ABSTRACT
Acute promyelocytic leukemia (APL) is characterized by the presence of promyelocytic leukemia-retinoic acid receptor α (PML-RARα) fusion gene, which is formed following the specific chromosomal translocation t(15;17)(q22;q21). However, cases with PML-RARα generated by occult t(15;17) which are negative by both cytogenetics and fluorescence in situ hybridization (FISH), are difficult to diagnose, leading to impaired treatment effectiveness. In the present study, we reported a case of a 66-year-old male patient, and bone marrow morphology, flow cytometry and cytogenetics did not support the diagnosis of APL. Molecular techniques, such as reverse-transcription polymerase chain reaction (RT-PCR), showed the existence of a cryptic PML-RARα fusion gene, and sequence analysis revealed a new variable isoform. Hotspot gene mutation analysis showed a biallelic CEBPA mutation. He received IA chemotherapy and all-trans retinoic acid (ATRA) treatment, and finally achieved complete remission. This case report provided valuable insights into the relevance of the correct identification of atypical PML–RARα fusion gene and biallelic CEBPA mutation. Moreover, combination of IA chemotherapy and ATRA treatment suggested a good clinical effect in this atypical PML–RARα.
Introduction
The promyelocytic leukemia-retinoic acid receptor α (PML-RARα) fusion gene, occurring in about 95% patients with acute promyelocytic leukemia (APL) by the chromosomal translocation t(15;17)(q22;q21), is considered to be responsible for the arrest of granulopoiesis by directly inhibiting the transcription of retinoic acid target genes.Citation1,Citation2 All-trans retinoid acid (ATRA), the first drug found to cause disease regression specifically in APL patients, interacts with the ligand-binding domain present on the RARα moiety of the chimeric oncoprotein and causes both its transcriptional activation and proteolytic degradation, leading to granulocyte differentiation of APL cells.Citation3,Citation4 Therefore, it is very important to detect the PML-RARα fusion gene in patients with acute myeloid leukemia (AML). In clinical diagnosis of APL, PML-RARα fusion gene can be detected by several molecular methods based on conventional karyotyping, fluorescence in situ hybridization (FISH) and reverse transcriptase-polymerase chain reaction (RT-PCR).Citation5,Citation6 However, rare cases with PML-RARα rearrangements, generated by occult t(15;17) which are negative by both cytogenetics and FISH, are difficult to diagnose, significantly impairing the treatment effectiveness.Citation7–Citation11
In the present study, we reported an AML patient with biallelic CEBPA mutation which was diagnosed as FAB M5 type by morphological analysis. In addition, the diagnosis of APL was supported by a cryptic PML-RARα translocation detected only by nested RT-PCR but not cytogenetics, FISH and real-time RT-PCR. The patient achieved complete remission (CR) after IA chemotherapy and ATRA-induced differentiation therapy.
Materials and methods
Patient and description
A 66-year-old Chinese male patient presented to our center due to petechiae or bruises on the lower limbs that lasted for half a month with intermittent fever and coughing. The computed tomography (CT) scan examination revealed slight high-density pulmonary parenchyma plaques in left lower lobe. The B type ultrasonography showed renal cyst (left) and benign prostate hyperplasia. He was diagnosed with pulmonary tuberculosis (TB) and given anti-TB drugs. Physical examination showed that he had mild anemia and multiple petechiae or bruises on the lower limbs. summarizes the peripheral blood examinations of the patient, including complete blood count, blood coagulation profile and blood chemistry analysis.
Table 1. Clinicopathologic features of the patient.
Bone marrow cell morphology
The bone marrow sample was spread on slides and stained by Wright-Giemsa stain (Baso Diagnostic Inc., China). A total of 200 bone marrow cells were analyzed by light microscopy (Olympus BX51, Olympus Corporation Tokyo, Japan). Cytochemical examinations, including peroxidase (POX) reaction, periodic acid Schiff (PAS) reaction and nonspecific esterase (NSE) reaction, were carried out according to manufacturer’s instructions (Baso Diagnostic Inc., China)
Flow cytometry
The immunophenotyping of bone marrow cells was conducted by a five-color flow cytometry (CYTOMICS FC500, Beckman Coulter, Inc., Brea, CA, USA). Each cluster of differentiation (CD) on gated cells was examined with monoclonal antibodies (Beckman Coulter, Inc., Brea, CA, USA), and it was defined as positive when over 20% of the gated cells showed fluorescence above the staining background of controls by a clinical laboratory physician.
