ABSTRACT
Background
MicroRNAs (miRNAs) as the subtype of non-coding RNAs are revealed to be crucial players in cellular activities. It has been reported that miR-3619-5p functions as a tumor inhibitor in several cancers. However, the connection between miR-3619-5p and stomach adenocarcinoma (STAD) remains to be discovered.
Aim of the study
The purpose of the study is to figure out the role and molecular regulation mechanism of miR-3619-5p in STAD.
Methods
The expression of miR-3619-5p was evaluated via qRT-PCR analysis. Gain-of-function experiments demonstrated the effects of miR-3619-5p on cellular functions. The upper-stream transcription factor STAT4 and downstream target gene TBC1D10B of miR-3619-5p were identified by bioinformatic analysis. The binding and interaction between the indicated molecules were verified by RNA pull-down and luciferase reporter assays.
Results
The expression of miR-3619-5p was prominently down-regulated in STAD cells and tissues. MiR-3619-5p suppresses cell proliferation, migration, invasion and tumor growth in STAD. Further, STAT4 bound with miR-3619-5p promoter and inhibited its transcription. MiR-3619-5p was also recognized to modulate STAD progression through the regulation of downstream target gene TBC1D10B.
Conclusion
STAT4-mediated miR-3619-5p controls STAD carcinogenesis and progression through modulating TBC1D10B expression, which may provide a novel insight for researching the STAD-related molecular mechanism.
Introduction
Globally, stomach cancer (SC) is one of the most common malignancies affecting the human digestive tract. SC is also one of the leading causes of cancer-associated deaths worldwide.Citation1 According to cancer statistics, there are 950,000 newly occurred SC cases and 720,000 SC-related deaths in 2012 worldwide.Citation2 Notably, SC is commonly diagnosed at an advanced stage, frequently accompanied by poor prognosis due to a lack of efficient diagnosis strategies.Citation3 Based on histopathologic characters, SC is classified into stomach adenocarcinoma (STAD) and stomach squamous cell carcinoma (SSCC), where STAD accounts for the vast majority.Citation3,Citation4
MicroRNAs (miRNAs) are short RNA molecules with 19–25 nucleotides at length. As a subtype of non-coding RNA family, miRNAs have limited capacity in protein coding and were considered as transcription noises for quite a long time.Citation5,Citation6 Over the past decades, significant efforts have been devoted to investigating miRNAs.Citation7 It was revealed that a single miRNA can target hundreds of messenger RNAs (mRNAs) and regulate the expression of targeted genes.Citation5 More and more studies have recognized that miRNAs are essential post-transcriptional regulators in gene expression and promising prognostic biomarkers in various diseases.Citation8-Citation10 For instance, miR-140-5p inhibits gastric cancer progression by targeting YES1.Citation11 MiR-373 attenuates gastric cancer migration and invasion by down-regulating Vimentin level.Citation12 Moreover, miR-26a and miR-148a are promising prognostic biomarkers of gastric cancer.Citation13
In our study, we detected the down-regulated expression of miR-3619-5p in STAD cells and tissues. Regarding miR-3619-5p, several reports have elucidated its tumor-suppressive role in cancers. For example, miR-3619-5p functions as a tumor suppressor to repress prostate cancer cell growth.Citation14 Overexpression of miR-3619-5p suppresses non-small cell lung cancer by binding to 3ʹ-UTR of β-catenin mRNA and inhibiting its translation.Citation15 MiR-3619-5p hampers cell proliferation and chemoresistance of cutaneous squamous cell carcinoma by targeting KPNA4.Citation16 However, after retrieving PubMed database, we did not obtain any information about exploring the function or underlying mechanism of miR-3619-5p in STAD.
Therefore, this study was aimed to investigate the key role of miR-3619-5p in STAD tumorigenesis and progression. Here, we reported that miR-3619-5p was a tumor suppressor in STAD, inhibited STAD proliferation and metastasis by binding to TBC1D10B, which might provide new thoughts for STAD studies.
