ABSTRACT
Long noncoding RNAs (lncRNAs) have been reported to play a significant role in the occurrence and progression of tumors. In different tumors, they can either act as an oncogene or tumor suppressor via modulating various target mRNAs. OIP5-AS1 belongs to lncRNA family. It has been reported to be involved in the tumorigenesis of some cancers, such as bladder cancer, gastric cancer, and multiple myeloma. However, the role it plays in hepatocellular carcinoma (HCC) remains unclear. This study aims to explore the inherent mechanism of lncRNA OIP5-AS1 in HCC. In the first place, qRT-PCR found that OIP5-AS1 and VEGFA expressions were significantly increased while miR-3163 was obviously reduced in HCC cells and tissues. Next, a series of functional experiments found that knockdown of OIP5-AS1 suppressed HCC cell proliferation, migration and angiogenesis abilities while promoting cell apoptosis simultaneously. Last but not least, miR-3163 inhibition or VEGFA overexpression can reverse the anti-tumor effect of OIP5-AS1. In summary, OIP5-AS1 affects HCC proliferation, metastasis, and angiogenesis in HCC by regulating VEGFA expression through sponging miR-3163.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common primary liver cancer with high mortality.Citation1 Though various therapeutic treatments have been utilized in recent years, HCC still takes possession of over 60,000 deaths and approximately 750,000 diagnoses every year.Citation2 Frequent intrahepatic propagation and extrahepatic metastasis are the main causes of the poor prognosis of HCC. Therefore, it is paramount to explore the molecular mechanism of HCC cell proliferation and metastasis, so as to provide a hopeful target for HCC diagnosis and therapy.
In human genome, approximately 2% of transcripts can be translated into proteins, while the other 98% are called non-coding RNAs, as they could not be translated into proteins. Micro-RNA (miRNA) and long non-coding RNA (lncRNA) belong to the latter category.Citation3 LncRNAs have been reported to work as ceRNAs to regulate the function of the target gene via sponging miRNA.Citation4,Citation5 Furthermore, lncRNAs can either function as oncogenes or cancer suppressor genes in various tumors, and are closely associated with various pathological activities, such as cell proliferation, apoptosis, invasion, and migration.Citation6–Citation9 OIP5-AS1 is a newly identified lncRNA. Its expression status and functional role in certain kinds of tumor were studied by some scholars. For instance, OIP5-AS1 was found to suppress cell viability and facilitate radio-induced apoptosis in colorectal cancer (CRC) cells.Citation10 Besides, in osteosarcoma, it was found to increase cisplatin-resistance (CR) by sponging miR-340-5p and activating the LPAATβ/PI3K/AKT/mTOR signaling pathway.Citation11 In multiple myeloma (MM), Yang et, al. explored that OIP5-AS1 inhibition could cause miR-410 accumulation and induce PTEN/PI3K/AKT pathway, thus promoting cell proliferation but suppressing apoptosis.Citation12 Zhang et al. recently published that knockdown of OIP5-AS1 expression inhibited proliferation, metastasis, and EMT progress in hepatoblastoma cells.Citation13 However, the functional role and underlying mechanism of OIP5-AS1 in HCC are still largely unknown. Therefore, this study was conducted to explore the biological role of OIP5-AS1 in HCC. We found that OIP5-AS1 impacted HCC progression via regulating miR-3163/VEGFA.
Materials and methods
Tissue samples
HCC and adjacent normal tissue (ANT) samples were collected from 48 patients at the Third Affiliated Hospital of Wenzhou Medical University between 2014 and 2018. Each patient offered the written informed consent for this study. None of them have received any preoperative therapy before. After surgical resection, samples were stored in liquid nitrogen at −80°C until required. This protocol was approved by the Ethics Committee of the Third Affiliated Hospital of Wenzhou Medical University.
Cell culture
Normal human liver cell (HL-7702) and human liver cancer cells (HepG2, HCCLM3, Huh7, Hep3B, and HCCLM6) were obtained from the Shanghai Cell Bank (Shanghai, China). All cells were maintained in RPMI-1640 (Gibco, Grand Island, NY, USA) containing 10% FBS (Gibco) and 1% streptomycin and penicillin, and cultivated in a moist incubator with 5% CO2 at 37°C.
Cell transfection
Cells were cultivated into 6-well plates at first until 70% of cell density. HepG2 and HCCLM3 cells were transfected with shRNAs for OIP5-AS1 or VEGFA (shOIP5-AS1 or shVEGFA) and their corresponding negative control (shNC) in accordance with instructions. The miR-3163 mimics/miR-3163 inhibitor and NC mimics/NC inhibitor were gained from Genechem (Shanghai, China). Besides, plasmids were transfected into cells by Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA). The cells were gathered for subsequent experiments after transfection for 48 h.
Quantitative real-time PCR
Total RNA in HCC cells was extracted using TRIzol method (Invitrogen). Later, RNA was reverse transcribed into cDNA by utilizing Reverse Transcription Kit or Taqman Advanced miRNA cDNA Synthesis Kit, and the SYBR Green method was utilized for PCR detection. The fold expression changes were counted by the method of 2−∆∆Ct. Besides, GAPDH or U6 was seen as internal control.
