ABSTRACT
Increasing evidence has revealed that long noncoding RNAs (lncRNAs) emerge as pivotal regulators in diverse cancers, including hepatocellular carcinoma (HCC). This study was conducted to investigate the role of lncRNA WWOX antisense RNA 1 (WWOX-AS1) in HCC progression. Our present study illustrated that WWOX-AS1 was lowly expressed in HCC tissues and cell lines. High WWOX-AS1 expression was further confirmed to predict a favorable prognosis in HCC patients. Through functional assays, we observed that upregulated WWOX-AS1 was correlated with decreased cell proliferation, migration, epithelial to mesenchymal transition (EMT) process and increased cell apoptosis, suggesting that WWOX-AS1 exerted anti-carcinogenic role in the development of HCC. Moreover, WWOX, the nearby gene of WWOX-AS1, was found at a low level in HCC tissues and cell lines. Furthermore, there was a positive relationship between WWOX-AS1 and WWOX. Additionally, WWOX overexpression hampered cell proliferation, migration, EMT process and induced cell apoptosis in HCC. Mechanically, WWOX-AS1 was identified as a cytoplasmic RNA in HCC cells and sponged miR-20b-5p to regulate WWOX expression. Rescue assays further indicated that WWOX knockdown counteracted WWOX-AS1 overexpression-mediated suppressive function on HCC progression. Collectively, WWOX-AS1/miR-20b-5p/WWOX axis suppresses HCC tumorigenesis, hinting a potential molecular mechanism for the therapy of HCC patients.
KEYWORDS:
Introduction
The malignancy of hepatocellular carcinoma (HCC) ranks fifth among the tumors worldwide and the mortality ranks third in terms of cancer-related death.Citation1 It has been reported that there are nearly 600,000 deaths occurred every year.Citation2Although great progress has been made in curative treatments, such as liver resection and liver transplant, the prognosis remains quite unsatisfactory.Citation3,Citation4 The unfavorable survival rate is largely caused by distant metastasis and easy recurrence after surgery.Citation5 A growing number of tumor-relevant genes have been identified; however, the underlying mechanisms in HCC have not been fully elucidated.Citation6
In recent years, long noncoding RNA (lncRNA), mainly composed of intergenomic transcription, has become a hotspot of tumor transcriptome research.Citation7 Existing studies have manifested that they participate in biological processes of various diseases through different mechanisms.Citation8,Citation9 The effect of lncRNAs on tumorigenesis has been researched by multiple researchers over the past years, revealing the oncogenic or tumor-suppressive role of lncRNAs in cancers.Citation10,Citation11 Up to now, a slew of lncRNAs have been confirmed to be a regulator in HCC progression, such as RGMB-AS1,Citation12 DLX6-AS1,Citation13 ANRILCitation14 and HULC.Citation15 For finding promising biomarkers in the development of HCC, it is still necessary to investigate the novel lncRNAs and new mechanisms. In previous report, lncRNA WWOX-AS1 acts as a tumor suppressor in osteosarcoma,Citation16 whereas its role and underlying mechanism in HCC still need to be elaborated.
In order to determine the mechanism pattern of WWOX-AS1 in HCC, the subcellular localization of WWOX-AS1 was detected in HCC cells. Result showed that WWOX-AS1 was a cytoplasmic RNA which worked as post-transcriptional regulator of gene expression, indicating that WWOX-AS1 could be a competing endogenous RNA (ceRNA) through competitively combining with miRNA to upregulate mRNA expression.Citation17,Citation18 Based on this hypothesis, functional and mechanical experiments were carried out and it was discovered that WWOX-AS1 contributed to HCC progression by downregulating its nearby gene WWOX via sequestering miR-20b-5p. This discovery provides a valuable enlightenment for improving the medical level of HCC.
Materials and methods
Tissue specimens
Forty cases of HCC tissues and paired adjacent non-tumor samples were gained from Hainan General Hospital, Hainan Medical University. Procedures of this study were licensed by the Institutional Review Board of Hainan General Hospital, Hainan Medical University. Written informed consent was attained from each participant. None of the patients underwent chemo- or radiation therapy prior to resection. Tissues were snap-frozen in liquid nitrogen and stored at −80°C till further use.