Materials and methods
Tissue samples
Sixty STAD tissue samples and 60 adjacent non-tumor tissue samples were obtained from Wannan Medical College First Affiliated Hospital, Yijishan Hospital. All tissues were collected during surgical resection of STAD patients at Wannan Medical College First Affiliated Hospital, Yijishan Hospital. Adjacent non-tumor tissues were obtained from areas ≥6 cm away from the tumor site. All of the patients did not receive radiation and chemotherapy treatments before surgery. All tissues were quickly frozen in liquid nitrogen following surgical removal and reserved at −80°C until being used. The study got the approval of the Ethical Committee of Wannan Medical College First Affiliated Hospital, Yijishan Hospital. All participants supplied written informed consent in this study.
Cell lines
STAD cell lines including MGC-803, HGC-27, SGC-7901, BGC-823 and AGS were preserved in Dulbecco’s Modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA, USA), from American Type Culture Collection (ATCC, Rockville, Maryland, USA). One percent penicillin-streptomycin solution (HyClone, Logan, UT, USA) and 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA) were utilized for cell culture at 37°C supplied with 5% CO2 atmosphere.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The extraction of total cellular RNA from AGS and SGC-7901 cells as well as STAD tissue and adjacent non-tumor tissue samples was achieved by TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The reverse transcription was conducted with SYBR Green PCR Master Mix (Takara, Kyoto, Japan) on ABI Step-One Plus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Gene expression was calculated by the comparative 2−ΔΔCt method, with U6 and GAPDH as the normalized genes.
Cell transfection
After cell density reached about 80%, confluent AGS and SGC-7901 cells were transfected with plasmids for 48 h by use of Lipofectamine 2000 (Invitrogen). For gene silencing analysis, the short hairpin RNAs (shRNAs) against STAT4 (sh-STAT4 #1/2/3) or TBC1D10B (sh-TBC1D10B#1/2/3) and control nonspecific shRNAs (sh-NC) were produced by Genepharma (Shanghai, China). For gene overexpression analysis, the miR-3619-5p mimics and miR-NC, pcDNA3.1/TBC1D10B (OE/TBC1D10B) and empty vector, were all procured from GeneCopoecia (Guangzhou, China).
Cell counting kit-8 (CCK-8)
The transfected AGS and SGC-7901 cells were incubated in 96-well plates for 24, 48, 72 and 96 h, with the addition of CCK-8 reagent (10 μL; Beyotime, Shanghai, China) for 2 h. Absorbance was examined at 450 nm by a microplate reader (Bio-Rad, Hercules, CA, USA).
Colony formation assay
Clonogenic AGS and SGC-7901 cells were cultivated for 14 d in 6-well plates (500 cells/well). Finally, colonies were dyed in 0.5% crystal violet and counted after fixing in 4% paraformaldehyde.
Flow cytometry of cell apoptosis
Cell apoptosis assay was detected using Annexin V apoptosis detection kit (Invitrogen). 2 × 105 AGS and SGC-7901 cells were harvested from cold phosphate-buffered saline (PBS) and cultured in 500 μL of binding buffer adding 10 μL of propidium iodide (PI) and 10 ml of Annexin V-FITC for 15 min. Flow cytometry (BD Biosciences, San Jose, CA) was applied finally.
Transwell assay
Twenty-four-well transwell inserts (Corning Incorporated, Corning, NY, USA) with matrigel (BD Biosciences) coating were applied for cell invasion or migration assay. AGS and SGC-7901 cells (1 × 105 cells/well) in serum-free medium were transferred to the upper chamber, while the complete culture medium was added to the lower chamber. After incubation for 48 h, the invading or migrating cells were subjected to 0.1% crystal violet after fixing with ice-cold methanol for counting and imaging.
Tumor xenograft assay
Seven-week-old male BALB/C athymic nude mice were provided by Wannan Medical College First Affiliated Hospital, Yijishan Hospital. AGS cells were injected into the fat pads of second mammary glands of mice (5 × 106 AGS cells/100 μl per point for each nude mouse). On the 10th d after AGS cell injection, we confirmed tumor transplantation. AGS xenograft-bearing mouse models were randomly divided into two groups (n = 5), five mice were injected with 5 nM micrONTM miR-3619-5p agomir, and five mice with NC agomir. The mice were artificially sacrificed on the 28th d, and the tumors were removed, photographed and weighed. Tumor volume was calculated as the formula “length × width2/2”. After killing mice, the excised tumors were weighed for subsequent assays.
