ABSTRACT
Globally, lung cancer is known as a major cause of cancer-associated death and non-small-cell lung cancer (NSCLC) accounts for majority of all cases. Growing evidence has emerged that long non-coding RNAs (lncRNAs) act as vital regulatory molecules in various malignancies. Nevertheless, the function of SLCO4A1 antisense RNA 1(SLCO4A1-AS1) in NSCLC is vague. This study intended to investigate the biological role and probable regulatory mechanism of SLCO4A1-AS1 in NSCLC. qRT-PCR revealed that SLCO4A1-AS1 level was upregulated in NSCLC. Function assays manifested that silence of SLCO4A1-AS1 attenuated NSCLC cell proliferation, migration and invasion but promoted NSCLC cell apoptosis. Furthermore, we disclosed that SLCO4A1-AS1 activated NF-κB pathway in NSCLC, and that IKKα, an NF-κB pathway-related gene, possessed an enhanced level in NSCLC tissues and cells. Importantly, miR-223-3p bound with SLCO4A1-AS1 and IKKα. Further, SLCO4A1-AS1 competitively bound with miR-223-3p to increase IKKα expression, thereby activating NF-κB signaling pathway. In conclusion, SLCO4A1-AS1 drove NSCLC progression by activating NF-κB signaling pathway via sponging miR-223-3p to enhance IKKα expression. Thus, SLCO4A1-AS1 might be a promising biomarker for NSCLC treatment.
Introduction
As one of the most prevalent malignancies, lung cancer is featured with high morbidity and fatality globally.Citation1 Non-small-cell lung cancer (NSCLC) accounts for the highest proportion among all lung cancer cases.Citation2 Unfortunately, the precise causes of NSCLC have not been thoroughly explored until now, although the biology and management of this disease has been developed to some extent.Citation3 Accumulating studies have demonstrated that long-term heavy smoking is a risk factor for NSCLC development.Citation4 Moreover, previous researches about biomarker susceptibility related to NSCLC have also validated the importance of genetic factors in NSCLC tumorigenesis. Due to limited available strategies for early diagnosis and treatment, only few NSCLC patients could survive for more than 5 y.Citation5 Taking these into account, it is of great significance to search for effective molecular markers for the prediction and treatment of NSCLC.
Long (lncRNAs) of over 200nts at length are a subtype of non-coding RNAs (ncRNAs).Citation6 In recent years, mounting evidence has shown that aberrant expression of certain lncRNAs is associated with the progress of various cancers. For example, lncRNA PTAR aggravated epithelial-to-mesenchymal transition and metastasis in ovarian cancer by serving as a competitive sponge of miR-101-3p to increase ZEB1 expression.Citation7 LncRNA SNHG20 accelerates osteosarcoma tumorigenesis via mitochondrial apoptosis pathway through modulating miR-139/RUNX2 pathway.Citation8 LncRNA HNF1A-AS1 participates in cell proliferation and migration and the tumorigenesis of esophageal adenocarcinoma.Citation9 The oncogenic role of lncRNA SLCO4A1-AS1 (SLCO4A1 antisense RNA 1) was identified in several cancers, too. SLCO4A1-AS1 triggers cell proliferation via increasing autophagy and targets miR-508-3p/PARD3 axis in colorectal cancer.Citation10 SLCO4A1-AS1 drives the progression of bladder cancer via competitively binding with miR-335-5p to enhance OCT4 expression.Citation11 SLCO4A1-AS1 facilitates colorectal cancer cell growth and migration by activating Wnt/β-catenin pathway.Citation12 Nonetheless, the specific expression pattern and function of SLCO4A1-AS1 in NSCLC are unknown to a great extent.
Our present work intended to probe into the biological function and possible regulatory mechanism of lncRNA SLCO4A1-AS1 in NSCLC, which might provide possible novel diagnostic markers and treatment targets for NSCLC.
