ABSTRACT
Colorectal cancer (CRC) is globally one of the most common malignant tumors. Increasing number of studies indicate that circular RNAs (circRNAs) play a significant role in the initiation and progression of CRC. However, the role of circRNA_100876 in CRC progression remains unclear. In this study, we investigated the expression and function of circRNA_100876 in CRC progression. The expression of circRNA_100876 and microRNA-516b (miR-516b) was compared in normal and CRC tissues using quantitative real-time polymerase chain reaction (RT-qPCR). In addition, proliferation, metastasis, and apoptosis of the cells were analyzed using Cell Counting Kit-8 (CCK-8) assay, Transwell assay, and flow cytometry, respectively. The relationship between circRNA_100876 and miR-516b was further verified using dual-luciferase reporter assay. Our data showed that circRNA_100876 was highly expressed in CRC tumor tissues, and the high expression gtransition (EMT)-related proteins. Furthermore, we found that the addition of miR-516b reversed the anti-tumor effect induced by the downregulation of circRNA_100876. In conclusion, this study revealed that circRNA_100876 is overexpressed in CRC tissues and represents a promising therapeutic target for CRC.
Introduction
Colorectal cancer (CRC) is the third most frequently diagnosed malignant tumor in the world and ranks second in terms of mortality.Citation1,Citation2 Due to the lack of early diagnostic methods, several patients are diagnosed with advanced CRC.Citation3 The main treatment modality for such patients includes radical gastrectomy combined with systemic chemotherapy.Citation4 In recent years, bacteria-mediated cancer therapy has become the focus of intense research.Citation5 A large number of natural compounds are also being used for the treatment of cancer.Citation6 Despite the scientific and clinical advances in treatment strategies, the total survival time of advanced CRC patients remains low. Therefore, there is an urgent need to identify novel biomarkers for CRC and elucidate their underlying mechanism.
Circular RNAs (circRNAs) are a newly-discovered class of endogenous non-coding RNAs with a closed loop structure.Citation7,Citation8 CircRNAs play a significant role in various diseases, including diabetesCitation9 and cancer.Citation10,Citation11 Furthermore, circRNAs can serve as microRNA (miRNA) sponges to regulate the biological function of miRNAs and participate in carcinogenesis and tumor progression.Citation12 Circ_0091570 has been identified as a tumor inhibitor of hepatocellular cancer through sponging of hsa-miR-1307.Citation13 In addition, Gao et al. reported that circ-PKD2 sponges miR-204-3p and suppresses carcinogenesis of oral squamous cell carcinoma.Citation14 However, the biological function of circRNA as miRNA sponges has not yet been clearly elucidated in CRC.
CircRNA_100876 has been confirmed as an oncogenic factor in the development of lung cancer,Citation15 esophageal squamous cell carcinoma,Citation16 and osteosarcoma.Citation17 Jin et al. showed that circRNA_100876 plays a significant role in the progression of osteosarcoma by regulating the function of miR-136.Citation17 However, the role and potential regulatory mechanism of circRNA_100876 in CRC remain unclear.
In this study, we first examined the relationship between the clinicopathological characteristic of patients with CRC and the expression level of circRNA_100876. Subsequently, we performed in vitro experiments to detect the biological function of circRNA_100876 in CRC progression. Further, based on the results of bioinformatics analysis, we postulate that circRNA_100876 sponges miR-516b to promote CRC. In summary, our data reveal that circRNA_100876 may be a novel diagnostic and therapeutic biomarker for CRC.