Immunohistochemistry (IHC) staining
Immunohistochemistry staining assay was carried out on paraformaldehyde-fixed paraffin sections, which were dewaxed and dehydrated later on. Following rehydration and antigen repairing in citrate buffer, 3.0% hydrogen peroxide was used to block endogenous peroxidase activity, which lasted for 10 min. In addition, 10% of goat plasma was utilized to block the sections for 30 min. Subsequently, sections were, respectively, cultured with the primary antibodies directed against STAT4 (1:400), Ki-67 (1:400) and cleaved caspase3 (1:200) at 4°C overnight. The primary antibody was measured with biotinylated secondary antibodies (Abcam, Cambridge, MA, USA) in line with the manufacturer’s proposals. The sections were visualized using diaminobenzidine, then counterstained by hematoxylin and dehydrated by alcohol and xylene. Finally, sections were mounted onto glass slides for the following treatments.
Western blot
Cell protein samples of AGS and SGC-7901 cells were acquired through RIPA lysis buffer and electrophoresed by 10% SDS-PAGE, followed by loading onto PVDF membranes. The specific primary antibodies to STAT4 (ab68156, Abcam, Cambridge, MA, USA) and GAPDH (ab128915; Abcam), as well as secondary antibody Goat Anti-Rabbit IgG (ab6940; Abcam), were treated with samples after blocking membranes with 5% nonfat milk. The protein band density was monitored by enhanced chemiluminescence reagent (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Chromatin immunoprecipitation (ChIP)
Magna ChIP Kit (Millipore, Bedford, MA, USA) was employed for ChIP assay of miR-3619-5p promoter in AGS and SGC-7901 cells. The crosslinked chromatin DNA was sonicated to 200-500-bp fragments, then immunoprecipitated with anti-STAT4 antibody. Normal IgG was taken as a negative control. The recovered DNA by magnetic beads was analyzed by qRT-PCR and agarose gel electrophoresis (AGE).
Dual-luciferase reporter vector analysis
For promoter assay, AGS and SGC-7901 cells were transfected with pGL3-Basic luciferase vector (Promega, Madison, WI, USA) containing miR-3619-5p promoter, pRL-TK-Renilla and sh-STAT4 or control. Besides, the binding sites of the wild type (Wt) or mutant (Mut) TBC1D10B 3ʹ-UTR and miR-3619-5p were inserted to pmirGLO Dual-Luciferase Vector (Promega), then co-transfected with miR-3619-5p mimics or NC mimics. The Dual-Luciferase Reporter Assay System (Promega) was applied for luciferase activity after 48 h of transfection.
RNA pull down
The protein extracts from AGS and SGC-7901 cells were subjected to 50 pmol of biotinylated miR-3619-5p (miR-3619-5p biotin probe) or control (miR-3619-5p non-biotin probe), in the presence of 50 μL of magnetic beads. The level of retrieved TBC1D10B was assayed by qRT-PCR.
Statistical analyses
All experimental procedures were independently repeated at least three times. Data were processed through Student’s t-test and one-way ANOVA applying GraphPad PRISM 6.0 (GraphPad, San Diego, CA, USA), showed as mean value ± SD. Correlation between miR-3619-5p and STAT4/TBC1D10B expression as well as between STAT4 and TBC1D10B expression in STAD tissues was confirmed by using Pearson’s correlation analysis. The survival rate curve was analyzed utilizing Kaplan–Meier method and log-rank test. The probability (p) less than 0.05 was taken as the cutoff value.