Fish
DCDF PML-RARα (Vysis Inc., Downers Grove, IL, USA) FISH was performed on bone marrow cells prepared as routine FISH methods. The result was determined by a clinical laboratory physician using an Olympus BX51 fluorescence microscope (Olympus Corporation Tokyo, Japan). At least 200 cells with well-delineated signals were evaluated. The cutoff level was set as 1% according to mean ± standard deviation in normal controls.
Molecular analysis
Acute non-lymphoblastic leukemia fusion genes, such as MLL/AF, MLL/AF9, MLL/AF10, MLL/AF17, MLL/ELL, AML/ETO, DEK/CAN, dupMLL, TLS/ERG, EVII, PML/RARA, PLZF/RARA, NPM/RARA, NPM/MLF1, BCR/ABL1(p190), BCR/ABL1(p210), CBFβ/MYH11 and HOX11, were detected using multiplex nested RT-PCR technique by Kindstar Global Co., Ltd., China. Hotspot mutation regions of genes, such as FLT3-ITD, NPM1 (exon 11), c-KIT-D816, DNMT3A-R882, CEBPA, RUNX1 (exon 2–6) and TP53 (exon 5–8), were detected by Sanger sequencing using an AB 3500XL sequencer by Kindstar Global Co., Ltd., China.
Results
Bone marrow cell morphology, flow cytometry and FISH do not support the diagnosis of APL
Bone marrow cell morphology analysis showed marked hyperplasia in nuclear cells. It contained 51.5% abnormal blast cells, including 6% monoblasts and 45.5% promonocytes. These cells were of uneven sizes and shapes, exhibiting irregular nuclear shapes and misty nucleoli (). Moreover, the blasts were positive for POX (data not shown). Chromosome analysis using GTG-banding indicated a karyotype of 46, XY (). FISH signals from the PML-RARA dual-color dual-fusion probe showed that normal cell patterns consisted of two red and two green signals (). Flow cytometry revealed that the AML cells positively expressed CD34, CD38, CD7, CD33, CD13, MPO, CD117 and HLA-DR, but negatively expressed CD19, CD10, CD3, CD4 and CD8 ().
Figure 1. The results of cell morphology, cytogenetics, FISH and flow cytometry of the bone marrow cells. (A) Bone marrow smear exhibiting abnormal, uneven cell sizes and shapes with irregular nuclear shapes and misty nucleoli blast cells. (B) G-banding karyotype of a bone marrow cell exhibiting 46, XY[20]. (C) Dual-color FISH analysis with PML/RARα-specific probes 15q22 (red) and 17q21 (green) exhibiting two red and two green signals as a normal signal type. (D) Flow cytometry indicated that the leukemia cells positively expressed CD34, CD38, CD13, CD33, CD117, CD7, HLA-DR and MPO.