Materials and methods
Tissue samples
Thirty-six NSCLC tissues and paired non-tumor tissues were gained from NSCLC patients who received surgery at the First Affiliated Hospital of Suzhou University. No patients underwent radiotherapy or chemotherapy and all of them signed the written informed consent before surgery. Fresh tissues collected from the surgery were preserved at −80°C after immediate frozen by liquid nitrogen. This study was supported by the First Affiliated Hospital of Suzhou University.
Cell culture and treatment
Human bronchial epithelial cell (16HBE) and NSCLC cells (A549, H1299, H1260 and H520) were bought from American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultivated with RPMI 1640 medium (Gibco, Rockville, MD, USA) containing 10% FBS (Gibco) as well as 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) in a humidified incubator with 5% CO2 at 37°C. Jagged1, CD40 L and LiCl were acquired from Sigma-Aldrich (St. Louis, MO, USA).
Cell transfection
ShRNAs specifically against SLCO4A1-AS1 (sh-SLCO4A1-AS1#1 and sh-SLCO4A1-AS1#2) and relevant negative control sh-NC, together with the pcDNA3.1 vector covering SLCO4A1-AS1 or IKKα and the empty vector, were gained from Genechem (Shanghai, China). Meanwhile, miR-223-3p mimics and NC mimics were obtained from GenePharma (Shanghai, China). Indicated plasmids were appropriately transfected into A549 or H1299 cells using Lipofectamine 3000 (Invitrogen) as per the manufacturer’s protocol as described before.Citation13
qRT-PCR
Extraction of total RNA was operated using the Trizol kit (Invitrogen). Subsequently, total RNA was reversely transcribed into cDNA via a Prime Script RT reagent Kit (Takara, Tokyo, Japan). qRT-PCR was implemented through a Fast SYBR Green PCR kit (Applied biosystems, Foster City, CA, USA) on ABI7500 real-time quantitative PCR instrument (Applied biosystems). Gene expression was relative to GAPDH or U6 (internal reference) with 2−ΔΔCt approach.
Cell proliferation assay
A549 or H1299 cells under diverse treatments were put into 96-well plates. Following 0, 24, 48, 72 and 96 h incubation, CCK-8 solution (Sigma-Aldrich) was supplemented. Upon incubation for 3 h, absorbance was detected at 450 nm with a microplate reader (Bio-Rad, Sunnyvale, CA, USA).
Colony formation assay
After transfection, the cultured A549 and H1299 cells at logarithmic growth phase were trypsinized, reaped and re-suspended, followed by inoculation at 6-well culture plates (800/well) under the condition of 37°C and 5% CO2. After incubated for 2 weeks, cells were carefully rinsed twice utilizing PBS (Sigma-Aldrich) and fixated for 15 min using 75% ethyl alcohol (Sigma-Aldrich), followed by 0.5 h of staining in 0.1% crystal violet (Sigma-Aldrich). After that, colonies containing ≥50 cells were manually counted.
Transwell assay
Transwell chambers, with or without Matrigel (Corning Costar, Cambridge, MA, USA), were applied for examination of cell migration and invasion, respectively. Transfected A549 or H1299 cells with or without treatment were suspended in serum-free medium and were placed onto the upper chambers of Transwell, whereas medium containing 10% FBS was plated into the lower chambers as a chemoattractant. Forty-eight hours later, non-migratory or noninvasive cells were wiped out. Cells were immobilized for 20 min with ethanol (Sigma-Aldrich) and stained for 10 min in 0.1% crystal violet. Migratory or invasive cells were counted in five different microscopic fields, randomly.
Annexin V-FITC/PI analysis
Indicated A549 or H1299 cells were added in 12-well plates. The Annexin V-FITC/PI apoptosis detection kit (BD Biosciences, Franklin Lakes, NJ, USA) was utilized to evaluate cell apoptosis. The rate of apoptotic cells was calculated via a FACS flow cytometer (Beckman Coulter, Brea, CA, USA).
