consent and the study was approved by the Research Ethics Committee of Taizhou University Hospital.
DNA extraction
CSF and plasma cell-free DNA (cfDNA) were extracted with the QIAamp Circulating Nucleic Acid Kit (Qiagen). DNA was quantified using the Qubit dsDNA assay (Life Technologies).
Capture-based targeted DNA sequencing
CSF and plasma samples were subjected to capture-based targeted sequencing using a panel consisting of 168 lung cancer-related genes. cfDNA fragments were selected by bead (Agencourt AMPure XP Kit, Beckman Coulter, California, US) followed by hybridization with capture probes baits, hybrid selection with magnetic beads and PCR amplification. A bioanalyzer high-sensitivity DNA assay was then performed to assess the quality and size of the fragments and indexed samples were sequenced on Nextseq500 sequencer (Illumina, Inc., California, US) with pair-end reads.
Sequence data analysis
Burrows-Wheeler aligner 0.7.10Citation19 was used for mapping the pair-end reads to the human genome (hg19). Local alignment optimization, variant calling and annotation were performed using GATK 3.2, MuTect, and VarScan. DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3. Variants were filtered using the VarScan filter pipeline. Loci with depth less than 100 were filtered out. White blood cells were sequenced to filter out germline mutations. At least 2 and 5 supporting reads were needed for INDELs in plasma and CSF samples, respectively; while 8 supporting reads were needed for SNVs to be called in both plasma and CSF samples. According to the ExAC, 1000 Genomes, dbSNP, ESP6500SI-V2 database, variants with population frequency over 0.1% were grouped as SNP and excluded from further analysis. Remaining variants were annotated with ANNOVAR and SnpEff v3.6. DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3.
Copy number variation was detected by in-house analysis scripts based on depth of coverage data of capture intervals. Coverage data was corrected against sequencing bias resulting from GC content and probe design. The average coverage of all captured regions was utilized to normalize the coverage of different samples to comparable scales. Copy number was calculated based on the ratio between depth of coverage in tumor samples and average coverage of an adequate number (n > 50) of samples without copy number variation as references as to each capture interval. Copy number variation is called if the coverage data of the gene region was quantitatively and statistically significantly different from its reference control. The limit of detection for CNVs is 1.5 for deletion and 2.64 for amplification.
Results
Patient characteristics
This study enrolled 92 advanced Chinese lung adenocarcinoma patients diagnosed with LM. The median age was 53.9 years old (range, 26–78 years). Among them, 53 patients were male and 39 were females. Cytology analysis was performed on all CSF samples and adenocarcinoma cells were found in 51 patients. Sixty-three patients had typical imaging for LM judged by 2 independent radiologists. Ten patients were treatment-naïve and diagnosed with LM at their initial diagnosis. The remaining 82 patients developed LM during treatment. 58 cases carried EGFR mutations, including 25 cases with EGFR 19 del, 26 cases with EGFR L858R, 5 cases with EGFR exon 20 insertions and 27 cases with other EGFR mutations.
CSF was superior to plasma in reflecting the genomic profile of brain metastases
We performed capture-based targeted sequencing to detect and quantify mutations in CSF and plasma with an average sequencing depth of 1,442x for CSF and 14,810x for plasma. We achieved detection rates, defined as having any mutation detected, of 81.5% (75/92) and 62.5% (45/72) for CSF and plasma, respectively. Our results revealed that the detection rate obtained from CSF was significantly higher than from plasma (p = 0.008) (). Next, we compared the maximum allelic fraction (MaxAF) of CSF and plasma. The average MaxAF of CSF and plasma were 43.64% and 4.58%, respectively, demonstrating that CSF had a statistically significantly higher MaxAF than plasma (p < 0.001) (). Collectively, our data demonstrate that CSF is superior in identifying mutations of LM than plasma.