Results
CircRNA_100876 overexpression is associated with poor prognosis
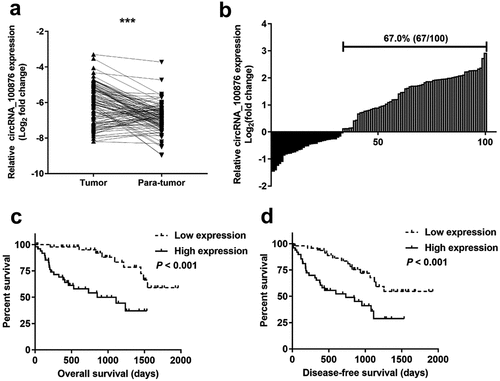
The expression of circRNA_100876 was compared in CRC and normal tissues using RT-qPCR. The RT-qPCR results revealed that circRNA_100876 was markedly upregulated in the CRC tissues (). Furthermore, 67% (67/100) of patient-derived CRC specimens showed overexpression of circRNA_100876 (). Based on the expression level of circRNA_100876, the samples were divided into high and low expression groups (). In addition, we analyzed the association of circRNA_100876 expression with CRC clinical features and found that circRNA_100876 expression was strongly associated with tumor differentiation (P = .000), vascular invasion (P = .005), tumor size (P = .044), T stage (P = .001), lymphatic metastasis (P = .007), and distant metastasis (P = .022). Our data also showed that CRC patients with circRNA_100876 overexpression were more likely to have shorter overall survival time () and shorter disease-free survival time (). Taken together, these results demonstrated that circRNA_100876 was upregulated in CRC tissues, and closely associated with a poor clinical outcome.
Table 1. Sequences of primers used in the present research.
Table 2. Correlation between circRNA_100876 expression and clinicopathological parameters of colorectal carcinoma.
Figure 1. CircRNA_100876 expression in CRC tissues, and correlation analyzes with post-operative survival. (a) CircRNA_100876 expression was significantly higher in CRC tissues than in adjacent normal tissues. (b) CircRNA_100876 expression was upregulated in 67% (67/100) of patients with CRC. (c) Correlation between circRNA_100876 expression and post-operative overall survival. (d) Correlation between circRNA_100876 expression and post-operative recurrence-free survival. All experiments were repeated at least three times. *** P < .001.
Downregulation of circRNA_100876 inhibits cell proliferation and increases cell apoptosis
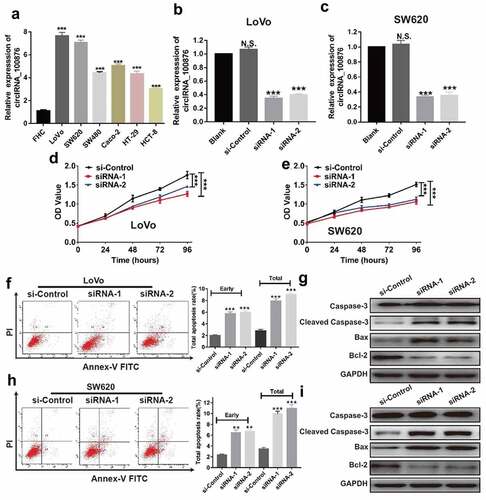
To explore the role of circRNA_100876 in CRC, we compared its expression level in CRC cell lines. RT-qPCR results revealed that circRNA_100876 was markedly upregulated in the CRC cell lines compared to a normal colon epithelial cell line, FHC. The cell lines, LoVo and SW620 showed highest expression levels (). Next, we depleted circRNA_100876 expression in LoVo and SW620 cells using lentivirus-mediated siRNA transfection and confirmed the knockdown using RT-qPCR (). Our data revealed that downregulation of circRNA_100876 significantly suppressed cell growth rate in both LoVo and SW620 cells, as evidenced by the lower OD values in the siRNA-1 and siRNA-2 transfected groups in CCK-8 assay (). Following the depletion of circRNA_100876, higher early and total apoptotic rates were observed in siRNA-1 and siRNA-2 groups than in the negative control group (si-Control) in LoVo or SW620 cells (). To further elucidate the molecular mechanism through which circRNA_100876 regulates the anti-apoptotic abilities in CRC cells, we analyzed the apoptosis-related markers in the CRC cells following depletion of circRNA_100876. The western blotting results revealed that the expression of Bcl-2 was inhibited, while that of cleaved caspase-3 and Bax was significantly upregulated following the depletion of circRNA_100876 ().