Cell viability and colony formation assays
The purpose of these two experiments was to monitor the proliferation abilities of cells.Citation14
Migration assay
For this experiment, Matrigel was not added to the upper chamber of the membrane. Each well in the upper chamber was incubated with serum-free medium and 10% FBS was placed into the lower chamber. After 24 h of cultivation, cells were fixed with paraformaldehyde (Solarbio, Beijing, China) and stained with crystal violet (Solarbio). Fields were randomly selected for observing and photographing cells under a microscope (Olympus, Tokyo, Japan).
TUNEL assay
TUNEL Apoptosis Kit (Invitrogen) was employed to assess the apoptosis of HepG2 and HCCLM3 cells based on the manufacturer’s guidance. All cells were stained with DAPI (Koritai Biotechnology, Beijing, China) or Merge (Thermo Fisher Scientific, Waltham, MA, USA). Then, cells were surveyed and captured by using fluorescence microscopy (Olympus).
Western blot assay
Total protein obtained from cells was isolated by SDS-PAGE and then moved onto PVDF membranes (Millipore, Billerica, MA, USA). After being blocked with skim milk, the membranes were incubated with primary antibodies including: anti-VEGFA (ab1316, Abcam, Cambridge, USA), anti-PARP (ab74290, Abcam), anti-cleaved PARP (ab32064, Abcam), anti-caspase 3 (ab13847, Abcam), anti-cleaved caspase 3 (ab2302, Abcam), anti-caspase 8 (ab108333, Abcam), anti-cleaved caspase 8 (MA5-15054, Thermo Fisher), anti-caspase 9 (ab219590, Abcam), anti-cleaved caspase 9 (ab2324, Abcam), anti-p-Akt (SAB4301414, Sigma Aldrich), anti-Akt (ab38449, Abcam), anti-p-mTOR (SAB4504476, Sigma Aldrich), anti-mTOR (ab2732, Abcam) and anti-GAPDH (ab8245, Abcam). Next, the membranes were cultivated with a secondary antibody for 1 h in a dark room. Eventually, the protein bands were examined by the chemiluminescence method (Thermo Fisher Scientific).
EdU incorporation assay
The experimental procedures were performed as before.Citation15
Flow cytometry assay
HepG2 and HCCLM3 cells transfected with miR-3163 mimics, or miR-3163 inhibitor were collected after 48 h transfection by trypsinization. After double staining with FITC-Annexin V and propidium iodide (PI) had been performed using the FITC Annexin V Apoptosis Detection Kit (BD Biosciences), conforming to the manufacturer’s directions, the cells were evaluated by flow cytometry (FACScan; BD Biosciences, Franklin Lakes, NJ, USA) with CellQuest software (BD Biosciences). Cells were divided into living cells, dying cells, early apoptotic cells, and apoptotic cells. The relative ratio of early apoptotic cells was compared with that of the control group.
Tube formation assay
Human umbilical vein endothelial cells (HUVECs, Corning Incorporated, NY., USA) and HepG2 or HCCLM3 cells were cultured with DMEM containing 10% FBS, respectively. After transfection for 48 h, the cells of HepG2 and HCCLM3 were centrifuged, with the supernatant collected afterward. Matrigel was added to each well of a 96-well plate and cultivated in a 37°C incubator for 30 min. Tumor-conditioned medium and HUVEC suspension were co-cultured in the Matrigel-coated plate after coagulation. The amount of tubes was calculated, and images were collected from four randomly selected fields via a microscope (Olympus).
Luciferase reporter assay
The wild-type and mutant binding sites of VEGFA or OIP5-AS1 to miR-3163 were sub-cloned into pmirGLO dual-luciferase vector to establish reporter plasmids, called VEGFA-WT or OIP5-AS1-MUT. Subsequently, the plasmids were co-transfected with miR-3163 mimics or NC mimics into HepG2 and HCCLM3 cells, separately. Finally, the luciferase activity was detected by (Dual-Luciferase Reporter Assay System (Promega, USA).
Subcellular fractionation assay
PARIS Kit (Life Technologies, Carlsbad, CA) was applied to collect fractions of nuclear and cytoplasmic in strict accordance with manufacturer’s guides and RT-qPCR was performed to evaluate the relative expression of OIP5-AS1. GAPDH and U6 were regarded as the endogenous controls for cytoplasm and nucleus, respectively.
FISH assay
Cy3-labeled OIP5-AS1 and Hoechst-labeled U6 probes were synthesized by RiboBio (Guangzhou, China). RNA-FISH assay was conducted under a fluorescent in situ hybridization kit (Thermo Fisher) based on the manufacturer’s recommendations.