![Figure 1. The results of cell morphology, cytogenetics, FISH and flow cytometry of the bone marrow cells. (A) Bone marrow smear exhibiting abnormal, uneven cell sizes and shapes with irregular nuclear shapes and misty nucleoli blast cells. (B) G-banding karyotype of a bone marrow cell exhibiting 46, XY[20]. (C) Dual-color FISH analysis with PML/RARα-specific probes 15q22 (red) and 17q21 (green) exhibiting two red and two green signals as a normal signal type. (D) Flow cytometry indicated that the leukemia cells positively expressed CD34, CD38, CD13, CD33, CD117, CD7, HLA-DR and MPO.](/cms/asset/65aa1655-68aa-4e6e-91e7-74ee4c3bdc74/kcbt_a_1702398_f0001_c.jpg)
Atypical PML-RARα and biallelic CEBPA mutation
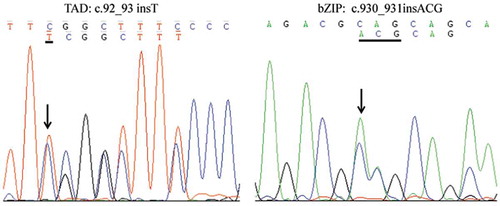
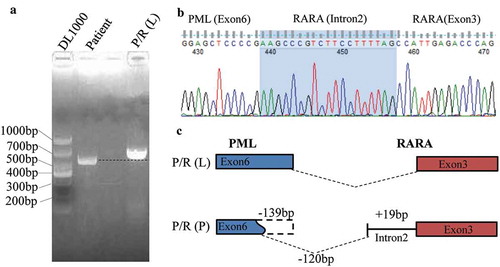
Although cytogenetic and FISH analyses demonstrated the absence of t(15;17)(q24;q21) and PML-RARα in the patient, nested RT-PCR with PML-RARα specific primer and electrophoresis showed a specific band, which was slightly smaller than L-type PML-RARα (). We speculated that it was a new variant of PML-RARα. To validate such a hypothesis, the sequence of PCR amplicon showed PML breakpoint within exon 6 and RARα breakpoint within intron 2 (). Compared with the L-type PML-RARα, the variant detected in our current patient lacked a 139-bp fragment in exon 6 of PML, and there was a 19-bp increase in length in intron 2 of RARα (). Myeloid leukemia-related hotspot gene mutation analysis showed that the patient had a biallelic mutation of CEBPA gene in the transactivation domains (TADs) at its N-terminus as well as a DNA-binding and dimerization bZIP structure at its C-terminus (c.92_93 insT in TAD region and c.930_931 insACG in bZIP region) ().
Figure 2. Detection of the PML-RARα using the semi-quantitative PCR and DNA sequencing method. (A) Agarose gel electrophoresis showed a specific slightly smaller band in the patient compared with L-type PML-RARα. (B) DNA sequencing analysis of PCR product. (C) The schema chart of the new PML-RARα transcript.
Therapy and clinical outcome
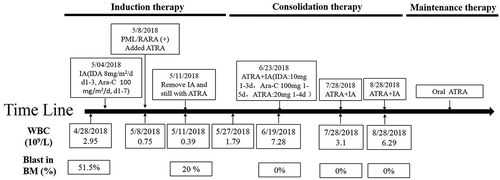
shows the detailed therapeutic regimen. This patient was first treated with IA chemotherapy (idarubicin, 8 mg/m2/day for 3 days and cytarabine, 100 mg/m2/day for 7 days). After treated with IA for 4 days, 40 mg/day ATRA was added to regimen when the atypical PML-RARα was detected. When the first IA regimen was completed, the patient continued to receive ATRA treatment for 30 days. His hemogram of peripheral blood returned to normal levels (Hb 108 g/L, WBC 7.28 × 109/L, PLT 153 × 109/L). The analysis of bone marrow cell morphology showed CR, and minimal residual disease (MRD) monitoring using flow cytometry was 0.00%. He remained CR until the last follow-up on December 25, 2018.
Discussion
To the best of our knowledge, the present case was the first case describing an AML containing a new cryptic atypical bcr2 transcript of PML-RARα and a biallelic CEBPA mutation. However, this case did not show typical morphological, immunological and cytogenetic features of APL, and the therapy of ATRA plus cytotoxic chemotherapy was effective. In recent years, some patients with atypical PML-RARα transcripts have been reported.Citation12–Citation16 In the reported cases, the cell morphological and immunological characteristics are consistent with typical APL. However, in our case, the bone marrow cell morphology of the patient showed FAB M5 type with irregular nuclear shapes and misty nucleoli, and weakly positive POX cytochemical staining. Flow cytometry analysis showed that the leukemia cells expressed CD34, CD7, CD13, CD33, CD117, CD38, HLA-DR and MPO, which was not inconsistent with the immune-phenotype of typical APL. Therefore, we speculated that these features might be related with CEBPA double mutation in the patient.