Cell culture
Normal liver cell line (THLE-3) and human HCC cells (HepG2, LM3, Hep3B, Huh7 and MHCC97H) were acquired from American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were incubated at 37°C with Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Rockville, MD, USA) supplying 1% penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA, USA) as well as 10% fetal bovine serum (FBS; Thermo Fisher Scientific), with a constant 5% CO2 air flow under the humidified condition.
Cell transfection
The pcDNA3.1 vector targeting WWOX-AS1 (pcDNA3.1/WWOX-AS1) or the pcDNA3.1 vector targeting WWOX (pcDNA3.1/WWOX) and the empty vector (pcDNA3.1), together with specific shRNAs against WWOX (sh-WWOX#1 and sh-WWOX#2) and their corresponding NC (sh-NC), were gained from Genechem (Shanghai, China). Simultaneously, miR-20b-5p mimics and NC mimics were gained from GenePharma (Shanghai, China). For the stable transfection, all these plasmids were cloned into the lentivirus expression vectors pCDH-CMV-MCS-EF1-Puro following the introduction of supplier (System Bioscience, Palo Alto, CA). Besides, different doses of pcDNA3.1/WWOX-AS1 (#1/2/3) resulted in varying increase of WWOX-AS1 in HCC cells, with pcDNA3.1/WWOX-AS1#3 also written as pcDNA3.1/WWOX-AS1 here. Finally, cells with stable transfection were screened out via puromycin treatment and used for further analyses.
qRT-PCR
Total RNA was isolated by adopting Trizol reagent (Invitrogen), after which was reversed transcribed into cDNAs employing a PrimeScript™ one step RT-PCR kit (TaKaRa, Tokyo, Japan). Gene level was assayed using a SYBR® Premix DimmerEraser™ kit (TaKaRa) plus the ABI7500 system (Applied Biosystems, Foster City, CA, USA). The relative expression changes were calculated as per 2−ΔΔCt method. GAPDH or U6 served as the internal control for normalization.
CCK-8 assay
Transfected HepG2 or Huh7 cells were plated into each well of the 96-well plate. Absorbance at 450 nm was recorded following addition of CCK-8 solution (Dojindo, Kumamoto, Japan).
Colony formation assay
Transfected HepG2 or Huh7 cells were trypsinized, and plated in 24‐well plates and incubated for 14 days. Colonies were immobilized for 10 min with methanol (Sigma-Aldrich, St. Louis, MO, USA) and dyed for 15 min utilizing 0.1% crystal violet (Sigma-Aldrich). Cell colonies were eventually counted and analyzed.
Western blot
After extraction and quantification, equal amounts of protein were resolved via 10% SDS-PAGE (Bio-Rad, Hercules, CA, USA), followed by transfer to PVDF membrane (Millipore, Billerica, MA, USA). The membrane was later blocked applying 5% skim milk, followed by incubated with primary antibodies against Bcl-2 (ab32124), Bax (ab32503), E-cadherin (ab40772), N-cadherin (ab76057), Vimentin (ab8978), WWOX (ab137726) and GAPDH (ab245356) which were gained from Abcam (Cambridge, UK). Upon incubation with secondary antibodies, ECL fluorescent detection reagent (Millipore) was applied.
TUNEL assay
The In Situ Cell Death Detection kit (Roche, Mannheim, Germany) was applied in this assay on the basis of its guide. For total cell count, the nucleus was counterstained utilizing DAPI fluorescent dye (Beyotime, Shanghai, China). The fluorescence microscope (Olympus, Tokyo, Japan) was applied so that images were obtained and the apoptotic cells were assessed.
Cell migration assay
Transwell chambers with a 24-well 8 μm pore filter (Corning, Cambridge, MA, USA) were adopted for transwell assay. Transfected HepG2 or Huh7 cells were suspended in serum-free medium, and seeded in the upper chamber. 600 mL serum-free medium with 10% FBS was plated in the lower chamber. Following 24 h of incubation, migratory cells were immobilized with methanol and dyed in crystal violet staining solution. An inverted microscope (Olympus) was at length employed for imaging migratory cells.
Subcellular fractionation
Cytoplasmic and nuclear RNA were isolated employing a PARIS™ Kit (Ambion, Austin, TX, USA). HepG2 or Huh7 cells were re-suspended in cell fraction buffer, followed by cultivation for 10 min on ice. The upper solution was removed following centrifugation. The nuclear pellet was gained, while RNA was kept to isolate via cell disruption buffer. All isolated RNA was studied with qRT-PCR.