RIP assay
RNA immunoprecipitation experiment was processed according to the requirements of the Magna RNA Binding Protein Immunoprecipitation Kit (Sigma-Aldrich).Citation16
Tumor growth in nude mice
In vivo animal study conformed to the Animal Care and Use Committee guidelines of the Third Affiliated Hospital of Wenzhou Medical University. The female nude mice (9 mice in each group) were gained from the Third Affiliated Hospital of Wenzhou Medical University. 2 × 106 HepG2 cells stably transfected with indicated plasmids were subcutaneously injected into the flanks of nude mice. After incubation for specific time points (7, 14, 21 and 28 days), the tumor volume was measured separately. Twenty-eight days later, the mice were killed and the tumors were resected and weighed. This study was permitted by the ethics committee of the Third Affiliated Hospital of Wenzhou Medical University.
Statistical analysis
All data were processed by GraphPad Prism 7 software package (Graph-Pad Software, La Jolla, CA, USA) and expressed as mean ± SD. Data between two groups and multiple groups were analyzed using Student’s t-test or ANOVA. Correlation between VEGFA and miR-3163 (miR-3163 and OIP5-AS1 or VEGFA and OIP5-AS1) expression in HCC tissues was evaluated by Pearson’s correlation analysis. Differences in the survival rate curve were assessed utilizing a log-rank test and made using Kaplan–Meier method. Differences were considered significant when p < .05. And the experiments were done at least thrice.
Results
MiR-3163 inhibits cell proliferation, metastasis, and angiogenesis but promotes cell apoptosis in HCC
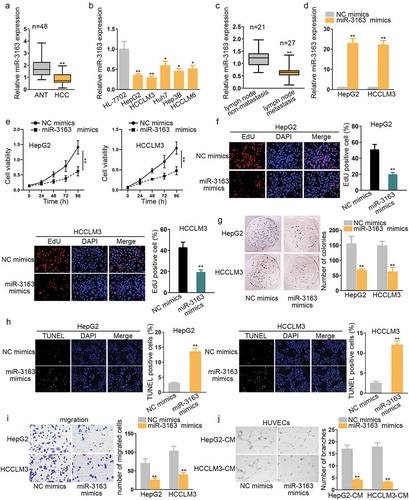
Mir-3163 has been reported to inhibit cell growth via suppressing the translation of Skp2 mRNA in non-small lung cancer.Citation17 In retinoblastoma cancer stem cells (RCSCs), it could suppress multidrug resistance and influence cell proliferation and apoptosis.Citation18 Nevertheless, we found there was very little research on its function in cancer on PubMed (https://www.ncbi.nlm.nih.gov/). We were interested in its role in HCC progression and chose it as candidate miRNA for research objective. Firstly, its expression status was examined in 48 paired HCC tissues and adjacent normal tissues (ANT). qRT-PCR showed that its expression was significantly reduced in HCC tissues (). And GEO database showed that miR-3163 was remarkably up-regulated in human normal liver tissues (Supplementary Figure 1a). Additionally, high expression of miR-3163 was closely related with a higher survival rate of HCC patients (Supplementary Figure 1b). The miR-3163 expression level was also significantly down-regulated in HCC cell lines (HepG2, HCCLM3, Huh7, Hep3B, and HCCLM6) compared with normal cell line (HL-7702) (). Lower expression of miR-3163 was linked to lymph node metastasis (). Therefore, we preliminarily supposed that miR-3163 may act as a tumor suppressor in HCC progression.
To further confirm the role of miR-3163, the gain-of-function experiment was conducted in HepG2 and HCCLM3, which were two cell lines we chose for the present study due to the lowest expression of miR-3163 in them. We ensured the transfection efficiency of miR-3163 mimics in the first place (). CCK8, EdU, and colony formation were then conducted, respectively, to evaluate cell viability (). All these three experiments revealed that overexpression of miR-3163 significantly impaired cell proliferation ability. Next, cell apoptosis capacity was measured by TUNEL assay, which showed a significantly higher ratio of apoptotic cells in miR-3163 mimics treated group than the negative control group (). Flow cytometry assay verified that miR-3163 overexpression stimulated cell apoptosis in HCC (Supplementary Figure 1c). Western blot tested the expression of apoptosis-related proteins, and the results exhibited that cleaved PARP and cleaved caspase3/8/9 (pro-apoptosis proteins) increased when overexpressing miR-3163 (Supplementary Figure 1d). Transwell assay manifested that migration ability was significantly reduced when the expression of miR-3163 was enriched compared with the negative control group (). To detect the influence of miR-3163 mimics in HCC tube formation ability, an angiogenesis experiment was performed in human vascular endothelial cells (HUVECs) incubated with the conditional medium (CM) containing HepG2 and HCCLM3 cells. The result demonstrated that overexpression of miR-3163 reduced the number of tube branches compared with the negative control group (), suggesting that overexpression of miR-3163 suppressed angiogenesis ability in HUVEC cells. On the contrary, when knocking down miR-3163, the expression of miR-3163 was downregulated obviously (Supplementary Figure 2a). Functionally, cell proliferation was activated and cell apoptosis was suppressed in miR-3163 inhibitor transfected HCC cells (Supplementary Figure 2b-g). Migration ability of HCC cells and tube formation ability of HUVECs were also enhanced by miR-3163 inhibitor (Supplementary Figure 2h-i). From the findings above, we elucidate that miR-3163 plays a tumor inhibitor role in HCC development.