JC-1 staining
Indicated A549 or H1299 cells were fixated in 24-well plates, followed by rinsing twice utilizing PBS. Cells were then stained using JC-1 (Beyotime, Shanghai, China). Stained cells were then analyzed via a fluorescence microscope (Olympus, Tokyo, Japan).
Xenograft model
A549 cells treated with sh-SLCO4A1-AS1#1 or sh-NC were injected subcutaneously into 6-week-old BALB/c nude mice acquired from SLAC (Shanghai, China). Tumor volumes were recorded every 4 d. Four weeks later, mice were sacrificed and tumors were weighted.
Western blot
Total protein extraction was carried out using RIPA Lysis Buffer (Solarbio, Beijing, China), followed by centrifugation for 10 min. Thereafter, proteins in cell lysates were separated using 10% SDS-PAGE (Bio-Rad) and transferred to nitrocellulose membranes (Millipore, Bedford, MA, USA). Membranes were then processed with primary antibodies against p-IkB-α (Ser32) (ab92700, Abcam, Cambridge, USA), p-IkB-α(Ser36) (ab133462, Abcam), IKKα (ab32041, Abcam), GAPDH (ab8245, Abcam) or Tubulin (ab7291, Abcam). After washing, membranes were incubated with secondary antibodies, followed by autoradiography using the ECL Detection Kit (Pierce, Rockford, IL, USA).
Subcellular fractionation
Extraction of cytoplasmic and nuclear RNA was undertaken utilizing a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, Shanghai, China) plus Trizol. Expression of p65 was carried out via qRT-PCR.
RNA immunoprecipitation (RIP)
RIP was implemented with a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) using anti-Ago2 antibody (Abcam) or anti-IgG antibody (Abcam). qRT-PCR was applied to assay the relative levels of SLCO4A1-AS1, miR-223-3p and IKKα in immunoprecipitates of each group.
Luciferase reporter assay
SLCO4A1-AS1-WT/Mut or IKKα-WT/Mut was sub-cloned into pmirGLO dual-luciferase vector (Promega, Madison, WI, USA) to generate pmirGLO-SLCO4A1-AS1-WT/Mut or pmirGLO-IKKα-WT/Mut. The pmirGLO-SLCO4A1-AS1-WT/Mut was co-transfected into A549 or H1299 cells with miR-223-3p mimics or NC mimics. The pmirGLO-IKKα-WT/Mut was co-transfected into A549 or H1299 cells with miR-223-3p mimics or miR-223-3p mimics+pcDNA3.1/SLCO4A1-AS1 or NC mimics. Luciferase activities were inspected via Dual-Luciferase reporter assay system (Promega).
Statistical analysis
SPSS 17.0 (SPSS, Chicago, IL, USA) was employed for statistical analysis. All assays were implemented in triplicate and data were presented as mean ± SD. Differences between groups were studied by the use of Student’s t-test or one-way ANOVA, with P < .05 as significant. Pearson’s correlation analysis was applied to verify the relationship among SLCO4A1-AS1, miR-223-3p and IKKα in expression.