CEBPA mutations are found in approximately 10% of AML cases.Citation17,Citation18 Most CEBPA-mutated AML cases exhibit two mutations, which frequently include TADs at its N-terminus as well as a DNA-binding and dimerization bZIP structure at its C-terminus.Citation18–Citation20 Recently, Mannelli F et al. have found that CEBPA-double-mutated AML displays a unique phenotypic profile.Citation19 They have analyzed 259 AML cases and found 16 patients with biallelic mutation. Moreover, the blast cells of these patients always express CD34, CD7, CD117 and HLA-DR. Furthermore, some other studies have also found that the expression of CD7 in AML is usually associated with loss of wild-type CEBPA due to mutations or silencing by epigenetic mechanisms.Citation21–Citation24 It suggested that the immune phenotype of our patient was related to the CEBPA mutation, even if no reports about the CEBPA mutation have been found in other APL patients.
Since no case reports of PML-RARα and CEBPA mutations have been described, no currently available treatment regimen can be used as reference. When an APL is suspected, the clinicians always give ATRA treatment to patients even before a definitive diagnosis is made. Once genetic diagnosis is confirmed, full induction treatment should be started. For the non-high risk patients (WBC count <10 × 109/L), the ATRA+ATO combination is the first choice for induction therapy. When the patient’s WBC is more than 10 × 109/L, induction therapy is always combined with anthracycline-based chemotherapy according to therapy-induced differentiation.Citation25,Citation26 In 2016, World Health Organization (WHO) included AML with double-mutated CEBPA as a provisional prognostic category.Citation27,Citation28 Since our patient had no typical APL specificity in cell morphology, immune-phenotype and cytogenetics, we treated the patient with anthracycline-based chemotherapy at the initial stage of diagnosis although the patient’s WBC was below 10 × 109/L. We added ATRA to the traditional chemotherapy to induce differentiation when the molecular diagnosis confirmed that the patient had PML-RARα fusion gene and CEBPA double mutations. After treated for 7 days, the patient continued to receive ATRA treatment until he achieved CR. Fortunately, the patient did not have DIC during the course of treatment, which provided a guarantee for our later use of ATRA treatment.
Recently, there were some other similar cases like the current report in the literature. The karyotype is normal, dual-fusion PML–RARα FISH is initially classified as normal, the presence of sis identified by one of the molecular techniques, and the ATRA treatment is effective in these patients. The new case in our present report did not show typical morphological, immunological and cytogenetic features of APL, allowing us to recognize the importance of morphology, immunophenotyping, cytogenetics, molecular cell biology (MICM) in leukemia diagnosis. Collectively, relying on a single diagnostic method often leads to misdiagnosis or even wrong diagnosis.
Authors’ contributions
Zhang Zhanglinand Li Feiconceived and designed the study. Zhang Zhanglin Xu Yawen and Jiang Mei performed the experiments. Zhang zhanglin and Li Fei wrote the paper. Zhang Zhanglin, Xu Yawen, Jiang Mei, Kong Fancong, Chen Zhiwei and Liu Shuyuan interpreted the data and provided final approval of the version to be published. All authors read and approved the manuscript.
Ethics approval and consent to participate
The patient provided written informed consent for the publication of this study and the study was approved by the Medical Ethics Committee of the First Affiliated Hospital of Nanchang University (Nanchang, China).
Acknowledgments
The authors would like to thank the members of the Department of Clinical Laboratory of the First Affiliated Hospital of Nanchang University (Nanchang, China) for their support.
Additional information
Funding
References
- Chen Z, Chen SJ. RARA and PML genes in acute promyelocytic leukemia. Leuk Lymphoma. 1992;8:253–260. doi:10.3109/10428199209051004.
- Goddard AD, Borrow J, Solomon E. A previously uncharacterized gene, PML, is fused to the retinoic acid receptor alpha gene in acute promyelocytic leukaemia. Leukemia. 1992;6 Suppl 3:117S–9S.