VEGFA is a potential target gene of miR-3163
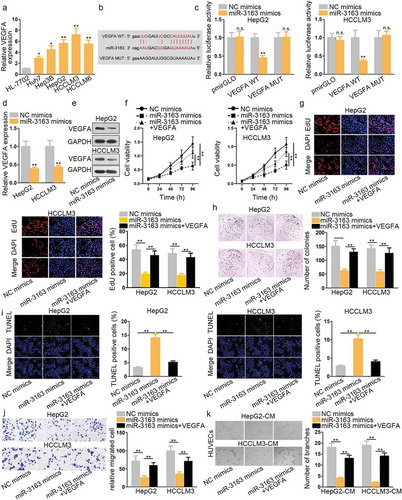
Accumulating evidence has proved that VEGFA is closely associated with cell proliferation and migration in certain cancers, such as myeloma.Citation19 It has been reported that the inhibition of VEGFA can suppress tumor growth, migration, and angiogenesis in colorectal cancer.Citation20 Furthermore, VEGFA has been extensively delineated to be up-regulated in various tumors and is positively related to distant migration and poor prognosis.Citation21 Since miR-3163 was downregulated in HCC, we supposed that it might interact with VEGFA in HCC cells. In order to verify this, qRT-PCR assay showed that VEGFA expression was significantly higher in HCC cell lines and tissues than normal cell and ANT (, Supplementary Figure 3a). Accordingly, VEGFA was negatively correlated with miR-3163 in regard with the expression in tissues (Supplementary Figure 3b). Further, HCC patients with higher expression of VEGFA possessed a shorter survival rate (Supplementary Figure 3c). Afterward, the potential binding site between VEGFA 3ʹUTR and miR-3163 was provided by Starbase (). Dual-luciferase reporters were applied to further verify the interaction between miR-3163 and VEGFA. Forty-eight hours later, we found that the relative luciferase activity of VEGFA 3ʹUTR was greatly weakened after co-transfecting with miR-3163 mimics, while no significant change was observed in VEGFA 3ʹUTR mutant type or miR-3163 negative control group (). Given VEGFA is a target gene of miR-3163, we intended to further determine the correlation between them. qRT-PCR result showed that the expression of VEGFA is negatively modulated by miR-3163. VEGFA expression was decreased when overexpressing miR-3163 (). Western blot assay result further validated this with the result, showing that VEGFA protein was remarkably downregulated after overexpression of miR-3163 compared with the negative control group ().
A series of rescue experiments were adopted to investigate the cellular function of the combination between miR-3163 and VEGFA. CCK8 was used to observe cell proliferation ability. The result revealed that miR-3163 mimics greatly weakened cell proliferation ability in vitro (). However, when miR-3163 mimics was co-transfected with VEGFA, cell proliferation capacity was enhanced to a large extent. This experiment conclusion was further confirmed by EdU and colony formation experiments (). TUNEL was used to detect HCC cell apoptosis capacity. The result showed that miR-3163 mimics increased HCC apoptosis cells in vitro, whereas the co-transfection of miR-3163 mimics with VEGFA can reduce the number of apoptotic HCC cells compared with miR-3163 mimics group (). Transwell assay result manifested that miR-3163 mimics significantly reduced migrated cells compared with the negative control group, while VEGFA reversed this anti-migration role of miR-3163 mimics largely (). From the angiogenesis experiment result, we found that miR-3163 overexpression reduced branch number effectively. Nevertheless, when supplementing with VEGFA, the tube formation ability of HUVECs was recovered instead (). These experimental results suggested that VEGFA could reverse the anti-tumor role of miR-3163 mimics. In the meantime, the downstream pathway of VEGFA in HCC interested us. Considering it has been reported that VEGFA could mediate Akt/mTOR pathway in colorectal cancer.Citation22 In this study, western blot assay detected that VEGFA knockdown (Supplementary Figure 3d) decreased the level of p-Akt and p-mTOR, indicating that Akt/mTOR pathway was suppressed when silencing VEGFA in HCC cells (Supplementary Figure 3e). That is to say, VEGFA is a direct downstream target gene of miR-3163.
OIP5-AS1 accelerates HCC progression via competitively interacting with miR-3163
Given the down-regulation of miR-3163 in HCC tissues and cell lines, we anticipated that lncRNA may sponge it via competing with VEGFA. We browsed Starbase to identify lncRNA that could bind with miR-3163. We limited the number of combinable lncRNA to seven (SNHG1, OIP5-AS1, NEAT1, MEG8, HCG11, XIST, and DSCAM-AS1) by narrowing the scope. Then, we knocked down their expressions, respectively, and only found that OIP5-AS1 inhibition could up-regulate the expression of miR-3163 most significantly (Supplementary Figure S4A). Besides, the role of OIP5-AS1 in HCC has not been reported yet. Hence, it was selected as candidate lncRNA.