Figure 2. CircRNA_100876 knockdown inhibited CRC cell proliferation and induced apoptosis in vitro. (a) CircRNA_100876 expression in CRC cell lines and in human normal intestinal cell line, FHC. (b) Expression of circRNA_100876 in circRNA_100876-knockdown LoVo cell model. (c) Expression of circRNA_100876 in circRNA_100876-knockdown SW620 cell model. (d) Stable knockdown of circRNA_100876 suppressed the viability of LoVo cells and (e) SW620 cells detected using a CCK-8 assay. (f,h) Increase in the percentage of apoptotic cells following circRNA_100876 knockdown in LoVo cells (f) and SW620 cells (h). (g,i) Stable knockdown of circRNA_100876 upregulated the expression cleaved caspase-3, Bax, and downregulated Bcl-2 expression in (g) LoVo cells and (i) SW620 cells. All experiments were repeated at least three times. ** P < .01, *** P < .001.
Downregulation of circRNA_100876 inhibits the metastasis ability through suppressing epithelial-mesenchymal transition (EMT) pathway
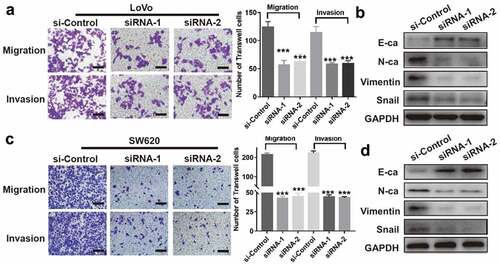
From the Transwell assay, we confirmed that the migration and invasion potentials of LoVo or SW620 cells were significantly reduced in both siRNA-1 and siRNA-2 groups, compared to si-Control group (). Furthermore, we analyzed the EMT-related proteins by western blotting. Interestingly, we found an increase in the epithelial marker, E-cadherin, and a decrease in the mesenchymal markers, N-cadherin, vimentin, and Snail in both LoVo and SW620 cell lines, following the downregulation of circRNA_100876 [].
Figure 3. CircRNA_100876 knockdown inhibited metastasis and epithelial-mesenchymal transition (EMT) in CRC cells. (a,c) Stable knockdown of circRNA_100876 significantly inhibited migration and invasion of (a) LoVo and (c) SW620 cells, measured using the Transwell assay, Scar bar, 50 μm. (b,d) CircRNA_100876 knockdown in (b) LoVo cells and (d) SW620 cells resulted in upregulation of the epithelial marker E-cadherin, downregulation of the mesenchymal markers N-cadherin and vimentin, and a decrease in the EMT–associated transcription factor, Snail. All experiments were repeated at least three times. *** P < .001.
Downregulation of miR-516b rescues cell proliferation and metastasis of CRC cells induced by depletion of circRNA_100876
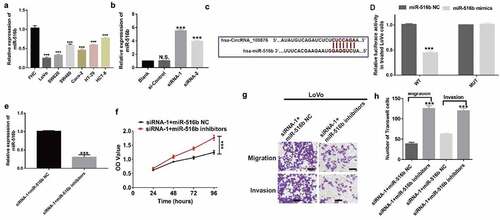
Based on the results of the bioinformatics analysis, we postulated that miR-516b may be a downstream target of circRNA_100876. Consequently, we evaluated the expression of miR-516b in CRC cell lines. RT-qPCR results showed that miR-516b was significantly downregulated in all the CRC cell lines (LoVo, SW620, SW480, Caco-2, HT-29 and HCT-8 cells), compared with that in the normal cell line (FHC). LoVo and SW620 cells showed the lowest expression of miR-516b (). Furthermore, depletion of circRNA_100876 in LoVo cells using siRNA-1 and siRNA-2 resulted in higher expression of miR-516b than that in the si-Control group (). Taken together, expression of miR-516b negatively correlated with the expression of circRNA_100876. Subsequently we predicted the potential binding sites of circRNA_100876 and miR-516b (). The results of luciferase reporter assay revealed that luciferase activity was markedly reduced in wild-type cells when treated with miR-516b mimics (). To explore the function of miR-516b, we reduced miR-516b expression in siRNA-1 LoVo cells using miR-516b specific inhibitors () and performed CCK-8 assays. The results of the CCK-8 assay revealed that miR-516b inhibitors rescued the cell proliferation ability of circRNA_100876-depleted LoVo cells (). Furthermore, the downregulation of miR-516b in LoVo cells also reduced cell migration and invasive abilities compared to the miR-516b NC group ().