Figure 1. CSF is superior to plasma in reflecting LM. A). Detection rates, defined as having any mutation detected from the panel, in CSF (red) and plasma (blue). (B) Average maximum allelic fractions in CSF and plasma. (C) Oncoprint of CSF. Each column represents a patient; each row represents a gene. Different types of mutations were denoted in different colors. The total number of mutations a patient has is summarized by the top bar. The frequency of a mutation in this cohort is summarized by the side bar. * denotes p values< 0.05. CSF, cerebrospinal fluid; PLA, plasma; CNVs, copy number variations.
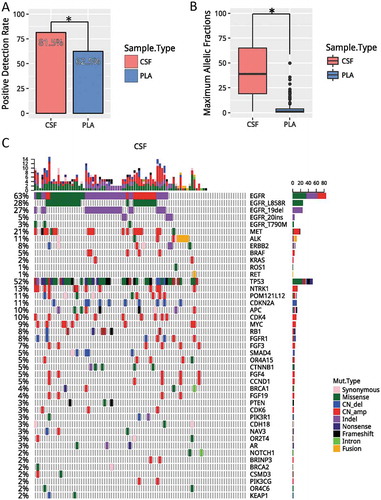
Next, we investigated the genomic profile associated with CSF. We identified at least one mutation from CSF of 75 patients (81.5%). The remaining 17 patients had no mutation detected from this panel. This can be potentially contributed to the fact that they were undergoing tyrosine kinase inhibitor treatment. Collectively, we identified 367 mutations, including 152 single-nucleotide variants (SNVs), 51 insertion/deletions (indels), 127 copy-number amplification (CNA), 29 copy number deletions and 8 translocations. The most frequent driven mutation was EGFR, occurring in 58% of patients, followed by TP53, occurring in 52% of patients (). In addition, other classic NSCLC driver mutations were also identified in CSF, including 6 cases with ALK fusion, 1 cases with ERBB2 amplification, 16 cases with MET amplification, and 2 cases with RET fusion.
CSF revealed a unique genomic profile
We compared and contrasted genomic profiles of CSF and plasma obtained from 72 patients with matched CSF and plasma samples (Fig. S1 and ). Collectively, we identified 280 and 137 genomic alterations from CSF and plasma samples, mapping to 60 and 33 genes, respectively. The pair-wise comparison indicated that mutations revealed by plasma and CSF were highly divergent, achieving a by-variant concordance rate of 25%, with 83 mutations shared between two media. There were 197 and 54 mutations that were only present in CSF and plasma, respectively (). EGFR was the most frequently mutated gene in both media, occurring in 58.33% and 44.44% of CSF and plasma, respectively. TP53 mutation, ALK fusion, and ERBB2 amplification were also frequently observed and occurring in 50%, 5.6%, 1.4% of CSF and 38.9%, 5.6%, 1.4% of plasma, respectively. Furthermore, alterations like MET amplification, CDKN2A, NTRK1 and CDK4 mutations were predominantly observed in CSF (p < 0.05).
Figure 2. A comparisons of genomic profiles obtained from CSF and plasma. (A) Using genomic profile obtained from CSF as a reference, mutations identified in CSF only were denoted in pink; mutations only identified in plasma were denoted in blue. Mutations identified from both media were denoted in green. (B) Unique/shared CNVs and non-CNVs were plotted. Mutations unique to CSF were plotted on top; mutations shared by CSF and plasma were plotted on the bottom. Dark and light red denote CNV; dark and light blue denote non-CNVs. PLA, plasma; CSF, cerebrospinal fluid; CNVs, copy number variations.