Results
MiR-3619-5p overexpression represses cell proliferation and metastasis in stomach adenocarcinoma
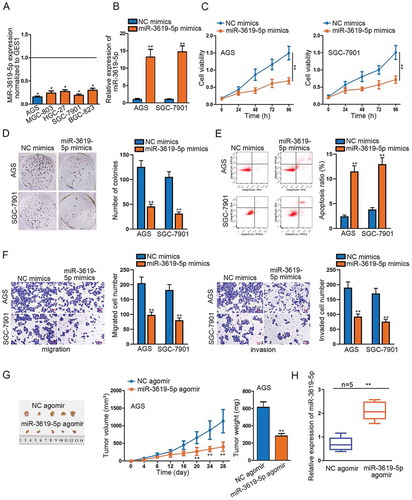
To comprehend the underlying mechanism of miR-3619-5p in STAD, we examined the relative expression of miR-3619-5p in STAD cells and tissues, detecting its down-regulated expression in cancerous cells and tumor tissues. Besides, AGS and SGC-7901 cells contained the least expression of miR-3619-5p among the indicated STAD cells ( and Supplementary Figure 1a). Meanwhile, the results of Kaplan–Meier analysis displayed that low expression of miR-3619-5p was vitally associated with a shorter survival rate of STAD patients (Supplementary Figure 1b). To determine the functional effects of miR-3619-5p in STAD cells, we conducted miR-3619-5p gain-of-function experiments next. Firstly, we transfected with miR-3619-5p mimics into AGS and SGC-7901 cells and observed an obvious up-regulation in miR-3619-5p expression (). Through CCK-8 and colony formation assays, cell viability and proliferation were significantly decreased in miR-3619-5p mimics’ transfected cells (–d). And the flow cytometry assay demonstrated that miR-3619-5p overexpression sharply increased apoptotic cell number (). Moreover, transwell assays presented a significant decrease in migrated and invaded cell number caused by miR-3619-5p mimics (). Additionally, in vivo experiments were performed as well. The AGS xenograft-bearing mice were randomly divided into two groups. Twenty-eight days after inoculation, we found that compared to the control group, the tumor size, volume and weight in the group injected with miR-3619-5p agomir were obviously smaller, suggesting the tumor-suppressing role of miR-3619-5p in STAD in vivo (). Further, qRT-PCR analysis illustrated that miR-3619-5p was up-regulated in tissues obtained from mice injected with miR-3619-5p agomir (). To sum up, the expression of miR-3619-5p is down-regulated in STAD cells and tissues, and miR-3619-5p overexpression represses STAD both in vitro and vivo.
STAT4 inhibits transcription of miR-3619-5p
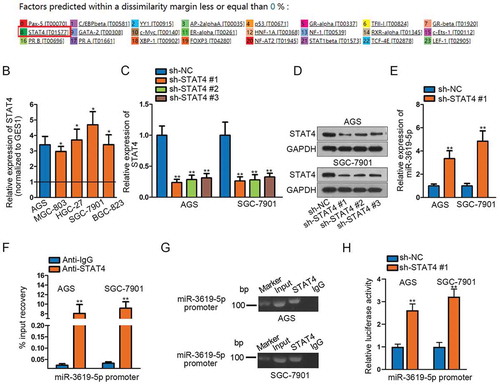
To seek for the possible transcription inhibitor of miR-3619-5p, we took advantage of UCSC (http://genome.ucsc.edu/) and PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) databases (). Among these candidates, STAT4 was reported to be a transcription repressor in previous studies. For example, STAT4 functions as a transcriptional repressor in response to IFN-α/β signaling in human memory Th2 cells.Citation17 To investigate whether STAT4 similarly had a repressive impact on miR-3619-5p transcription, we examined the expression of STAT4 first. The qRT-PCR results demonstrated the elevated expression of STAT4 in STAD cells and tissues ( and Supplementary Figure 1c). High expression of STAT4 was detrimental to the prognosis of STAD patients (Supplementary Figure 1d). Based on the low expression of miR-3619-5p in tissues, we conducted Pearson’s correlation analysis and concluded that STAT4 was negatively correlated with miR-3619-5p (Supplementary Figure 1e). Next, we examined the inhibited STAT4 mRNA and protein levels after silencing STAT4 through qRT-PCR and Western blotting (–d). In addition, the expression of miR-3619-5p significantly increased after silencing STAT4, suggesting the negative regulation of STAT4 to miR-3619-5p (). Furthermore, ChIP assay demonstrated the obvious input recovery of miR-3619-5p promoter in anti-STAT4 group, indicating the binding between miR-3619-5p promoter and STAT4 (). Additionally, agarose gel electrophoresis assay confirmed the PCR product of miR-3619-5p promoter in anti-STAT4 group, further elucidating the binding relation between the indicated molecules (). Finally, we conducted luciferase reporter assay and found that silencing STAT4 caused a significant increase in luciferase activity of miR-3619-5p promoter, suggesting the negative modulation role of STAT4 in miR-3619-5p transcription (). Additionally, IHC staining assay detected that the expression of STAT4 and Ki-67 was down-regulated, but that of cleaved caspase3 was up-regulated, when injecting miR-3619-5p agomir into AGS xenograft-bearing mice, ulteriorly supported the tumor-suppressor role of miR-3619-5p in STAD and the interactions between miR-3619-5p and STAT4 (Supplementary figure 1f). Collectively speaking, we verify that STAT4 binds with miR-3619-5p promoter and STAT4 represses the transcription of miR-3619-5p.