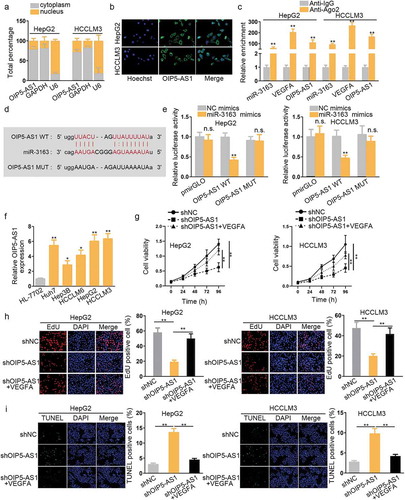
We checked whether OIP5-AS1 could act as ceRNA mechanistically to competitively combine with miR-3163. To begin with, subcellular fractionation assay and FISH assay were performed to determine the location of OIP5-AS1 in cells. Results of both cells revealed that OIP5-AS1 mainly located in the cytoplasm (). RNA immunoprecipitation (RIP) assay was further performed to verify the interaction between OIP5-AS1 and miR-3163 using anti-IgG and anti-Ago, for that Ago2 is a component protein of RISC, mature miRNAs can be integrated into RISC, and then miRNAs can be combined with lncRNA or mRNA. Therefore, there is an enrichment of miRNA, lncRNA, miRNA in the precipitate pulled down by anti-Ago2, which indicates that miRNA may coexist with mRNA or lncRNA In RISC, and their combination was also indirectly demonstrated. Herein, significant high enrichment of OIP5-AS1 and VEGFA were observed in anti-Ago2 group compared with anti-IgG group. Also, miR-3163 enrichment was found in anti-Ago2 group compared with IgG control group. These findings proved the binding relation among OIP5-AS1, miR-3163 and VEGFA, indicating that OIP5-AS1 could competitively bind with miR-3163 (). In order to further verify it, Starbase was utilized to search the binding site of miR-3163 and OIP5-AS1, as shown in . Dual-luciferase reporters conducted later also confirmed the physical interaction. Mir-3163 mimics and negative control were co-transfected with OIP5-AS1 wild type and mutant type. Forty-eight hours later, the relative luciferase activity of OIP5-AS1WT was impaired by miR-3163 mimics, while no significant change was observed in that of OIP5-AS1 MUT. NC mimics served as negative control (). These findings demonstrated that OIP5-AS1 could competitively bind with miR-3163.
Knockdown of OIP5-AS1 suppresses cell proliferation, migration, and angiogenesis while promoting apoptosis in HCC cells in vitro
To determine the expression status of OIP5-AS1 in HCC, qRT-PCR was performed. It was discovered to be up-regulated in HepG2, Huh7, HCCLM6, Hep3B, and HCCLM3 HCC cell lines compared with that of normal cell line HL-7702 (). OIP5-AS1 was also highly expressed in HCC tissues compared with ANT (Supplementary Figure S4B). Based on the above researching results, we concluded that miR-3163/VEGFA was negatively/positively correlated with OIP5-AS1 (Supplementary Figure S4C). Furthermore, higher expression of OIP5-AS1 led to the shorter survival rate of HCC patients (Supplementary Figure S4D). Judging from this, we presumed that OIP5-AS1 may act as an oncogene in HCC. Hence, loss-of-function experiments were carried out. We knocked down OIP5-AS1 by transfecting shOIP5-AS1 in HepG2 and HCCLM3 cell lines, and conducted a series of cellular functional experiments to further explore this assumption. CCK8 was used to examine cell proliferation ability. The result showed that shOIP5-AS1 decreased HCC cell viability () EdU assay also proved this result (). We observed that the downregulation of OIP5-AS1 also facilitated HCC cell apoptosis ability, as shown in .
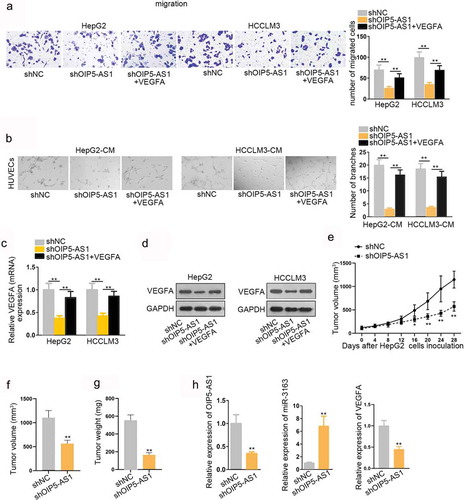
Next, the effect of OIP5-AS1 knocking down on HCC cell migration was evaluated by transwell assay. According to the result, inhibition of OIP5-AS1 largely restrained cell migration capacity (). Angiogenesis experiment result manifested that knockdown of OIP5-AS1 dampened tube formation capacity (). However, VEGFA reversed the function of shOIP5-AS1 in inhibiting proliferation as well as promoting migration and angiogenesis. In other words, VEGFA rescued the tumor suppressor role of OIP5-AS1 knockdown through mutual interaction. To further prove this, qRT-PCR assay was performed to determine the effects of knockdown of OIP5-AS1 on VEGFA. The result revealed that knockdown of OIP5-AS1 significantly reduced VEGFA expression (). Western blot assay result also validated that VEGFA protein expression was significantly decreased in group transfected with shOIP5-AS1 compared with the negative control group ().