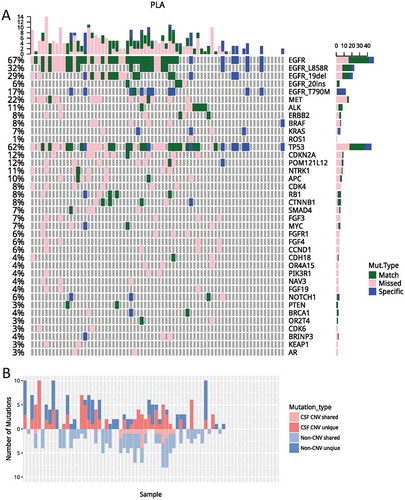
Next, we performed a detailed analysis on EGFR mutations. EGFR activating mutations were detected in 51.4% (37/72) of CSF samples and 38.9% (28/72) plasma sample, resulting in a concordance rate of 47.7%, which was marginally higher than the overall concordance rate (32.7%) observed for all mutations (p = 0.06). Interestingly, EGFR T790M was the least concordant, with a concordance rate of 8.3%. It was the only EGFR mutation which had a higher rate of detection in plasma. It was detected in 15.3% (11/72) plasma samples, but only in 2.8% (2/72) of the CSF samples (p = 0.017) (Fig. S1 and ). This observation was in an agreement of previous reports which also reported a much lower frequency of T790M in CSF and CNS lesions.Citation20–Citation22
Copy number variations in CSF ctdna
Copy number variations have the potential to underlie diseases by altering the diploid status of DNA. Next, we performed CNV analysis to compare and contrast the genomic variability generated by deletions and amplifications. Overall, a total of 121 CNVs were detected in CSF, and a majority of them (81.8%) were specific to CSF. Only 18.2% CNV events were shared with plasma (Fig. S1 and ). EGFR copy number gain was the most frequently observed CNV, occurring in 18 patients (25%) followed by MET and NTRK1 amplification, occurring in 13 and 7 patients, respectively. Other frequently amplified genes included, CDK4, POM121L12, BRAF, FGF3 and MYC. Furthermore, amplification of NTRK1, CDK4, BRAF, and FGF3 were only occurred in CSF. In addition to amplification, we also observed copy number deletions, including CDKN2A, SMAD4, FGFR1 and TP53, occurring in 9, 4, 3 and 2 patients, respectively. Collectively, our data demonstrated that CSF harbored a significant number of unique CNVs.
TP53 loss of heterozygosity in CSF ctdna
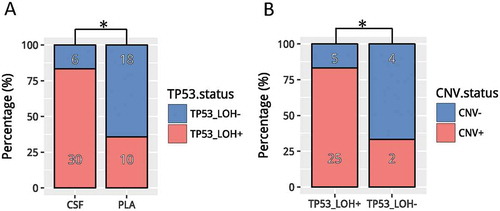
In our cohort, TP53 loss of heterozygosity (LOH) was identified in 41.7% (30/72) of CSF ctDNA, which was much higher than in matched plasma (10/72, 13.9%; p < 0.001) (). Among patients with TP53 mutation, we observed marginally higher mutation frequencies in EGFR (p = 0.073) and MET (p = 0.062). Mutations in CDKN2A, FGFR1, EGFR T790M and NTRK1 were only detected in patients with TP53 LOH (Fig. S1A). Furthermore, patients with TP53 LOH (25/30, 83.3%) were likely to harbor CNVs in CSF than those without TP53 LOH (2/6, 33.3%; p = 0.024) (). Collectively, our data revealed distinct genomic profiles between patients with and without TP53 LOH.