MiR-3619-5p modulates stomach adenocarcinoma by modulating TBC1D10B expression
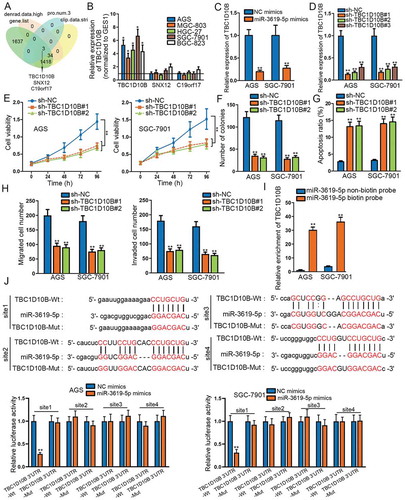
MiRNA as a non-coding RNA could regulate relevant cellular functions by modulating the expression of mRNA.Citation18 In our study, miR-3619-5p was confirmed to suppress STAD progression. However, its target gene that ultimately functions had not been identified. We screened out three candidate mRNAs in StarBase database (http://starbase.sysu.edu.cn/; clip data: strict; denrad data: high; program number: 3□gene. List) (). The relative expression of the three candidate mRNAs was detected in STAD cells, and only the expression of TBC1D10B was up-regulated in STAD cells compared to that in normal cells (). Moreover, TBC1D10B was up-regulated in STAD tissues rather than in adjacent non-tumor tissues (Supplementary Figure 2a). High expression of TBC1D10B was linked to the bad prognosis of STAD patients (Supplementary Figure 2b). Afterward, data of Pearson’s correlation analysis revealed that TBC1D10B was negatively/positively correlated with miR-3619-5p/STAT4 (Supplementary Figure 2c–d). Also, overexpression of miR-3619-5p led to significant down-regulation in TBC1D10B expression (). TBC1D10B was a rarely explored mRNA, and there are limited related investigations. We applied TBC1D10B loss-of-function experiments subsequently. Knockdown efficiency of TBC1D10B was examined first (). Next, CCK-8 and colony formation assays measured that knockdown of TBC1D10B attenuated cell viability and proliferation ability (–f). Apoptotic cell number increased after silencing TBC1D10B by flow cytometry assay (). Moreover, we could observe the decrease in cell migration and invasion when down-regulating TBC1D10B by transwell assays (). Furthermore, to confirm the binding relation between TBC1D10B and miR-3619-5p, we carried out RNA pull-down and luciferase reporter assays. According to pull-down assay, we could see the biotinylated miR-3619-5p could pull down TBC1D10B, suggesting the binding between the two molecules (). Via StarBase, the four potential binding sites between TBC1D10B and miR-3619-5p were exhibited. Then, by luciferase reporter assay, an obvious decrease was observed in luciferase activity of TBC1D10B wild type (site1) in miR-3619-5p mimics transfected STAD cells, while that of TBC1D10B mutant type presented no significant change (). Overall, TBC1D10B acts as the target gene miR-3619-5p, and knockdown of TBC1D10B suppresses proliferation, migration and invasion of STAD cells.