RNA immunoprecipitation (RIP) assay
A Magna RNA-binding protein immunoprecipitation kit (Millipore) was adopted in RIP. HepG2 or Huh7 cells were harvested and subjected to complete RNA lysis buffer. Acquired cell lysates were incubated together with anti-Ago2 (Millipore) and anti-IgG (Millipore), and later incubated with Proteinase K buffer in order to digest proteins. qRT-PCR was undertaken to reveal expression levels of WWOX-AS1, miR-20b-5p and WWOX.
Luciferase reporter assay
The wild-type or mutant sequences of miR-20b-5p in WWOX-AS1 or WWOX were sub-cloned into pmirGLO dual-luciferase vector (Promega, Madison, WI, USA) to establish pmirGLO-WWOX-AS1-WT/Mut or pmirGLO-WWOX-WT/Mut which was co-transfected into HepG2 or Huh7 cells with miR-20b-5p mimics, NC mimics, or miR-20b-5p mimics+pcDNA3.1/WWOX-AS1. Dual-luciferase reporter assay system (Promega) was adopted for examination of luciferase activities.
Statistical analysis
Statistical analysis was accomplished with GraphPad Prism 5 software (GraphPad Software, San Diego, CA, USA). Data were listed as mean ± SD from experiments conducted in triplicate. Besides, Student’s t-test, one-way or two-way ANOVA was adopted for demonstrating significance of differences in groups. Determination of correlations among WWOX-AS1, miR-20b-5p and WWOX was done via Pearson’s correlation analysis. P < .05 indicated statistical significance. Overall survival was plotted by use of the Kaplan Meier method and compared applying log-rank test.
Results
Upregulated WWOX-AS1 hampers HCC progression
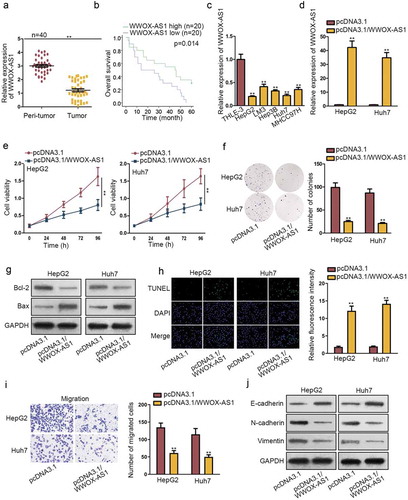
Firstly, WWOX-AS1 expression level was detected in tissue samples obtained from 40 HCC patients. As shown in , compared with adjacent normal tissues, the expression of WWOX-AS1 was remarkably downregulated in HCC tissues. To analyze the effect of WWOX-AS1 on the prognosis of HCC patients who did not undergo any chemo- or radiation therapy prior to resection, we generated Kaplan–Meier surviving curve and it demonstrated that high WWOX-AS1 level was closely correlated with high overall survival of patients with HCC (). Consistently, WWOX-AS1 expression was found to be lower in HCC cell lines (HepG2, LM3, Hep3B, Huh7, MHCC97H) than that in normal liver cell line THLE-3 (). These data suggested that WWOX-AS1 might participate in the regulation of HCC development. To further study the biological function of WWOX-AS1 in HCC, the functional assays were designed in two HCC cells. HepG2 and Huh7 cells displayed relative lower expression of WWOX-AS1; hence, we upregulated WWOX-AS1 expression in these two cells (). At first, CCK-8 and colony formation assays were carried out to detect cell proliferation with transfection of pcDNA3.1/WWOX-AS1. Results manifested that overexpressed WWOX-AS1 notably restrained the proliferative ability of HepG2 and Huh7 cells (–f). Subsequently, we wondered whether WWOX-AS1 mediated cell proliferation via inducing cell apoptosis. Through western blot assay, we observed a reduced Bcl-2 expression and an increased Bax expression in pcDNA3.1/WWOX-AS1-transfected HepG2 and Huh7 cells (), indicating that WWOX-AS1 upregulation could promote cell apoptosis in HCC cells. Later, results from TUNEL assay also showed an induced cell apoptosis caused by WWOX-AS1 overexpression (). To explore whether the exogenous WWOX-AS1 could also induce cell apoptosis in normal cells at this high dose, we applied this assay again in THLE3 cells. After elevating WWOX-AS1 expression in THLE3 cells, cell apoptosis capability was enhanced (Fig. S1A-B). Moreover, transwell assay was conducted to test cell migration. Prior to that, we discovered that cell viability was not affected by upregulation of WWOX-AS1 after 24 h of incubation. However, upregulated WWOX-AS1 markedly diminished the number of migrated cells (), confirming that WWOX-AS1 overexpression could inhibit cell migration. Furthermore, enhanced expression of E-cadherin and declined