- Degos L. All-trans-retinoic acid treatment and retinoic acid receptor alpha gene rearrangement in acute promyelocytic leukemia: a model for differentiation therapy. Int J Cell Cloning. 1992;10:63–69. doi:10.1002/stem.v10:2.
- Nasr R, Lallemand-Breitenbach V, Zhu J, Guillemin MC, de The H. Therapy-induced PML/RARA proteolysis and acute promyelocytic leukemia cure. Clin Cancer Res. 2009;15:6321–6326. doi:10.1158/1078-0432.CCR-09-0209.
- Kwon WK, Lee JY, Mun YC, Seong CM, Chung WS, Huh J. Clinical utility of FISH analysis in addition to G-banded karyotype in hematologic malignancies and proposal of a practical approach. Korean J Hematol. 2010;45:171–176. doi:10.5045/kjh.2010.45.3.171.
- Chauffaille ML, Figueiredo MS, Beltrani R, Antunes SV, Yamamoto M, Kerbauy J. Acute promyelocytic leukemia: the study of t(15;17) translocation by fluorescent in situ hybridization, reverse transcriptase-polymerase chain reaction and cytogenetic techniques. Braz J Med Biol Res. 2001;34:735–743. doi:10.1590/s0100-879x2001000600006.
- Blanco EM, Curry CV, Lu XY, Sarabia SF, Redell MS, Lopez-Terrada DH, Roy A. Cytogenetically cryptic and FISH-negative PML/RARA rearrangement in acute promyelocytic leukemia detected only by PCR: an exceedingly rare phenomenon. Cancer Genet. 2014;207:48–49.
- Goyal M, Dattatreya PS, Goud I, Murthy SS. Cryptic PML-RARalpha positive acute promyelocytic leukemia with unusual morphology and cytogenetics. Indian J Pathol Microbiol. 2010;53:817–819. doi:10.4103/0377-4929.72097.
- Han JY, Kim KE, Kim KH, Park JI, Kim JS. Identification of PML-RARA rearrangement by RT-PCR and sequencing in an acute promyelocytic leukemia without t(15;17) on G-banding and FISH. Leuk Res. 2007;31:239–243. doi:10.1016/j.leukres.2006.05.011.
- Huh J, Moon H, Chi H, Chung W. Acute promyelocytic leukemia with i(17)(q10) on G-banding and PML/RARA rearrangement by RT-PCR without evidence of PML/RARA rearrangement on FISH. Int J Lab Hematol. 2009;31:372–374. doi:10.1111/j.1751-553X.2008.01040.x.
- Lewis C, Patel V, Abhyankar S, Zhang D, Ketterling RP, McClure RF, Persons DL. Microgranular variant of acute promyelocytic leukemia with normal conventional cytogenetics, negative PML/RARA FISH and positive PML/RARA transcripts by RT-PCR. Cancer Genet. 2011;204:522–523. doi:10.1016/j.cancergen.2011.09.001.
- Walz C, Grimwade D, Saussele S, Lengfelder E, Haferlach C, Schnittger S, Lafage-Pochitaloff M, Hochhaus A, Cross NC, Reiter A. Atypical mRNA fusions in PML-RARA positive, RARA-PML negative acute promyelocytic leukemia. Genes Chromosomes Cancer. 2010;49:471–479.
- Park TS, Kim JS, Song J, Lee KA, Yoon S, Suh B, Lee J-H, Lee H-J, Kim J-K, Choi JR, et al. Acute promyelocytic leukemia with insertion of PML exon 7a and partial deletion of exon 3 of RARA: a novel variant transcript related to aggressive course and not detected with real-time polymerase chain reaction analysis. Cancer Genet Cytogenet. 2009;188:103–107. doi:10.1016/j.cancergencyto.2008.09.002.
- Jeziskova I, Razga F, Gazdova J, Doubek M, Jurcek T, Koristek Z, Mayer J, Dvoráková D. A case of a novel PML/RARA short fusion transcript with truncated transcription variant 2 of the RARA gene. Mol Diagn Ther. 2010;14:113–117. doi:10.1007/BF03256361.