TBC1D10B restoration reverses cellular functions affected by miR-3619-5p mimics
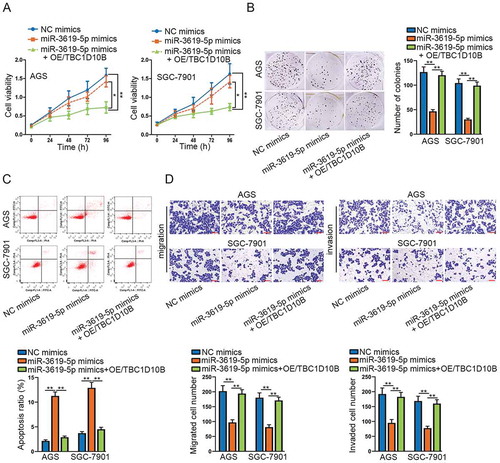
At last, we performed rescue experiments to verify whether TBC1D10B up-regulation could reverse the inhibitory function caused by overexpressing miR-3619-5p in the development of STAD. Cell viability was resumed when overexpressing TBC1D10B according to CCK-8 assay (). Also, cell proliferation ability nearly recovered to the previous level after overexpressing TBC1D10B by detection of colony formation assay (). Through flow cytometry assay, apoptotic cell number in miR-3619-5p mimics + OE/TBC1D10B transfected group decreased and reached closely to the control group (). Similarly, transwell assays demonstrated that the number of migrated and invaded cells recovered after transfection with overexpressed TBC1D10B (). Taken together, TBC1D10B up-regulation could save the loss of cellular function caused by overexpressing miR-3619-5p. In other words, miR-3619-5p modulates STAD progression by controlling the expression of TBC1D10B.
Discussion
Stomach adenocarcinoma (STAD) is one of the most common cancers in the digestive tract. Lack of effective biomarkers is one of the crucial causes of delayed diagnosis.Citation19 Therefore, the identification of efficient biomarkers is urgent. MiRNAs as important cancer biomarkers play a vital role in gene expression and control of cellular functions.Citation20-Citation22 MiR-3619-5p has been reported as a tumor inhibitor in various cancers including bladder cancer, retinoblastoma, infantile hemangioma and hepatocellular carcinoma.Citation23-Citation26 However, the connection between miR-3619-5p and STAD has not been explored yet. In our study, we detected the low expression of miR-3619-5p in STAD cells and tissues. Meanwhile, the patients with lower expression of miR-3619-5p had shorter survival time. The further gain-of-function experiments demonstrated the tumor-suppressive role of miR-3619-5p in STAD, such as the inhibition of STAD cell proliferation, migration and invasion as well as tumor growth.
To explore whether there was an upstream transcription inhibitor gene of miR-3619-5p, we retrieved PROMO database and identified the possible transcription factors for miR-3619-5p. Among these transcription factors, STAT4 was previously reported as a transcriptional repressor of IL5 gene in human memory Th2 cells.Citation17 To confirm whether STAT4 functions in the same way, we examined the expression of STAT4 first and detected the elevated expression of STAT4 in STAD cells and tissues. High expression of STAT4 was also closely related to the unsatisfactory prognosis of STAD patients. STAT4 was negatively correlated with miR-3619-5p about the expression in STAD tissues. Moreover, we observed knockdown of STAT4 caused a significant increase in miR-3619-5p expression. Through ChIP and agarose gel electrophoresis assays, we confirmed that STAT4 bound with miR-3619-5p promoter. Luciferase reporter assay verified the interaction between STAT4 and miR-3619-5p promoter. Thus, we can make sure that STAT4 suppresses transcription of miR-3619-5p by binding to miR-3619-5p promoter, which directly leads to the down-regulated expression of miR-3619-5p.