Pathway enrichment analysis
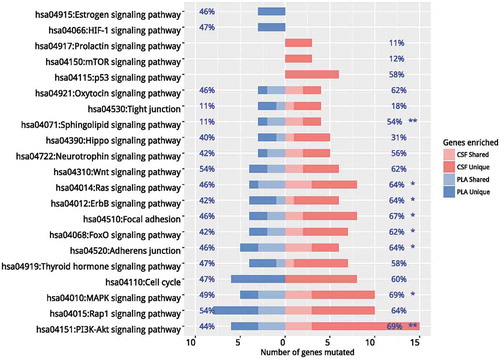
Our data revealed divergent mutation profiles with limited commonality between CSF and plasma. Next, we performed independent pathway enrichment analyses for CSF and plasma based on mutations occurring in more than 2 patients to identify functionally aberrant pathways. Collectively, 33 genes for CSF and 18 genes for plasma were used for the analysis. Among them, 14 genes were overlapped. The enrichment analysis was performed using DAVID software. The cut-off criteria were chosen as default included the EASE score (a modified Fisher Exact p-value proposed by the software) of 0.1 and a minimum of 2 genes belonging to a pathway. Twenty pathways with p values < 0.05 were shown in . Estrogen and HIF-1 signaling pathways, occurring in 46% and 47% of patients respectively, were plasma specific. In contrast, prolactin (PRL), mTOR and p53 pathways, occurring in 11%, 12%, and 58% patients respectively, were CSF specific. Other pathways with a significant difference in prevalence between the two media included Ras (p = 0.044), ErbB (p = 0.012), FOXO (p = 0.019), MAPK P = 0.017), PI3K-Akt signaling pathways (p = 0.004) and focal adhesion (p = 0.018). Pathways with comparable frequencies included but not limited to Hippo, neurotrophin, Wnt and cell cycle signaling pathways. Taken together, we have identified pathways which may facilitate the development of LM.
Figure 4. Pathway enrichment analyses. Independent pathway enrichment analyses for CSF and plasma based on mutations occurring in more than 2 patients were performed using DAVID. The cut-off criteria were chosen as default included the EASE score (a modified Fisher Exact p-value proposed by the software) of 0.1 and a minimum of 2 genes belonging to a pathway. Pathways with p values < 0.05 were listed. Percentages on the left of the figure indicate the frequency of a specific pathway mutated in plasma samples and percentages on the right of the figure indicate the frequency of a specific pathway mutated in CSF samples. * denotes p values< 0.05. ** denotes p values< 0.01. CSF, cerebrospinal fluid; PLA, plasma.
Discussion
The spread of tumor cells to the leptomeninges is a devastating complication of lung cancer, as well as other solid tumor types, including breast cancer and melanoma with exceptionally high mortality.Citation23–Citation26 There are formidable challenges associated with treating LM, including poor penetration of most available therapies across the BBB and poorly understood underlying biology due to inaccessibility. CSF is in intimate contact with tumor cells in CNS tumors and, recently, ctDNA has been shown to be present in the CSF of patients with brain tumors.Citation27 Owing to the BBB, a majority of plasma ctDNA is unable to circulate in the brain; thus, CSF ctDNA is potentially much more comprehensive than that of plasma in reflecting the genomic profile of LM.Citation11,Citation28
In this study, we identified and characterized mutations identified from CSF of patients with LM and compared it with mutation profiles obtained from matched plasma. To the best of our knowledge, this is the largest study comparing the mutation profiles of LM obtained from CSF to plasma by performing capture-based targeted sequencing on 72 matched CSF and plasma samples. We confirmed that CSF ctDNA is more representative of mutational spectrum of LM than plasma evident by the identification of significant number of unique mutations and CNVs events in CSF. Furthermore, the MaxAF was significantly higher in CSF than plasma (p < 0.001). Such phenomenon can be explained by the fact that ctDNA constitutes a very small fraction of cfDNA in plasma due to the presence of DNA released by normal cells. In CSF, significantly fewer normal cells are present, resulting in an increase in the percentage of ctDNA. The overall concordance in mutations identified between CSF and plasma was 32.7%; however, the concordance for EGFR activating mutations was significantly higher (47.7%, p = 0.06), potentially attributing to the positive correlation between EGFR mutation and the development of LMCitation29,Citation30. Other selectively enriched mutations in CSF included TP53 (p = 0.24), MET (p < 0.001), CDKN2A (p = 0.017), NTRK1 (p = 0.033), which all have a role in the development of metastases. Interestingly, comparing to plasma, we observed a much lower frequency (2.78%, 2/72) of EGFR_T790M in CSF, which was consistent with previous reports demonstrating that EGFR T790M are more likely to occur at extracranial sites.Citation22,Citation31–Citation33 This can be explained partially by the low penetrance of first-generation EGFR-TKIs to brain tumors due to low BBB permeabilityCitation33 thus preventing them from developing EGFR T790M.