Results
High expression of SLCO4A1-AS1 drove the progression of NSCLC
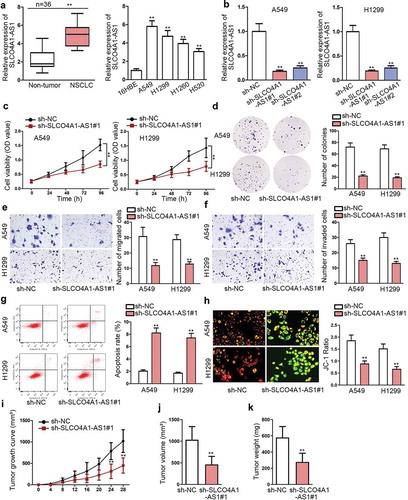
In order to investigate SLCO4A1-AS1 in NSCLC, qRT-PCR was applied to analyze SLCO4A1-AS1 level in NSCLC. Based on the results, SLCO4A1-AS1 expression was strikingly boosted in NSCLC tissues and cells (). Next, we investigated the function of SLCO4A1-AS1 in NSCLC through performing loss-of-function assays in A549 and H1299 cells since they contained a higher expression of SLCO4A1-AS1. To begin with, SLCO4A1-AS1 expression was suppressed by sh-SLCO4A1-AS1#1 and sh-SLCO4A1-AS1#2 vectors, resulting in an evident reduction of SLCO4A1-AS1 expression in NSCLC cells (). We chose sh-SLCO4A1-AS1#1 for the subsequent experiments as it presented a robust knockdown efficiency of SLCO4A1-AS1. As a result, cell proliferation was alleviated in two cells owing to SLCO4A1-AS1 depletion (, d). Consistently, transwell assay indicated that cell migration and invasion were impeded due to the silence of SLCO4A1-AS1 (, f). Conversely, cell apoptosis was increased by inhibition of SLCO4A1-AS1 based on the results from flow cytometry (). Likewise, JC-1 assay showed the decreased mitochondrial membrane potential in sh-SLCO4A1-AS1#1 group (). In addition, in vivo assays uncovered that tumor growth was obstructed, with the final volume and weight both abated, on account of SLCO4A1-AS1 ablation (–k). Taken together, SLCO4A1-AS1 was overexpressed in NSCLC tissues and cells, and depletion of SLCO4A1-AS1 impaired the progression of NSCLC.
Figure 1. High expression of SLCO4A1-AS1 drove the progression of NSCLC.
SLCO4A1-AS1 activated NF-κB signaling
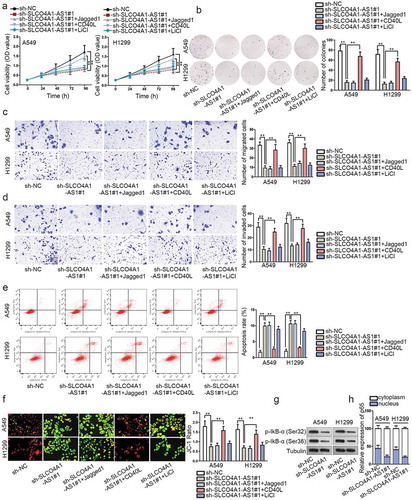
Multiple evidences suggested that Wnt/β-catenin, Notch and NF-κB signaling pathways exert function on cancer development.Citation14–Citation16 However, whether SLCO4A1-AS1 regulated these signaling pathways in NSCLC remains to be elucidated. Hence, we separately treated A549 and H1299 cells with activators LiCl, Jagged1 or CD40 L to activate Wnt/β-catenin, Notch and NF-κB signaling pathways, respectively. Interestingly, we revealed that the reduced NSCLC cell proliferation by SLCO4A1-AS1 deficiency was reversed by NF-κB signaling pathway activator (CD40 L), while LiCl, Jagged1 had no evident effects (, b). Similarly, only CD40 L treatment abolished the repressive impact on cell motility induced by SLCO4A1-AS1 depression (, d). Furthermore, SLCO4A1-AS1-mediated promotion on cell apoptosis was reversed by CD40 L treatment (, f). Besides, SLCO4A1-AS1 downregulation reduced the phosphorylation of IkB-α (Ser32) and IkB-α (Ser36), thereby inhibiting the nuclear translocation of p65 (, h). In sum, SLCO4A1-AS1 activated NF-κB signaling pathway in NSCLC.
Figure 2. SLCO4A1-AS1 activated NF-κB signaling pathway.