As what we cited above, miRNA modulates cellular functions via controlling target mRNA expression, which in our case, is to find a target gene of miR-3619-5p. We screened out three mRNAs that might share binding sites with miR-3619-5p utilizing StarBase database. Expression analysis excluded two mRNAs which showed no significant change in STAD cells. The remaining mRNA TBC1D10B was left for our further research. We transfected with miR-3619-5p mimics into AGS and SGC-7901 cells and observed significant down-regulation of TBC1D10B expression, suggesting TBC1D10B was negatively modulated by miR-3619-5p. Besides, the negative/positive correlation between TBC1D10B and miR-3619-5p/STAT4 was confirmed. There were no studies about TBC1D10B involving in miRNA modulation; thus, we decided to conduct TBC1D10B loss-of-function experiments to confirm the oncogenic role of TBC1D10B in STAD. The results verified our conjecture that STAD cell proliferation, migration and invasion were suppressed by silencing TBC1D10B. Furthermore, the mechanistic experiments confirmed the binding and interaction between TBC1D10B and miR-3619-5p as well. The final rescue experiments further confirmed that TBC1D10B up-regulation could inversely modulate repressive functions of miR-3619-5p overexpression in STAD development.
In summary, as shown in Supplementary Figure 3, STAT4-repressed miR-3619-5p affected cell proliferation, apoptosis, invasion and metastasis in STAD via modulating the downstream target gene TBC1D10B. Our study investigated the function and mechanism of miR-3619-5p in STAD and came up with a novel pathway STAT4/miR-3619-5p/TBC1D10B which regulated STAD tumorigenesis and progression. However, during the selection of miR-3619-5p transcription factors, we only explored the transfection effects of STAT4 in STAD cells while the other screened potential transcription factors have not been explored. In the future study, we will further explore the modulation role of other potential factors in miR-3619-5p transcription and other possible pathways involved in STAD.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download Zip (2.3 MB)Acknowledgments
We appreciate the technical supports of laboratory members.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Wang Z, Wang K, Dang Y, Ouyang X, Zhang F, Wang W, Wang L, Huang Q. Evaluation of the expression and clinical value of lncRNA AC010761.9 in human gastric adenocarcinoma. World J Surg Oncol. 2018;16:40. doi:10.1186/s12957-017-1289-y.
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi:10.3322/caac.20138.
- Ma Y, Liu L, Yan F, Wei W, Deng J, Sun J. Enhanced expression of long non-coding RNA NEAT1 is associated with the progression of gastric adenocarcinomas. World J Surg Oncol. 2016;14:41. doi:10.1186/s12957-016-0799-3.
- Gu J, Li Y, Fan L, Zhao Q, Tan B, Hua K, Wu G. Identification of aberrantly expressed long non-coding RNAs in stomach adenocarcinoma. Oncotarget. 2017;8:49201–49216. doi:10.18632/oncotarget.17329.
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–1207. doi:10.1016/j.jaci.2017.08.034.
- Wu W. MicroRNA, noise, and gene expression regulation. Methods Mol Biol (Clifton, NJ). 2018;1699:91–96.
- Backes C, Meese E, Keller A. Specific miRNA disease biomarkers in blood, serum and plasma: challenges and prospects. Mol Diagn Ther. 2016;20:509–518. doi:10.1007/s40291-016-0221-4.
- Bernardo BC, Ooi JY, Lin RC, McMullen JR. miRNA therapeutics: a new class of drugs with potential therapeutic applications in the heart. Future Med Chem. 2015;7:1771–1792. doi:10.4155/fmc.15.107.
- Cao T, Zhen XC. Dysregulation of miRNA and its potential therapeutic application in schizophrenia. CNS Neurosci Ther. 2018;24:586–597. doi:10.1111/cns.12840.
- Tan YL, Bai ZG, Zou WL, Ma XM, Wang TT, Guo W, Liu J, Li JS, Jie Y, Zang YJ, et al. miR-744 is a potential prognostic marker in patients with hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2015;39:359–365. doi:10.1016/j.clinre.2014.09.010.
- Fang Z, Yin S, Sun R, Zhang S, Fu M, Wu Y, Zhang T, Khaliq J, Li Y. miR-140-5p suppresses the proliferation, migration and invasion of gastric cancer by regulating YES1. Mol Cancer. 2017;16:139. doi:10.1186/s12943-017-0708-6.