CNVs resulted from genomic instability have been shown to underlie tumorigenesis including NSCLC. Previous studies have reported the identification of unique CNVs in CSF, suggesting that CNVs may be predominately responsible for LM. We observed a significant number of CNV events frequently occurs in CSF, including but not limited to MET, NTRK1, CDK4, BRAF, and FGF3, suggesting that CNVs may act as key events in LM. Such theory has been supported by several lines of evidence. Studies have reported that MET amplification, occurring in 18.1% of our cohort, is associated with the development of NSCLC brain metastasis and selectively enriched in brain lesions comparing to paired primary lung tumors.Citation34 Another study has shown reproducible CNV events in circulating tumor cells of individual patient regardless of cancer type, suggesting CNVs occurring at certain loci are responsible for metastases.Citation35
Pathway enrichment analyses showed that PI3K-Akt signaling pathway is the most enriched pathway in CSF, significantly higher than its enrichment in plasma (p = 0.004), suggesting an important role of this pathway in the development of LM. Other significantly enriched pathways in CSF included but not limited to PRL, mTOR, p53 pathways, Ras, ErbB, MAPK and focal adhesion pathways. Studies have shown these pathways participated in metastases. PRL, also known as luteotropic hormone, secreted from the pituitary gland, has been reported to be aberrantly activated in lung tumors, especially in neuroendocrine tumors,Citation36 suggesting the enrichment of alterations in PRL pathway might be a shared feature of tumors arise from endocrine and nervous systems. In addition, PRL may play several other active roles in tumor development by stimulating angiogenesisCitation37 and activating pro-proliferation and anti-apoptotic pathways, including but not limited to PI3K/Akt/mTOR, MEK1/2/ERK1/2, p38MAPK, STAT3, and p53.Citation38–Citation40
In conclusion, our study interrogating 72 matched CSF and plasma samples, demonstrate that CSF is a superior liquid biopsy median than plasma for genomic analysis of LM, evident by associating with a significantly higher detection rate and MaxAF. Furthermore, it also provided a more comprehensive profile of LM because it identified significant number of unique CNV events and mutations. To the best of our knowledge, this is the largest study comparing the performance of CSF and plasma in reflecting the genomic profile of LM. Furthermore, we also elucidated pathways that are important to the development of LM.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Research Ethics Committee of Taizhou University Hospital. This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Author Contributions
Concept and design: BY, JX, XM, WH, LC; Data collection: SY, HK, YD, YL, XT, DY; Data analysis and interpretation: ML, SY, HK; Manuscript writing: JL, HZ; Final approval of manuscript: All authors; Accountable for all aspects of the work: All authors.
Supplemental Material
Download Zip (3.7 MB)Supplemental Material
Download Zip (3.7 MB)Supplementary Material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Li Y-S, Jiang B-Y, Yang -J-J, Tu H-Y, Zhou Q, Guo W-B, Yan -H-H, Wu Y-L. Leptomeningeal metastases in patients with nsclc with egfr mutations. J Thorac Oncol. 2016;11:1962–1969. doi:10.1016/j.jtho.2016.06.029.
- Kuiper JL, Hendriks LE, van der Wekken AJ, de Langen AJ, Bahce I, Thunnissen E, Heideman DAM, Berk Y, Buijs EJM, Speel E-JM, et al. Treatment and survival of patients with egfr-mutated non-small cell lung cancer and leptomeningeal metastasis: A retrospective cohort analysis. Lung Cancer. 2015;89:255–261. doi:10.1016/j.lungcan.2015.05.023.