Based on researching results above, we conclude that OIP5-AS1-overexpression induced VEGFA up-regulation promotes cell proliferation, migration, and angiogenesis, yet inhibits cell apoptosis in HCC. In other words, OIP5-AS1 exerts its oncogenic effects on HCC progression via regulation of miR-3163/VEGFA.
Knockdown of OIP5-AS1 inhibits tumor growth in vivo
Finally, we investigated the biological function of OIP5-AS1 in vivo, so as to testify the conclusion from vitro experiments. ShOIP5-AS1 and its negative control (shNC) were stably transfected into HepG2 cells using lentiviruses. Then, 6 × 106 cells were subcutaneously injected in the flank of female nude mice. These mice were divided into two groups. One group was injected with shOIP5-AS1-transfected cells, while another group injected with shNC-transfected cells. After 4 weeks of inoculation, the isolated tumors were isolated from xenograft mice. The experiment result showed that the tumor growth speed was much slower in shOIP5-AS1 group compared with the negative control group (). Also, the tumor volume was considerably smaller in shOIP5-AS1 group than the negative control group at the end of inoculation (). The tumor weight was lighter in shOIP5-AS1 group than the negative control group after measurement (). In addition, VEGFA overexpression reversed the obstructive function of shOIP5-AS1 on tumor growth, volume, and weight (Supplementary Figure S5A-C). Finally, we evaluated the expression of OIP5-AS1, miR-3163 and VEGFA in tissues obtained from the mice injected with OIP5-AS1 or shNC transfected HepG2 cells, and found that the expression of OIP5-AS1 and VEGFA were downregulated, but that of miR-3163 was upregulated (). Therefore, we confirmed that knockdown of OIP5-AS1 deterred tumor growth in vivo. All these in vivo experiments complied that OIP5-AS1 is an oncogene in HCC.
OIP5-AS1 promotes HCC progression via inhibiting miR-3163
Similarly, in vitro, miR-3163 downregulation could neutralize the inhibiting influence of shOIP5-AS1 on cell proliferation, migration, and angiogenesis, as well as the encouraging influence of that on cell apoptosis in HCC (Supplementary Figure S5D-H). In vivo, miR-3163 inhibitor relieved the hindering effects of shOIP5-AS1 on tumor growth, volume, and weight (Supplementary Figure S5I-K). In a word, OIP5-AS1 promotes HCC progression via inhibiting miR-3163 in vitro and in vivo.
Discussion
In recent years, lncRNAs have become an important theoretical research object, as they play a vital role in the occurrence and development of various tumors. Some known lncRNAs, including HOST2,Citation23 HCCL5,Citation24 CCAT1,Citation25 and ZFAS1,Citation26 were reported to be involved in HCC evolvement. In our research, OIP5-AS1 was identified to be an up-regulated lncRNA in HCC tissue, which led to poor prognosis of HCC patients, and it was chosen for further investigation in HCC.
miRNA can modulate the target gene via stimulating or silencing its function. It was found that miR-3163 could reduce multidrug resistance in retinoblastoma cancer stem cells.Citation18 And overexpression of miR-3163 was shown to enhance the sensitivity of HCC cells sorafenib.Citation27 In this paper, we found that miR-3163 was significantly decreased in HCC tissues and cells. And miR-3163 overexpression was closely related to the higher survival rate of HCC patients. Then, we presumed that it exerted its function by modulating its target gene. We confirmed that VEGFA was a target gene of miR-3163 through multiple channels. Besides, VEGFA was down-regulated in HCC tissues, which was negatively/positively correlated with miR-3163/OIP5-AS1. Meanwhile, the high expression of VEGFA was associated with the unsatisfactory prognosis of HCC patients.
Increasing evidence has demonstrated that lncRNA can play a competing endogenous RNA role to regulate mRNA via modulating miRNA expression in various tumors, hepatocellular carcinoma included.Citation28 For example, Tang et al. found that lncBC032469 sponged target miR-1207-5p and up-regulated hTERT expression in gastric cancer.Citation29 In the current study, we found that OIP5-AS1 was aberrantly upregulated in HCC tissues and cells, which restrained cell apoptosis, while contributed to cell proliferation, migration, and angiogenesis in vitro and tumor growth in vivo. We observed a negative correlation between the expression of OIP5-AS1 and miR-3163 and proved that OIP5-AS1 could sponge miR-3163.