Figure 4. Downregulation of miR-516b rescues cell proliferation and metastasis of CRC cells induced by depletion of circRNA_100876. (a) MiR-516b expression in CRC and the human normal intestinal cell line, FHC. (b) MiR-516b expression in circRNA_100876-knockdown LoVo cells. (c) The predicted binding region of circRNA_100876 in miR-516b. (d) Relative luciferase activity in 293 T cells after co-transfection with pmirGLO-circRNA_100876-WT or pmirGLO-circRNA_100876-MUT, along with miR-516b specific mimics or NC. (e) Expression of miR-516b in circ_100786-downregulated LoVo cells (siRNA-1), and downregulation of miR-516b in siRNA-1 LoVo cells, following the transfection with miR-516b inhibitors or NC. (f) Growth of siRNA-1 LoVo cells with or without depletion of miR-516b, detected using a CCK-8 assay. (g,h) Migration and invasion of siRNA-1 LoVo cells, with or without depletion of miR-516b (g), and the graphical representation of the data (h). All experiments were repeated at least three times. Scar bar, 50 μm. *** P < .001.
Discussion
CRC is one of the most common malignant tumors worldwide.Citation18,Citation19 Due to the lack of effective biomarkers and therapeutic targets, majority of patients are diagnosed with advanced CRC and their 5-year survival rate remains low. Recently, several studies have suggested that circRNAs serve a significant role in various cancer-associated biological processes, including promotion of cell proliferation, reducing cell apoptosis, and enhancing metastasis.Citation20 An oncogenic role for circ_0005075 has been reported in hepatocellular carcinoma.Citation21 Therefore, identifying novel circRNAs associated with the development of CRC and elucidating their potential regulatory mechanisms are of great importance.
Recently, the critical role of circRNA_100876 in the progression of various cancers and shown that its aberrant expression contributes to enhanced cell proliferation and metastasis.Citation16,Citation17 Yang et al. demonstrated that circRNA_100876 promotes cell proliferation and metastasis in breast cancer by sponging miR-361-3p.Citation22 To confirm the biological role of circRNA_100876 in CRC progression, we performed RT-qPCR assay to examine the expression pattern of circRNA_100876 in CRC tissues. Our data showed that circRNA_100876 was markedly upregulated in CRC tissues relative to normal tissues. In addition, overexpression of circRNA_100876 contributed to poorer tumor differentiation, vascular invasion, larger tumor size, advanced T stage, lymphatic metastasis, distant metastasis, as well as shorter survival time. Taken together, these data suggest that circRNA_100876 may be a tumor promoter in CRC.