expression of N-cadherin and Vimentin were discovered in HepG2 and Huh7 cells transfected with pcDNA3.1/WWOX-AS1 (), implying that EMT process was repressed by WWOX-AS1 upregulation. Based on the above findings, we wondered whether changes on WWOX-AS1 expression could determine the malignant phenotypes of HCC cells. As indicated in Fig. S1D, WWOX-AS1 expression was elevated in different degrees in Huh7 cells after transfection. Accordingly, the proliferation ability of Huh7 cells transfected with pcDNA3.1/WWOX-AS1#1/2/3 was attenuated gradually whereas the corresponding apoptosis capability was enhanced (Fig. S1E-F). Moreover, cell viability detected after 24 h of incubation showed no significant changes among different groups (Fig. S1G). Meanwhile, cell migration capability was gradually weakened by elevating WWOX-AS1 expression (Fig. S1H). Taken together, WWOX-AS1 is downregulated in HCC tissues and cells, and upregulation of WWOX-AS1 restrains HCC tumorigenesis.
Figure 1. Upregulated WWOX-AS1 hampers HCC progression. (a) WWOX-AS1 expression in HCC tissues and adjacent non-tumor tissues was detected via qRT-PCR. (b) Overall survival rate of HCC patients with high or low level of WWOX-AS1 was analyzed via Kaplan Meier method. (c) WWOX-AS1 expression in HCC cell lines (HepG2, LM3, Hep3B, Huh7, MHCC97H) and one control cell line (THLE-3) was examined via qRT-PCR. (d) qRT-PCR was used to detect WWOX-AS1 expression in HepG2 and Huh7 cells transfected with pcDNA3.1/WWOX-AS1 or pcDNA3.1 after 48 h of transfection. (e-f) Proliferative ability of HepG2 and Huh7 cells transfected with pcDNA3.1/WWOX-AS1 or pcDNA3.1 was evaluated via CCK-8 and colony formation assays. Cell viability was detected at indicated time points (0, 24, 48, 72 or 96 h). The number of colonies was detected after 14 days of incubation. (g) The expression levels of apoptosis-related proteins (Bcl-2 and Bax) were detected in transfected cells via western blot after 48 h of transfection. (h) The apoptosis of transfected cells was tested by TUNEL assay after 24 h of cultivation. (i) Transwell assay was applied to assess cell migration in transfected cells after 24 h of incubation. (j) The effect of WWOX-AS1 overexpression on EMT process was detected through western blot analysis of EMT-related proteins after 48 h of transfection. **P < .01
WWOX is positively associated with WWOX-AS1 and exhibits anti-carcinogenic property in HCC
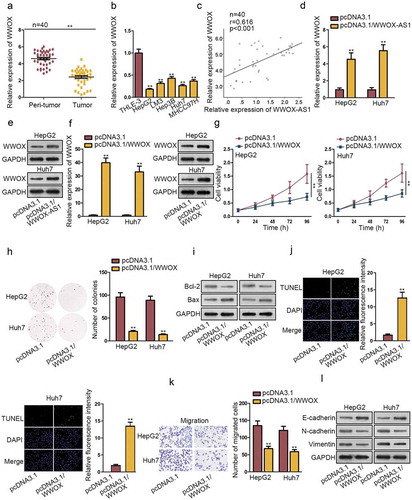
Through bioinformatics analysis tool, the nearby gene of WWOX-AS1 (WWOX) was found. WWOX has been reported to play anti-carcinogenic role in some cancers. In our study, we found that WWOX was obviously downregulated in HCC tissues (). Consistently, the relative low expression of WWOX was evaluated in HCC cells compared to control cells (). Furthermore, Pearson’s correlation analysis showed a positive correlation between WWOX-AS1 and WWOX (). In addition, WWOX-AS1 was found to positively regulate WWOX mRNA and protein level by qRT-PCR and western blot analyses (-e). To research the potential regulatory role of WWOX in HCC tumorigenesis, we upregulated its expression in HepG2 and Huh7 cells by transfecting with pcDNA3.1/WWOX (). Subsequently, CCK-8 and colony formation assays were executed to estimate the importance of WWOX in the proliferation of HCC cells. Interestingly, overexpressed WWOX inhibited cell proliferation (-h). Consistent with WWOX-AS1, WWOX upregulation also boosted the apoptosis of HepG2 and Huh7 cells (-j). In addition, upregulation of WWOX did not affect cell viability detected after 24 h of cultivation, whereas decreased the number of migrated cells (Fig. S2A, ), suggesting that overexpressed WWOX could suppress cell migration. Importantly, EMT process was also repressed by the overexpression of WWOX through western blot analysis of EMT-related proteins (). In brief, WWOX is positively associated with WWOX-AS1 and exhibits anti-carcinogenic property in HCC.