To further verify the mechanism of OIP5-AS1 in HCC, functional experiments were carried out. On the one hand, knockdown of OIP5-AS1 could accelerate tumor cell apoptosis, while inhibiting proliferation, metastasis, and angiogenesis formation. On the other hand, knockdown of OIP5-AS1 reduced the VEGFA and protein expressions. While it was fully established that VEGFA could trigger tumor cell proliferation, metastasis, and angiogenesis.Citation30 Meanwhile, miR-3163 inhibition or VEGFA overexpression reversed the anti-tumor effect of OIP5-AS1. In other words, OIP5-AS1 influenced HCC progression via regulating VEGFA expression through miR-3163.
In summary, this study revealed that OIP5-AS1 down-regulation accelerated tumor cell apoptosis, while inhibited cell proliferation, metastasis, and angiogenesis by suppressing VEGFA through up-regulating miR-3163 in HCC for the first time. Hence, we suggested that OIP5-AS1 might provide some novel thoughts for the medical treatment of HCC in the future.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download Zip (4.9 MB)Acknowledgments
This study was supported by research members for their meaningful experiment results.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- Ziogas IA, Sioutas G, Mylonas KS, Tsoulfas G. Role of microRNA in the diagnosis and management of hepatocellular carcinoma. MicroRNA (Shariqah, United Arab Emirates). 2020;9:25–40. DOI:10.2174/2211536608666190619155406.
- Liu Y, Zhao L, Ma W, Cao X, Chen H, Feng D, Liang J, Yin K, Jiang X. The blockage of KCa3.1 channel inhibited proliferation, migration and promoted apoptosis of human hepatocellular carcinoma cells. J Cancer. 2015;6:643–651.
- Lu S, Zhang J, Lian X, Sun L, Meng K, Chen Y, Sun Z, Yin X, Li Y, Zhao J, et al. A hidden human proteome encoded by ‘non-coding’ genes. Nucleic Acids Res. 2019;47(15):8111–8125. doi:10.1093/nar/gkz646.
- Gao J, Yin X, Yu X, Dai C, Zhou F. Long noncoding LINC01551 promotes hepatocellular carcinoma cell proliferation, migration, and invasion by acting as a competing endogenous RNA of microRNA-122-5p to regulate ADAM10 expression. J Cell Biochem. 2019;120(10):16393–16407. doi:10.1002/jcb.v120.10.
- Chen W, Peng R, Sun Y, Liu H, Zhang L, Peng H, Zhang Z. The topological key lncRNA H2k2 from the ceRNA network promotes mesangial cell proliferation in diabetic nephropathy via the miR-449a/b/Trim11/Mek signaling pathway. FASEB J. 2019;33:11492–11506. DOI: 10.1096/fj.201900522R.
- Huang L, Wang Y, Chen J, Wang Y, Zhao Y, Wang Y, Ma Y, Chen X, Liu W, Li Z, Zhao L, Shan B, Dong X, Li D, Shao S, Song Y, Zhan Q, Liu X. Long noncoding RNA PCAT1, a novel serum-based biomarker, enhances cell growth by sponging miR-326 in oesophageal squamous cell carcinoma. Cell Death Dis. 2019;10:513. doi:10.1038/s41419-019-1745-4.
- Wang J, Li B, Wang C, Luo Y, Zhao M, Chen P. Long noncoding RNA FOXD2-AS1 promotes glioma cell cycle progression and proliferation through the FOXD2-AS1/miR-31/CDK1 pathway. J Cell Biochem. 2019. doi:10.1002/jcb.29284.
- Zhang X, Wu N, Wang J, Li Z. LncRNA MEG3 inhibits cell proliferation and induces apoptosis in laryngeal cancer via miR-23a/APAF-1 axis. J Cell Mol Med. 2019;23:6708–6719. DOI:10.1111/jcmm.14549.
- Zheng Z-Q, Li Z-X, Zhou G-Q, Lin L, Zhang -L-L, Lv J-W, Huang X-D, Liu R-Q, Chen F, He X-J, et al. Long non-coding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79(18):4612–4626. doi:10.1158/0008-5472.CAN-19-0799.
- Zou Y, Yao S, Chen X, Liu D, Wang J, Yuan X, Rao J, Xiong H, Yu S, Yuan X, et al. LncRNA OIP5-AS1 regulates radioresistance by targeting DYRK1A through miR-369-3p in colorectal cancer cells. Eur J Cell Biol. 2018;97(5):369–378. doi:10.1016/j.ejcb.2018.04.005.
- Song L, Zhou Z, Gan Y, Li P, Xu Y, Zhang Z, Luo F, Xu J, Zhou Q, Dai F, et al. Long noncoding RNA OIP5-AS1 causes cisplatin resistance in osteosarcoma through inducing the LPAATβ/PI3K/AKT/mTOR signaling pathway by sponging the miR-340-5p. J Cell Biochem. 2019;120(6):9656–9666. doi:10.1002/jcb.v120.6.
- Yang N, Chen J, Zhang H, Wang X, Yao H, Peng Y, Zhang W. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017;8(8):e2975. doi:10.1038/cddis.2017.358.