- Morris PG, Reiner AS, Szenberg OR, Clarke JL, Panageas KS, Perez HR, Kris MG, Chan TA, DeAngelis LM, Omuro AM. Leptomeningeal metastasis from non-small cell lung cancer: survival and the impact of whole brain radiotherapy. J Thorac Oncol. 2012;7:382–385. doi:10.1097/JTO.0b013e3182398e4f.
- Gwak H-S, Joo J, Kim S, Yoo H, Shin SH, Han J-Y, Kim HT, Lee JS, Lee SH. Analysis of treatment outcomes of intraventricular chemotherapy in 105 patients for leptomeningeal carcinomatosis from non–small-cell lung cancer. J Thorac Oncol. 2013;8:599–605. doi:10.1097/JTO.0b013e318287c943.
- Remon J, Le Rhun E, Besse B. Leptomeningeal carcinomatosis in non-small cell lung cancer patients: A continuing challenge in the personalized treatment era. Cancer Treat Rev. 2017;53:128–137. doi:10.1016/j.ctrv.2016.12.006.
- Yang JC-H, Cho BC, Kim D-W, Kim S-W, Lee J-S, Su W-C, John T, Kao SC-H, Natale R, Goldman JW, et al. Osimertinib for patients (pts) with leptomeningeal metastases (lm) from egfr-mutant non-small cell lung cancer (nsclc): updated results from the bloom study. J Clin Oncol. 2017;35:2020–2020.
- Ahn M-J, Kim D-W, Cho BC, Kim S-W, Lee J-S, Ahn JS, Kim TM, Lin -C-C, Kim H, John T, et al. Phase i study (bloom) of azd3759, a bbb penetrable egfr inhibitor, in patients with tki-naive, egfrm nsclc with cns metastases. J Clin Oncol. 2017;35:2006-2006. doi:10.1200/JCO.2017.35.15_suppl.2006.
- Lee E, Keam B, Kim D-W, Kim TM, Lee S-H, Chung DH, Heo DS. Erlotinib versus gefitinib for control of leptomeningeal carcinomatosis in non–small-cell lung cancer. J Thorac Oncol. 2013;8:1069–1074. doi:10.1097/JTO.0b013e318294c8e8.
- Lee SJ, Lee J-I, Nam D-H, Ahn YC, Han JH, Sun J-M, Ahn JS, Park K, Ahn M-J. Leptomeningeal carcinomatosis in non–small-cell lung cancer patients: impact on survival and correlated prognostic factors. J Thorac Oncol. 2013;8:185–191. doi:10.1097/JTO.0b013e3182773f21.
- Wang Y, Springer S, Zhang M, McMahon KW, Kinde I, Dobbyn L, Ptak J, Brem H, Chaichana K, Gallia GL, et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Nat Acad Sci. 2015;112:9704–9709. doi:10.1073/pnas.1511694112.
- De Mattos-Arruda L, Mayor R, Ng CK, Weigelt B, Martínez-Ricarte F, Torrejon D, Oliveira M, Arias A, Raventos C, Tang J, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. doi:10.1038/ncomms9839.
- Vanden Bempt I, Wauters E, Vansteenkiste J. Genetic profiling of cell-free DNA from cerebrospinal fluid: opening the barrier to leptomeningeal metastasis in egfr-mutant nsclc. Ann Oncol. 2018;29:789–791. doi:10.1093/annonc/mdy053.
- Crowley E, Di NF, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472. doi:10.1038/nrclinonc.2013.110.
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi:10.1126/scitranslmed.3007094.
- Pan W, Gu W, Nagpal S, Gephart MH, Quake SR. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem. 2015;61:514–522. doi:10.1373/clinchem.2014.235457.
- Li YS, Jiang BY, Yang JJ, Zhang XC, Zhang Z, Ye JY, Zhong WZ, Tu HY, Chen HJ, Wang Z, et al. Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of egfr-mutant non-small cell lung cancer: A new medium of liquid biopsy. Ann Oncol Off J Eur Soc Med Oncol. 2018;29:945–952. doi:10.1093/annonc/mdy009.