SLCO4A1-AS1 competitively bound with miR-233-3p to regulate IKKα
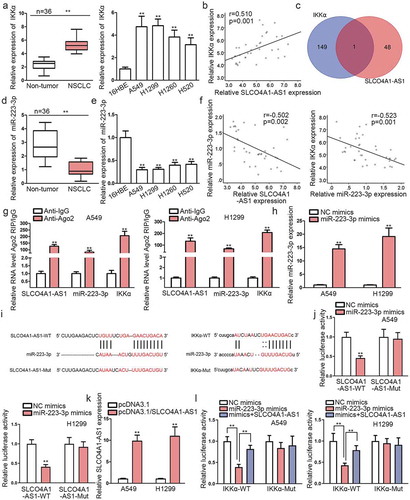
As prior studies have confirmed that SLCO4A1-AS1 activated NF-κB signaling pathway in NSCLC cells, specific regulatory mechanism of SLCO4A1-AS1 and NF-κB signaling pathway needed to be clarified. IKKα is an important regulator of NF-κB signaling pathway, so we speculated that SLCO4A1-AS1 might regulate NF-κB signaling pathway through IKKα. qRT-PCR manifested the elevation of IKKα level in NSCLC samples and cells (). Besides, IKKα expression was positively associated with SLCO4A1-AS1 expression (), suggesting there might be some mechanisms between SLCO4A1-AS1 and IKKα. A large body of studies have evidenced the acting of lncRNAs as a competing endogenous RNA (ceRNA) via absorbing miRNAs to regulate mRNAs.Citation17,Citation18 Therefore, starBase algorithm (http://starbase.sysu.edu.cn/) and DIANA tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex) were utilized for the prediction of potential miRNAs which could bind with SLCO4A1-AS1 or IKKα. Results displayed that only miR-223-3p could bind with SLCO4A1-AS1 and targeted IKKα simultaneously (). qRT-PCR unveiled an obvious decreased expression of miR-223-3p in NSCLC tissues and cells (-e). Also, miR-223-3p was negatively related to SLCO4A1-AS1 or IKKα in expression (). Further, we unveiled that the expression levels of SLCO4A1-AS1 and IKKα were both lessened in in vivo tumors originated from A549 cells with depleted SLCO4A1-AS1, whereas that of miR-223-3p nearly unaltered (). Moreover, IkB-α phosphorylation was largely obstructed in the above tumors due to SLCO4A1-AS1 inhibition-declined IKKα protein (), further validating that SLCO4A1-AS1 affected NF-κB pathway through targeting IKKα. Then, we planned to figure out whether SLCO4A1-AS1 modulated IKKα through its sponging effect on miR-223-3p. As expected, RIP assay revealed that SLCO4A1-AS1, miR-223-3p and IKKα were all co-immunoprecipitated by anti-Ago2 but never by anti-IgG (). MiR-223-3p was overexpressed in A549 and H1299 cells (). The predicted binding sites for miR-223-3p at SLCO4A1-AS1 or IKKα were shown in . Expectedly, elevated miR-223-3p expression decreased fluorescence of SLCO4A1-AS1-WT rather than that of SLCO4A1-AS1-Mut (). Of note, after SLCO4A1-AS1 upregulation in A549 and H1299 cells (), the decreased activity of IKKα-WT under miR-223-3p upregulation was recovered due to SLCO4A1-AS1 upregulation (). On the whole, SLCO4A1-AS1 sequestered miR-233-3p to boost IKKα level and therefore activate NF-κB signaling in NSCLC.
Figure 3. SLCO4A1-AS1 competitively bound with miR-233-3p to regulate IKKα.