- Shi Y, Shi H, Zhang B, Yan Y, Han X, Jiang W, Qian H, Xu W. miR-373 suppresses gastric cancer metastasis by downregulating vimentin. Mol Med Rep. 2018;17:4027–4034. doi:10.3892/mmr.2017.8291.
- Qiu X, Zhu H, Liu S, Tao G, Jin J, Chu H, Wang M, Tong N, Gong W, Zhao Q, et al. Expression and prognostic value of microRNA-26a and microRNA-148a in gastric cancer. J Gastroenterol Hepatol. 2017;32:819–827. doi:10.1111/jgh.13533.
- Huang H, Liao W, Zhu X, Liu H, Cai L. Knockdown of long noncoding RNA GHET1 inhibits cell activation of gastric cancer. Biomed Pharmacother. 2017;92:562–568. doi:10.1016/j.biopha.2017.05.088.
- Niu X, Liu S, Jia L, Chen J. Role of MiR-3619-5p in beta-catenin-mediated non-small cell lung cancer growth and invasion. Cel. Physiol Biochem. 2015;37:1527–1536. doi:10.1159/000438520.
- Zhang M, Luo H, Hui L. MiR-3619-5p hampers proliferation and cisplatin resistance in cutaneous squamous-cell carcinoma via KPNA4. Biochem Biophys Res Commun. 2019;513:419–425. doi:10.1016/j.bbrc.2019.03.203.
- Gonzales-van Horn SR, Estrada LD, van Oers NS, Farrar JD. STAT4-mediated transcriptional repression of the IL5 gene in human memory Th2 cells. Eur J Immunol. 2016;46:1504–1510. doi:10.1002/eji.201546050.
- Jin K, Zhao W, Xie X, Pan Y, Wang K, Zhang H. MiR-520b restrains cell growth by targeting HDAC4 in lung cancer. Thoracic Cancer. 2018;9:1249–1254. doi:10.1111/1759-7714.12825.
- Li Y, Wen X, Wang L, Sun X, Ma H, Fu Z, Li L. LncRNA ZEB1-AS1 predicts unfavorable prognosis in gastric cancer. Surg Oncol. 2017;26:527–534. doi:10.1016/j.suronc.2017.09.008.
- Sanjay S, Girish C. Role of miRNA and its potential as a novel diagnostic biomarker in drug-induced liver injury. Eur J Clin Pharmacol. 2017;73:399–407. doi:10.1007/s00228-016-2183-1.
- Ma Y. The Challenge of microRNA as a Biomarker of Epilepsy. Curr Neuropharmacol. 2018;16:37–42. doi:10.2174/1570159X15666170703102410.
- Bekris LM, Leverenz JB. The biomarker and therapeutic potential of miRNA in Alzheimer’s disease. Neurodegener Dis Manag. 2015;5:61–74. doi:10.2217/nmt.14.52.
- Zhang Q, Miao S, Han X, Li C, Zhang M, Cui K, Xiong T, Chen Z, Wang C, Xu H. MicroRNA-3619-5p suppresses bladder carcinoma progression by directly targeting beta-catenin and CDK2 and activating p21. Cell Death Dis. 2018;9:960. doi:10.1038/s41419-018-0986-y.
- Yan G, Su Y, Ma Z, Yu L, Chen N. Long noncoding RNA LINC00202 promotes tumor progression by sponging miR-3619-5p in retinoblastoma. Cell Struct Funct. 2019;44:51–60. doi:10.1247/csf.18033.
- Liu Z, Kang Z, Dai Y, Zheng H, Wang Y. Long non-coding RNA LINC00342 promotes growth of infantile hemangioma by sponging miR-3619-5p from HDGF. Am J Physiol Heart Circ Physiol. 2019. doi:10.1152/ajpheart.00188.2019.
- Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/beta-catenin pathway. Arch Biochem Biophys. 2019;661:196–202. doi:10.1016/j.abb.2018.11.020.