- Ismail S, Ababneh N, Awidi A. Identification of atypical PML-RARA breakpoint in a patient with acute promyelocytic leukemia. Acta Haematol. 2007;118:183–187. doi:10.1159/000109471.
- Chillon MC, Gonzalez M, Garcia-Sanz R, Balanzategui A, Gonzalez D, Lopez-Perez R, Mateos MV, Alaejos I, Rayón C, Arbeteta J, et al. Two new 3ʹ PML breakpoints in t(15;17)(q22;q21)-positive acute promyelocytic leukemia. Genes Chromosomes Cancer. 2000;27:35–43. doi:10.1002/(SICI)1098-2264(200001)27:1<35::AID-GCC5>3.0.CO;2-W.
- Rubio P, Campos B, Digiorge JA, Gallego MS, Medina A, Rossi JG, Felice MS, Alonso CN. NPM1, FLT3 and CEBPA mutations in pediatric patients with AML from Argentina: incidence and prognostic value. Int J Hematol. 2016;104:582–590. doi:10.1007/s12185-016-2064-5.
- Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, Behre G, Hiddemann W, Tenen DG. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–270. doi:10.1038/85820.
- Mannelli F, Ponziani V, Bencini S, Bonetti MI, Benelli M, Cutini I, Gianfaldoni G, Scappini B, Pancani F, Piccini M, et al. CEBPA-double-mutated acute myeloid leukemia displays a unique phenotypic profile: a reliable screening method and insight into biological features. Haematologica. 2017;102:529–540. doi:10.3324/haematol.2016.151910.
- Su L, Tan Y, Lin H, Liu X, Yu L, Yang Y, Liu S, Bai O, Yang Y, Jin F, et al. Mutational spectrum of acute myeloid leukemia patients with double CEBPA mutations based on next-generation sequencing and its prognostic significance. Oncotarget. 2018;9:24970–24979. doi:10.18632/oncotarget.v9i38.
- Wouters BJ, Lowenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113:3088–3091. doi:10.1182/blood-2008-09-179895.
- Lin LI, Chen CY, Lin DT, Tsay W, Tang JL, Yeh YC, Shen HL, Su FH, Yao M, Huang SY, et al. Characterization of CEBPA mutations in acute myeloid leukemia: most patients with CEBPA mutations have biallelic mutations and show a distinct immunophenotype of the leukemic cells. Clin Cancer Res. 2005;11:1372–1379. doi:10.1158/1078-0432.CCR-04-1816.
- Rohrs S, Scherr M, Romani J, Zaborski M, Drexler HG, Quentmeier H. CD7 in acute myeloid leukemia: correlation with loss of wild-type CEBPA, consequence of epigenetic regulation. J Hematol Oncol. 2010;3:15. doi:10.1186/1756-8722-3-15.
- Wouters BJ, Jorda MA, Keeshan K, Louwers I, Erpelinck-Verschueren CA, Tielemans D, Langerak AW, He Y, Yashiro-Ohtani Y, Zhang P, et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 2007;110:3706–3714. doi:10.1182/blood-2007-02-073486.
- Martinez-Cuadron D, Montesinos P, Vellenga E, Bernal T, Salamero O, Holowiecka A, Brunet S, Gil C, Benavente C, Ribera JM, et al. Long-term outcome of older patients with newly diagnosed de novo acute promyelocytic leukemia treated with ATRA plus anthracycline-based therapy. Leukemia. 2018;32:21–29. doi:10.1038/leu.2017.178.
- Mi JQ, Li JM, Shen ZX, Chen SJ, Chen Z. How to manage acute promyelocytic leukemia. Leukemia. 2012;26:1743–1751. doi:10.1038/leu.2012.57.
- Arber DA,Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi:10.1182/blood-2016-03-643544.
- Taskesen E, Bullinger L, Corbacioglu A, Sanders MA, Erpelinck CA, Wouters BJ, van der Poel-van de Luytgaarde SC, Damm F, Krauter J, Ganser A, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117:2469–2475. doi:10.1182/blood-2010-09-307280.