SLCO4A1-AS1 facilitated NSCLC progression by regulating IKKα
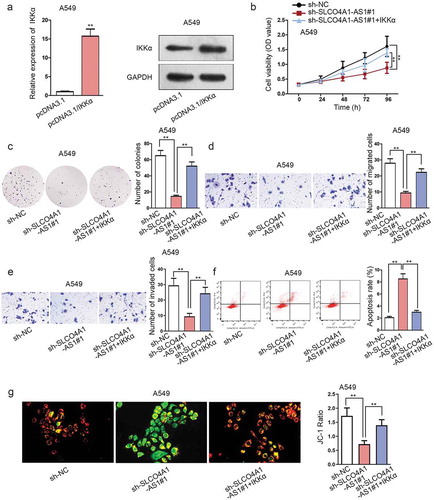
Finally, rescue assays were carried out to explore whether SLCO4A1-AS1 facilitated NSCLC progression by regulating IKKα in A549 cells. Prior to this, IKKα was first overexpressed in A549 cells at both mRNA and protein levels under transfection with pcDNA3.1/IKKα (). As expected, SLCO4A1-AS1 depletion-mediated suppression of cell proliferation was abrogated by IKKα upregulation (, c). Repressive influence on cell motility in NSCLC cells by SLCO4A1-AS1 depletion was also countervailed because of IKKα overexpression (, e). Conversely, cell apoptosis was elevated by SLCO4A1-AS1 deficiency, while IKKα overexpression counteracted apoptosis (, g). On the whole, our studies demonstrated that SLCO4A1-AS1 facilitated NSCLC progression by regulating IKKα.
Figure 4. SLCO4A1-AS1 facilitated NSCLC progression by regulating IKKα.
Discussion
NSCLC is the main type of devastating malignant lung cancer globally. Accumulating researches have indicated that lncRNAs play vital parts in NSCLC tumorigenesis. For instance, LINC01234 is identified as a pro-tumor lncRNA in NSCLC by binding to HNRNPA2B1 and modulating miR-106b Biogenesis.Citation19 LncRNA XIST contributed to oncogenesis and chemo-resistance in NSCLC.Citation20 PCNA-AS1 facilitates oncogenic activity in NSCLC by targeting CCND1.Citation21 LncRNA SLCO4A1-AS1 was implicated in bladder cancerCitation11 and colorectal cancer,Citation10,Citation12 but it has not been investigated in NSCLC till now. Presently, SLCO4A1-AS1 presented an enhanced expression in NSCLC. Also, SLCO4A1-AS1 silencing hindered cell proliferation and motility in NSCLC. In addition, in vivo assays disclosed that SLCO4A1-AS1 depletion blocked tumor growth of NSCLC. The data implied SLCO4A1-AS1 contributed to the progression of NSCLC.
Nuclear Factor-kappa B (NF-κB) pathway has been uncovered to control carcinogenesis processes in a variety of cancers. In this pathway, various stimuli drive the phosphorylation of IKKα/β and NF-κB p65 (p65), which finally triggers the translocation of p65 to the nucleus, and activates transcription of multiple NF-κB-associated tumor-promoting genes.Citation22 In this study, by utilizing the activator of NF-κB signaling pathway, we observed increased cell proliferation, migration and invasion and attenuated cell apoptosis. SLCO4A1-AS1 suppression declined the phosphorylation of IkB-α (Ser32) and IkB-α (Ser36), which further inhibited the nuclear import of p65. These data indicated SLCO4A1-AS1 activated NF-κB pathway in NSCLC.
IKKα is a regulator of NF-κB signaling pathway and has been verified to serve as a tumor promoter in liver cancer.Citation23 Currently, IKKα was uncovered as strongly expressed in NSCLC. Moreover, IKKαhad a positive correlation with SLCO4A1-AS1 in expression.
MicroRNAs (miRNAs) are a class of small non-coding RNAs that can affect various vital biological functions.Citation24 Previous reports proved that lncRNAs can sequester miRNAs to serve as ceRNAs and thereby regulate target mRNA expression.Citation25–Citation27 Herein, we disclosed that miR-223-3p shared by SLCO4A1-AS1 and IKKα. In addition, miR-223-3p had a positive correlation with SLCO4A1-AS1 or IKKα in expression. Rescue experiments demonstrated that ectopic IKKαexpression rescued the inhibitory role of SLCO4A1-AS1 deficiency in the malignant behaviors of NSCLC cells.