- Pentsova EI, Shah RH, Tang J, Boire A, You D, Briggs S, Omuro A, Lin X, Fleisher M, Grommes C, et al. Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J Clin Oncol. 2016; 34:2404–2415. doi:10.1200/JCO.2016.66.6487.
- Fan Y, Zhu X, Xu Y, Lu X, Xu Y, Wang M, Xu H, Ding J, Ye X, Fang L, et al. Cell-cycle and DNA-damage response pathway is involved in leptomeningeal metastasis of non-small cell lung cancer. Clin Cancer Res. 2018;24:209–216. doi:10.1158/1078-0432.CCR-17-1582.
- Li H, Durbin R. Fast and accurate long-read alignment with burrows–wheeler transform. Bioinf. 2010;26:589–595. doi:10.1093/bioinformatics/btp698.
- Yang H, Cai L, Zhang Y, Tan H, Deng Q, Zhao M, Xu X. Sensitive detection of egfr mutations in cerebrospinal fluid from lung adenocarcinoma patients with brain metastases. J Mol Diagn JMD. 2014;16:558–563. doi:10.1016/j.jmoldx.2014.04.008.
- Nanjo S, Arai S, Wang W, Takeuchi S, Yamada T, Hata A, Katakami N, Okada Y, Yano S. Met copy number gain is associated with gefitinib resistance in leptomeningeal carcinomatosis of egfr-mutant lung cancer. Mol Cancer Ther. 2017;16:506–515. doi:10.1158/1535-7163.MCT-16-0522.
- Hata A, Katakami N, Yoshioka H, Kaji R, Masago K, Fujita S, Imai Y, Nishiyama A, Ishida T, Nishimura Y, et al. Spatiotemporal t790m heterogeneity in individual patients with egfr-mutant non-small-cell lung cancer after acquired resistance to egfr-tki. J Thoracic Oncol: off Publ Int Assoc Study Lung Cancer. 2015;10:1553–1559. doi:10.1097/JTO.0000000000000647.
- Geukes Foppen M, Brandsma D, Blank C, van Thienen J, Haanen J, Boogerd W. Targeted treatment and immunotherapy in leptomeningeal metastases from melanoma. Ann Oncology Official Journal Eur Soc Med Oncol. 2016;27:1138–1142. doi:10.1093/annonc/mdw134.
- Le Rhun E, Taillibert S, Chamberlain MC. Carcinomatous meningitis: leptomeningeal metastases in solid tumors. Surg Neurol Int. 2013;4:S265. doi:10.4103/2152-7806.111304.
- Scott BJ, Kesari S. Leptomeningeal metastases in breast cancer. Am J Cancer Res. 2013;3:117.
- Hoffknecht P, Tufman A, Wehler T, Pelzer T, Wiewrodt R, Schütz M, Serke M, Stöhlmacher-Williams J, Märten A, Maria Huber R, et al. Efficacy of the irreversible erbb family blocker afatinib in epidermal growth factor receptor (egfr) tyrosine kinase inhibitor (tki)–pretreated non–small-cell lung cancer patients with brain metastases or leptomeningeal disease. J Thorac Oncol. 2015;10:156–163. doi:10.1097/JTO.0000000000000380.
- Pan W, Gu W, Nagpal S, Gephart MH, Quake SR. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem. 2015;61:514–522. doi:10.1373/clinchem.2014.235457.
- Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016. doi: 10.1158/2159-8290.CD-15-1483.
- Hsu F, De Caluwe A, Anderson D, Nichol A, Toriumi T, Ho C. Egfr mutation status on brain metastases from non-small cell lung cancer. Lung Cancer. 2016;96:101–107. doi:10.1016/j.lungcan.2016.04.004.