Both, infinite proliferative capability and loss of apoptotic ability are key characteristics of malignant cells. To assess the role of circRNA_100876 in these biological processes in CRC, the expression of circRNA_100876 was downregulated in CRC cell lines using circRNA_100876 siRNA and negative control siRNA. The circRNA_100876 knockdown cells were used to elucidate its biological function and the underlying mechanism. Our data showed that silencing of circRNA_100876 suppressed cell growth and increased cell apoptosis. Recent studies indicate that overexpression of Bax, a pro-apoptotic protein from the Bcl-2-family of proteins, induces the release of cytochrome C and subsequent caspase activation, which induces apoptosis.Citation23 Gao et al. demonstrated that circRNA_0007059 triggers apoptosis by enhancing the expression of Bax and cleaved caspase 3.Citation24 In addition, Luo et al. reported that circRNA_0000064 promotes apoptosis in lung cancer cells by activating the functions of caspase 3 and 9.Citation25 Consistent with these data, we also found that upregulation of circRNA_100876 led to an increase in the expression of apoptotic markers, including Bax and cleaved caspase-3, while it downregulated the expression of the anti-apoptotic protein, Bcl-2. Therefore, these results suggest that downregulation of circRNA_100876 induces cell apoptosis by activating the caspase-dependent apoptotic pathway.
Further, the Transwell assay revealed that downregulation of circRNA_100876 resulted in reduced migration and invasion of CRC cells. However, the mechanism through which circRNA_100876 mediates the development of CRC remains unclear. EMT, which involves the transformation of epithelial cells into a mesenchymal state and enables enhanced invasion and metastasis of cancer cells, plays a pivotal role in tumor progression.Citation26,Citation27 The EMT signaling pathway can be triggered through dysfunction of circRNAs.Citation28,Citation29 For instance, circRNA_0023642 functions as a tumor suppressor in gastric cancer development by suppressing the EMT signaling pathway.Citation30 In addition, a previous study reported that circRNA_0009361 inhibits the EMT signaling pathway resulting in inhibition of cell proliferation and metastasis in CRC.Citation31 In the present study, silencing of circRNA_100876 resulted in upregulation of the epithelial-like marker, E-cadherin, and downregulation of the mesenchymal markers, N-cadherin, vimentin, and Snail, suggesting that circRNA_100876 may mediate carcinogenesis by regulating the EMT signaling pathway.
Accumulating evidence indicates that circRNAs can directly inhibit the function of miRNAs through complex mechanisms and cause tumor suppression or promotion. For example, circRNA-CACTIN acts as a competing endogenous RNAs (ceRNA), and contributes to the progression of gastric cancer by regulating miR-331-3p expression.Citation32 Another study demonstrated that circRNA_0025202 functions as a tumor suppressor in breast cancer by regulating the miR-182-5p/FOXO3a axis.Citation33 Therefore, based on the results of the bioinformatics analysis, we postulated that miR-1516 may be a downstream target of circRNA_100876. Further, since miRNAs have been shown to play a significant role in the development and progression of CRC,Citation34,Citation35 we confirmed the relationship between circRNA_100876 and miR-516b using dual-luciferase reporter assay. Furthermore, previous studies demonstrated that miR-516b participates in the procession of various cancers, including esophageal squamous cell carcinomaCitation36 and lung cancer.Citation37 In present study, our data revealed that the expression of miR-516b was negatively correlated with the expression of circRNA_100876 in CRC cells. Moreover, our data revealed that the addition of miR-516b reversed the tumor inhibitory effects induced by the silencing of circRNA_100876. Taken together, these data suggest that circRNA_100876 is involved in the progression of CRC through miRNA sponging of miR-516b.
In conclusion, circRNA_100876 may act as a tumor promoter in CRC, and its overexpression is closely associated with poor prognosis in CRC patients. In addition, downregulation of circRNA_100876 directly regulates the expression of miR-516b, suppresses cell proliferation, and increases cell apoptosis through activation of the caspase-dependent apoptotic pathway. Silencing of circRNA_100876 also inhibits cell migration and invasion by interfering with the EMT signaling pathway. However, this study has some limitations. Firstly, the molecules acting downstream of the circRNA_100876/miR-516b axis are unknown. Secondly, in vivo experiments are needed to further corroborate these findings. Overall, circRNA_100876 is a potentially promising target for the treatment of CRC.