To sum up, our work demonstrated that SLCO4A1-AS1 drove progression of NSCLC through activating NF-κB signaling pathway in pair via sequestering miR-223-3p to enhance IKKα expression. This finding might contribute to NSCLC treatment in the future.
Conflicts of interest
The authors declare that there are no competing interests in this study.
Supplemental Material
Download Zip (92.5 KB)Acknowledgments
We appreciate all the lab members.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, Xia C, Sun K, Yang Z, Li H, et al. 2018. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Global Health. 6:e555–e67. doi:10.1016/S2214-109X(18)30127-X.
- Hoffman PC, Mauer AM, Vokes EE. 2000. Lung cancer. The Lancet. 355:479–485. doi:10.1016/S0140-6736(00)82038-3.
- Herbst RS, Morgensztern D, Boshoff C. 2018. The biology and management of non-small cell lung cancer. Nature. 553:446–454. doi:10.1038/nature25183.
- Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. 2002. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 21:7435–7451. doi:10.1038/sj.onc.1205803.
- Akhtar-Danesh N, Finley C. 2015. Temporal trends in the incidence and relative survival of non-small cell lung cancer in Canada: A population-based study. Lung Cancer (Amsterdam, Netherlands). 90:8–14. doi:10.1016/j.lungcan.2015.07.004.
- Wang D, Dai J, Hou S, Qian Y. 2019. LncRNA SNHG20 predicts a poor prognosis and promotes cell progression in epithelial ovarian cancer. Biosci Rep. 39(4).
- Liang H, Yu T, Han Y, Jiang H, Wang C, You T, Zhao X, Shan H, Yang R, Yang L, et al. 2018. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer. 17:119. doi:10.1186/s12943-018-0870-5.
- Wang W, Luo P, Guo W, Shi Y, Xu D, Zheng H, Jia L. 2018. LncRNA SNHG20 knockdown suppresses the osteosarcoma tumorigenesis through the mitochondrial apoptosis pathway by miR-139/RUNX2 axis. Biochem Biophys Res Commun. 503:1927–1933. doi:10.1016/j.bbrc.2018.07.137.
- Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, et al. 2014. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 63:881–890. doi:10.1136/gutjnl-2013-305266.
- Wang Z, Jin J. 2019. LncRNA SLCO4A1-AS1 promotes colorectal cancer cell proliferation by enhancing autophagy via miR-508-3p/PARD3 axis. Aging. 11(14):4876–4889.
- Yang Y, Wang F, Huang H, Zhang Y, Xie H, Men T. 2019. lncRNA SLCO4A1-AS1 promotes growth and invasion of bladder cancer through sponging miR-335-5p to upregulate OCT4. Onco Targets Ther. 12:1351–1358. doi:10.2147/OTT.S191740.
- Yu J, Han Z, Sun Z, Wang Y, Zheng M, Song C. 2018. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through beta-catenin-dependent Wnt pathway. J Exp Clin Cancer Res: CR. 37:222. doi:10.1186/s13046-018-0896-y.
- Rao S, Morales AA, Pearse DD. 2015. The Comparative Utility of Viromer RED and Lipofectamine for Transient Gene Introduction into Glial Cells. Biomed Res Int. 2015:458624. doi:10.1155/2015/458624.
- Wang G, Zhang ZJ, Jian WG, Liu PH, Xue W, Wang TD, Meng YY, Yuan C, Li HM, Yu YP, et al. 2019. Novel long noncoding RNA OTUD6B-AS1 indicates poor prognosis and inhibits clear cell renal cell carcinoma proliferation via the Wnt/beta-catenin signaling pathway. Mol Cancer. 18:15. doi:10.1186/s12943-019-0942-1.
- Zhu LF, Song LD, Xu Q, Zhan JF. 2019. Highly expressed long non-coding RNA FEZF1-AS1 promotes cells proliferation and metastasis through Notch signaling in prostate cancer. Eur Rev Med Pharmacol Sci. 23:5122–5132. doi:10.26355/eurrev_201906_18176.