- Zhang Z, Liu F, Yang F, Liu Y. Knockdown of OIP5-AS1 expression inhibits proliferation, metastasis and EMT progress in hepatoblastoma cells through up-regulating miR-186a-5p and down-regulating ZEB1. Biomed Pharmacother. 2018;101:14–23. doi:10.1016/j.biopha.2018.02.026.
- Wang X, Kan J, Han J, Zhang W, Bai L, Wu H. LncRNA SNHG16 functions as an oncogene by sponging MiR-135a and promotes JAK2/STAT3 signal pathway in gastric cancer. J Cancer. 2019;10(4):1013–1022. doi:10.7150/jca.29527.
- Li D, Jiang X, Zhang X, Cao G, Wang D, Chen Z. Long noncoding RNA FGD5-AS1 promotes colorectal cancer cell proliferation, migration, and invasion through upregulating CDCA7 via sponging miR-302e. In Vitro Cell Dev Biol Anim. 2019;55(8):577–585. doi:10.1007/s11626-019-00376-x.
- Dong YX, Pang ZG, Zhang JC, Hu JQ, Wang LY. Long non-coding RNA GClnc1 promotes progression of colorectal cancer by inhibiting p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:5705–5713. doi:10.26355/eurrev_201907_18308.
- Su L, Han D, Wu J, Huo X. Skp2 regulates non-small cell lung cancer cell growth by Meg3 and miR-3163. Tumor Biology. 2016;37(3):3925–3931. doi:10.1007/s13277-015-4151-2.
- Jia M, Wei Z, Liu P, Zhao X. Silencing of ABCG2 by microRNA-3163 inhibits multidrug resistance in retinoblastoma cancer stem cells. J Korean Med Sci. 2016;31(6):836–842. doi:10.3346/jkms.2016.31.6.836.
- Han KY, Chang JH, Azar DT. MMP14-containing exosomes cleave VEGFR1 and promote VEGFA-induced migration and proliferation of vascular endothelial cells. Invest Ophthalmol Vis Sci. 2019;60:2321–2329. doi:10.1167/iovs.18-26277.
- Chen X, Zeng K, Xu M, Liu X, Hu X, Xu T, He B, Pan Y, Sun H, Wang S, et al. P53-induced miR-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting VEGFA and HMGA2. Cell Death Dis. 2019;10(2):131. doi:10.1038/s41419-018-1188-3.
- Ye J, Zhu J, Chen H, Qian J, Zhang L, Wan Z, Chen F, Sun S, Li W, Luo C. A novel lncRNA-LINC01116 regulates tumorigenesis of glioma by targeting VEGFA. Int J Cancer. 2020;146:248–261. DOI:10.1002/ijc.32483.
- Chen X, Zeng K, Xu M, Hu X, Liu X, Xu T, He B, Pan Y, Sun H, Wang S, et al. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 2018;9(10):982. doi:10.1038/s41419-018-0962-6.
- Wu Y, Yuan T, Wang WW, Ge PL, Gao ZQ, Zhang G, Tang Z, Dang X-W, Zhao Y-F, Zhang J-Y, et al. Long noncoding RNA HOST2 promotes epithelial-mesenchymal transition, proliferation, invasion and migration of hepatocellular carcinoma cells by activating the JAK2-STAT3 signaling pathway. Cell Physiol Biochem. 2018;51:301–314. doi:10.1159/000495231.
- Peng L, Jiang B, Yuan X, Qiu Y, Peng J, Huang Y, Zhang C, Zhang Y, Lin Z, Li J, et al. Super-enhancer-associated long noncoding RNA HCCL5 is activated by ZEB1 and promotes the malignancy of hepatocellular carcinoma. Cancer Res. 2019;79(3):572–584. doi:10.1158/0008-5472.CAN-18-0367.
- Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi:10.1186/s13046-015-0136-7.
- Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B,Peng C, et al. Amplification of long noncoding rna zfas1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75(15):3181–3191. doi:10.1158/0008-5472.CAN-14-3721.
- Yang B, Wang C, Xie H, Wang Y, Huang J, Rong Y, Zhang H, Kong H, Yang Y, Lu Y, et al. MicroRNA-3163 targets ADAM-17 and enhances the sensitivity of hepatocellular carcinoma cells to molecular targeted agents. Cell Death Dis. 2019;10:784. doi:10.1038/s41419-019-2023-1.
- Liao X, Wang X, Huang K, Han C, Deng J, Yu T, Yang C, Huang R, Liu X, Yu L, et al. Integrated analysis of competing endogenous RNA network revealing potential prognostic biomarkers of hepatocellular carcinoma. J Cancer. 2019;10:3267–3283. doi:10.7150/jca.29986.
- Lu MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY, Wang S-M, He F-T, Yang S-M. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524–3534. doi:10.1038/onc.2015.413.
- Lin J, Cao S, Wang Y, Hu Y, Liu H, Li J, Chen J, Li P, Liu J, Wang Q, et al. Long non-coding RNA UBE2CP3 enhances HCC cell secretion of VEGFA and promotes angiogenesis by activating ERK1/2/HIF-1alpha/VEGFA signalling in hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:113. doi:10.1186/s13046-018-0727-1.