Materials and methods
Patient tissue samples
A total of 64 pairs of CRC and normal adjacent tissues were obtained from CRC patients admitted to the Wuwei People’s Hospital for radical surgery between 2015 to 2017. All tissue samples were verified by experienced pathologists and stored frozen at −80°C until use. The inclusion criteria were age 18–80 years; provision of written informed consent; and pathologically confirmed CRC as assessed by veteran pathologists. The exclusion criteria included patients with other malignant diseases and patients with previous neoadjuvant chemotherapy or radiotherapy. The medical ethics committee of the Wuwei People’s Hospital approved the study, and the study was conducted in accordance with the Declaration of Helsinki.
Cell culture
Six human CRC cell lines including LoVo, SW620, SW480, Caco-2, HT-29, and HCT-8, and the human normal colon epithelial cell line (FHC) were obtained from the Shanghai Institute for Biological Sciences, China. All the cell lines were cultured in Dulbecco’s modified eagle’s medium (DMEM; Gibco, California, USA) containing 10% fetal bovine serum (Gibco) in a humidified incubator with 5% CO2 at 37°C.
Cell transfection
CRC cell lines with knock down of circRNA_100876 expression were constructed using circRNA_100876-specific siRNA lentivirus (Gene Pharma, Jiangsu, China) at a multiplicity of infection [MOI] of 100 using 5 ug/mL polybrene. Puromycin were used to select for cells with stable knockdown of circRNA_100876. These circRNA_100876 stable knockdown cells were further transfected with miR-516b inhibitor or negative control sequence (miR-516b NC) (Gene Pharma) using Lipofectamine 3000 transfection reagent (Thermo Fisher, MA, USA). The sequences of siRNAs used in the study are listed in ().
RT-qPCR assay
Total RNA was isolated from both the tissue samples and the CRC cell lines using TRIzol reagent (Takara, Dalian, China). A total of 500 ng of RNA was reverse transcribed into cDNA using the Prime Script RT Master Mix (Takara) following manufacturer’s protocol. Subsequently, the SYBR-Green PCR kit (Roche) was used to amplify these cDNA, and the 2−ΔΔCt method was used to evaluate the relative expression of the target genes. The sequences of the PCR primers used in the study are shown in ().
Cell proliferation assay
The treated cells were plated in a 96-well plate. Diluted CCK-8 solution was added to each well following incubation for 24, 48, 72, and 96 h. After further incubation for 2 h, the absorbance of each well was read at 450 nm with the microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Transwell assay
To evaluate cell migration and invasion, Transwell chambers (0.8 μm; Corning, NY, USA) with or without Matrigel coating were used for the experiment. Briefly, the treated cells were collected and suspended in serum-free medium. Then, 100 μL of cell suspension containing 4 × 104 cells was added to the upper chamber, while 500 μL of DMEM supplemented with 20% FBS was added to the lower chamber. Subsequently, the cells that migrated and invaded through the membrane of Transwell chambers were fixed, stained, observed, and imaged with a light microscope.
Flow cytometric analysis
The treated cells were collected and fixed in 70% Ealcohol at 4°C overnight. The percentages of cells in various stages of the cell cycle were detected using a cell cycle detection kit (Keygen, Nanjing, China) according to the manufacturer’s instructions.
Dual-luciferase reporter assay
First, 293 T cells were collected and plated in a 24-well plate. Following culturing for 24 h, the cells were transfected with pmirGLO-circRNA_100876-WT or pmirGLO-circRNA_100876-MUT plasmid, with miR-516b mimics or miR-516b NC. After 48 h of incubation, the relative luciferase activity was detected using a dual-luciferase reporter assay system (Promega).
Western blotting
Cell pellets were treated with RIPA lysis buffer (Beyotime, Shanghai, China) for protein extraction. BCA protein Assay Kit (Beyotime) was used to determine the protein concentration. Equal amounts of proteins were separated on a 10% gel using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were then transferred onto a polyvinylidene fluoride (PVDF) membrane, followed by incubation with 10% BSA, primary antibody, and secondary antibody. Finally, the protein bands were visualized using the super sensitive ECL luminescence reagent (Meilunbio, Dalian, China).