- Wang RK, Shao XM, Yang JP, Yan HL, Shao Y. 2019. MicroRNA-145 inhibits proliferation and promotes apoptosis of HepG2 cells by targeting ROCK1 through the ROCK1/NF-kappaB signaling pathway. Eur Rev Med Pharmacol Sci. 23:2777–2785. doi:10.26355/eurrev_201904_17551.
- Lan X, Liu X. 2019. LncRNA SNHG1 functions as a ceRNA to antagonize the effect of miR-145a-5p on the down-regulation of NUAK1 in nasopharyngeal carcinoma cell. J Cell Mol Med. 23:2351–2361. doi:10.1111/jcmm.13497.
- Chen Y, Shen Z, Zhi Y, Zhou H, Zhang K, Wang T, Feng B, Chen Y, Song H, Wang R, et al. 2018. Long non-coding RNA ROR promotes radioresistance in hepatocelluar carcinoma cells by acting as a ceRNA for microRNA-145 to regulate RAD18 expression. Arch Biochem Biophys. 645:117–125. doi:10.1016/j.abb.2018.03.018.
- Chen Z, Chen X, Lei T, Gu Y, Gu J, Huang J, Lu B, Yuan L, Sun M, Wang Z. 2020. Integrative Analysis of NSCLC Identifies LINC01234 as an Oncogenic lncRNA that Interacts with HNRNPA2B1 and Regulates miR-106b Biogenesis. Mol Ther. 28(6):1479–1493.
- Xu X, Zhou X, Chen Z, Gao C, Zhao L, Cui Y. 2020. Silencing of lncRNA XIST inhibits non-small cell lung cancer growth and promotes chemosensitivity to cisplatin. Aging (Albany NY). 12:4711–4726. doi:10.18632/aging.102673.
- Wu C, Zhu X-T, Xia L, Wang L, Yu W, Guo Q, Zhao M, Lou J. 2020. High expression of long noncoding RNA PCNA-AS1 promotes non-small-cell lung cancer cell proliferation and oncogenic activity via upregulating CCND1. J Cancer. 11:1959–1967. doi:10.7150/jca.39087.
- Liu X, Chen W, Liu Q, Dai J. 2019. Abietic acid suppresses non-small-cell lung cancer cell growth via blocking IKKbeta/NF-kappaB signaling. Onco Targets Ther. 12:4825–4837. doi:10.2147/OTT.S199161.
- Ji DG, Guan LY, Luo X, Ma F, Yang B, Liu HY. 2018. Inhibition of MALAT1 sensitizes liver cancer cells to 5-flurouracil by regulating apoptosis through IKKalpha/NF-kappaB pathway. Biochem Biophys Res Commun. 501:33–40. doi:10.1016/j.bbrc.2018.04.116.
- Ambros V. 2003. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 113:673–676. doi:10.1016/S0092-8674(03)00428-8.
- Sun W, Lv J, Duan L, Lin R, Li Y, Li S, Fu C, Zhao L, Xin S. 2019. Long noncoding RNA H19 promotes vascular remodeling by sponging let-7a to upregulate the expression of cyclin D1. Biochem Biophys Res Commun. 508:1038–1042. doi:10.1016/j.bbrc.2018.11.185.
- Huang Y, Ni R, Wang J, Liu Y. 2019. Knockdown of lncRNA DLX6-AS1 inhibits cell proliferation, migration and invasion while promotes apoptosis by downregulating PRR11 expression and upregulating miR-144 in non-small cell lung cancer. Biomed Pharmacother. 109:1851–1859. doi:10.1016/j.biopha.2018.09.151.
- Zhang L, Wang L, Wang Y, Chen T, Liu R, Yang W, Liu Q, Tu K. 2019. LncRNA KTN1-AS1 promotes tumor growth of hepatocellular carcinoma by targeting miR-23c/ERBB2IP axis. Biomed Pharmacother. 109:1140–1147. doi:10.1016/j.biopha.2018.10.105.