- Rangachari D, Yamaguchi N, VanderLaan PA, Folch E, Mahadevan A, Floyd SR, Uhlmann EJ, Wong ET, Dahlberg SE, Huberman MS, et al. Brain metastases in patients with egfr-mutated or alk-rearranged non-small-cell lung cancers. Lung Cancer. 2015;88:108–111. doi:10.1016/j.lungcan.2015.01.020.
- Sasaki S, Yoshioka Y, Ko R, Katsura Y, Namba Y, Shukuya T, Kido K, Iwakami S, Tominaga S, Takahashi K. Diagnostic significance of cerebrospinal fluid egfr mutation analysis for leptomeningeal metastasis in non-small-cell lung cancer patients harboring an active egfr mutation following gefitinib therapy failure. Respir Investig. 2016;54:14–19. doi:10.1016/j.resinv.2015.07.001.
- Hata A, Katakami N, Yoshioka H, Takeshita J, Tanaka K, Nanjo S, Fujita S, Kaji R, Imai Y, Monden K, et al. Rebiopsy of non–small cell lung cancer patients with acquired resistance to epidermal growth factor receptor‐tyrosine kinase inhibitor: comparison between t790m mutation‐positive and mutation‐negative populations. Cancer. 2013; 119:4325–4332. doi:10.1002/cncr.28364.
- Zhao J, Ye X, Xu Y, Chen M, Zhong W, Sun Y, Yang Z, Zhu G, Gu Y, Wang M. Egfr mutation status of paired cerebrospinal fluid and plasma samples in egfr mutant non-small cell lung cancer with leptomeningeal metastases. Cancer Chemother Pharmacol. 2016;78:1305–1310. doi:10.1007/s00280-016-3155-y.
- Benedettini E, Sholl LM, Peyton M, Reilly J, Ware C, Davis L, Vena N, Bailey D, Yeap BY, Fiorentino M, et al. Met activation in non-small cell lung cancer is associated with de novo resistance to egfr inhibitors and the development of brain metastasis. Am J Pathol. 2010;177:415–423. doi:10.2353/ajpath.2010.090863.
- Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z, Zong C, Bai H, Chapman AR, Zhao J, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proceedings of the National Academy of Sciences 2013;110:21083–21088. doi:10.1073/pnas.1320659110.
- Le Ba, Vitte A-L, Debernardi A, Curtet S, Buchou T, Vayr J, Reyniès A, Ito A, Guardiola P, Brambilla C, et al. Receptor-independent ectopic activity of prolactin predicts aggressive lung tumors and indicates hdaci-based therapeutic strategies. Antioxid Redox Signal. 2015;23:1–14. doi:10.1089/ars.2013.5581.
- Muralidhar K, Lee J. Prolactin and angiogenesis: biological implications of microheterogeneity. In: Prolactin. InTech. 2013. doi:10.5772/54318.
- Aksamitiene E, Achanta S, Kolch W, Kholodenko BN, Hoek JB, Kiyatkin A. Prolactin-stimulated activation of erk1/2 mitogen-activated protein kinases is controlled by pi3-kinase/rac/pak signaling pathway in breast cancer cells. Cell Signal. 2011;23:1794–1805. doi:10.1016/j.cellsig.2011.06.014.
- Yan A, Chen Y, Chen S, Li S, Zhang Y, Jia J, Yu H, Liu L, Liu F, Hu C, et al. Leptin stimulates prolactin mrna expression in the goldfish pituitary through a combination of the pi3k/akt/mtor, mkk3/6/p38mapk and mek1/2/erk1/2 signalling pathways. Int J Mol Sci. 2017;18:2781. doi:10.3390/ijms18122781.
- Hartwell HJ, Petrosky KY, Fox JG, Horseman ND, Rogers AB. Prolactin prevents hepatocellular carcinoma by restricting innate immune activation of c-myc in mice. Proc Natl Acad Sci U S A. 2014;111:11455–11460. doi:10.1073/pnas.1404267111.