Statistics
Data are presented as mean ± standard deviation (SD), and analyzed using GraphPad Prism v7.0 software. The difference between two groups was evaluated using Student’s t test or Chi-square test. P < .05 was considered significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Huan Zeng and Jianrong Zhang conceived the study. Huaiming Wang and Fajie Zhan performed the experiments and wrote the manuscript. Huan Zeng, Ketong Wu and Jianrong Zhang participated in the revision of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
We thank Editage for its linguistic assistance during the preparation of this manuscript.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
- Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691.
- Garcia-Larsen V, Morton V, Norat T, Moreira A, Potts JF, Reeves T, Bakolis I. Dietary patterns derived from principal component analysis (PCA) and risk of colorectal cancer: a systematic review and meta-analysis. Eur J Clin Nutr. 2019;73(3):366–386.
- Mirzaei R, Mirzaei H, Alikhani MY, Sholeh M, Arabestani MR, Saidijam M, Karampoor S, Ahmadyousefi Y, Moghadam MS, Irajian GR, et al. Bacterial biofilm in colorectal cancer: what is the real mechanism of action? Microb Pathog. 2020;142:104052.
- Honari M, Shafabakhsh R, Reiter RJ, Mirzaei H, Asemi Z. Resveratrol is a promising agent for colorectal cancer prevention and treatment: focus on molecular mechanisms. Cancer Cell Int. 2019;19:180.
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 495(7441):333–338.
- Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–692.
- Abbaszadeh-Goudarzi K, Radbakhsh S, Pourhanifeh MH, Khanbabaei H, Davoodvandi A, Fathizadeh H, Sahebkar A, Shahrzad MK, Mirzaei H. Circular RNA and diabetes: epigenetic regulator with diagnostic role. Curr Mol Med. 2020;20(7);516–526.
- Naeli P, Pourhanifeh MH, Karimzadeh MR, Shabaninejad Z, Movahedpour A, Tarrahimofrad H, Mirzaei HR, Bafrani HH, Savardashtaki A, Mirzaei H, et al. Circular RNAs and gastrointestinal cancers: epigenetic regulators with a prognostic and therapeutic role. Crit Rev Oncol Hematol. 2020;145:102854.
- Shabaninejad Z, Vafadar A, Movahedpour A, Ghasemi Y, Namdar A, Fathizadeh H, Pourhanifeh MH, Savardashtaki A, Mirzaei H. Circular RNAs in cancer: new insights into functions and implications in ovarian cancer. J Ovarian Res. 2019;12(1):84.
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
- Wang YG, Wang T, Ding M, Xiang SH, Shi M, Zhai B. hsa_circ_0091570 acts as a ceRNA to suppress hepatocellular cancer progression by sponging hsa-miR-1307. Cancer Lett. 2019;460:128–138.
- Gao L, Zhao C, Li S, Dou Z, Wang Q, Liu J, Ren W, Zhi K. circ-PKD2 inhibits carcinogenesis via the miR-204-3p/APC2 axis in oral squamous cell carcinoma. Mol Carcinog. 2019;58(10):1783–1794.
- Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ, Nan KJ. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–456.
- Cao S, Chen G, Yan L, Li L, Huang X. Contribution of dysregulated circRNA_100876 to proliferation and metastasis of esophageal squamous cell carcinoma. Onco Targets Ther. 2018;11:7385–7394.
- Jin J, Chen A, Qiu W, Chen Y, Li Q, Zhou X, Jin D. Dysregulated circRNA_100876 suppresses proliferation of osteosarcoma cancer cells by targeting microRNA-136. J Cell Biochem. 2019;120(9):15678–15687.
- Collaborators GCC. The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol Hepatol. 2019;4(12):913–933.
- Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, Abdel-Rahman O, Abdelalim A, Abdoli A, Abdollahpour I, Abdulle ASM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study. JAMA oncol. 2019;5(12);1749–1768.
- Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881 e813.
- Yang X, Song H, Zi Z, Kou J, Chen S, Dai Y, Wang J, Yuan L, Gao K. Circ_0005075 promotes hepatocellular carcinoma progression by suppression of microRNA-335. J Cell Physiol. 2019;234(12):21937–21946.
- Yang CY, Zhang FX, He JN, Wang SQ. CircRNA_100876 promote proliferation and metastasis of breast cancer cells through adsorbing microRNA-361-3p in a sponge form. Eur Rev Med Pharmacol Sci. 2019;23(16):6962–6970.
- Finucane DM, Bossy-Wetzel E, Waterhouse NJ, Cotter TG, Green DR. Bax-induced caspase activation and apoptosis via cytochrome c release from mitochondria is inhibitable by Bcl-xL. J Biol Chem. 1999;274(4):2225–2233.
- Gao S, Yu Y, Liu L, Meng J, Li G. Circular RNA hsa_circ_0007059 restrains proliferation and epithelial-mesenchymal transition in lung cancer cells via inhibiting microRNA-378. Life Sci. 2019;233:116692.
- Luo YH, Zhu XZ, Huang KW, Zhang Q, Fan YX, Yan PW, Wen J. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892–898.
- Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–134.
- Feng YX, Sokol ES, Del Vecchio CA, Sanduja S, Claessen JH, Proia TA, Jin DX, Reinhardt F, Ploegh HL, Wang Q, et al. 2014. Epithelial-to-mesenchymal transition activates PERK-eIF2alpha and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 4(6):702–715.
- Xue YB, Ding MQ, Xue L, Luo JH. CircAGFG1 sponges miR-203 to promote EMT and metastasis of non-small-cell lung cancer by upregulating ZNF281 expression. Thorac Cancer. 2019;10(8):1692–1701.
- Wei S, Zheng Y, Jiang Y, Li X, Geng J, Shen Y, Li Q, Wang X, Zhao C, Chen Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193.
- Zhou LH, Yang YC, Zhang RY, Wang P, Pang MH, Liang LQ. CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci. 2018;22(8):2297–2303.
- Geng Y, Zheng X, Hu W, Wang Q, Xu Y, He W, Wu C, Zhu D, Wu C, Jiang J. Hsa_circ_0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin Sci. 2019;133(10):1197–1213.
- Zhang L, Song X, Chen X, Wang Q, Zheng X, Wu C, Jiang J. Circular RNA CircCACTIN Promotes Gastric Cancer Progression by Sponging MiR-331-3p and Regulating TGFBR1 Expression. Int J Biol Sci. 2019;15(5):1091–1103.
- Sang Y, Chen B, Song X, Li Y, Liang Y, Han D, Zhang N, Zhang H, Liu Y, Chen T, et al. 2019. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther. 27(9):1638–1652.
- Savardashtaki A, Shabaninejad Z, Movahedpour A, Sahebnasagh R, Mirzaei H, Hamblin MR. miRNAs derived from cancer-associated fibroblasts in colorectal cancer. Epigenomics. 2019;11(14):1627–1645.
- Moridikia A, Mirzaei H, Sahebkar A, Salimian J. MicroRNAs: potential candidates for diagnosis and treatment of colorectal cancer. J Cell Physiol. 2018;233(2):901–913.
- Zhao Y, Wang Y, Xing G. miR-516b functions as a tumor suppressor by directly modulating CCNG1 expression in esophageal squamous cell carcinoma. Biomed Pharmacother. 2018;106:1650–1660.
- Zhu J, Zhang Y, Yang X, Jin L. Clinical significance and tumor-suppressive function of miR-516b in nonsmall cell lung cancer. Cancer Biother Radiopharm. 2017;32(4):115–123.