Figure 2. WWOX is positively associated with WWOX-AS1 and exhibits anti-carcinogenic property in HCC. (a) WWOX expression in HCC tissues and adjacent non-tumor tissues was examined through qRT-PCR. (b) WWOX expression in HCC cell lines and one control cell line was detected via qRT-PCR after 48 h of transfection. (c) Pearson’s correlation analysis showed a positive association between WWOX-AS1 expression and WWOX expression in HCC tissues. (d-e) qRT-PCR and western blot were conducted to estimate the effect of WWOX-AS1 overexpression on WWOX mRNA and protein levels after 48 h of transfection. (f) WWOX expression in transfected cells was detected by qRT-PCR and western blot after 48 h of transfection. (g-h) CCK-8 and colony formation assays were utilized to test proliferative ability of pcDNA3.1/WWOX-transfected cells. Cell viability was detected at indicated time points (0, 24, 48, 72 or 96 h). The number of colonies was detected after 14 days of incubation. (i-j) Western blot (after 48 h of transfection) and TUNEL (after 24 h of incubation) were applied to estimated cell apoptosis in transfected cells. (k) Cell migration was measured in WWOX-overexpressed HepG2 and Huh7 cells by transwell assay after 24 h of incubation. (l) The effect of upregulated WWOX on EMT process was tested via western blot analysis of the protein levels of E-cadherin, N-cadherin and Vimentin after 48 h of transfection. **P < .01
WWOX-AS1 acts as a ceRNA to sponge miR-20b-5p and upregulate WWOX expression in HCC
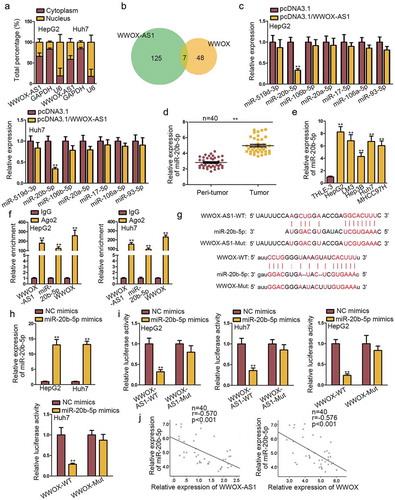
To research the mechanism pattern of WWOX-AS1 in HCC, we firstly performed subcellular fractionation assay to detect the subcellular distribution of WWOX-AS1. The result revealed that WWOX-AS1 was mainly distributed in cytoplasm of HepG2 and Huh7 cells (), indicating that WWOX-AS1 might exert function through ceRNA pattern which is a common mode of post-transcriptional regulation of lncRNAs. Based on the above data, we assumed that WWOX-AS1 might regulate WWOX expression through sequestering a certain miRNA. Through bioinformatics analysis, seven miRNAs were predicted to have complementary sites with both WWOX-AS1 and WWOX (). Next, qRT-PCR analysis delineated that only the expression of miR-20b-5p was significantly inhibited by WWOX-AS1 overexpression (). Hence, miR-20b-5p was selected for the following experiments. Later, miR-20b-5p expression in HCC tissues and cell lines was detected by qRT-PCR. As expected, its expression was markedly upregulated in HCC tissues and cell lines (-e). Besides, RIP assay implied that WWOX-AS1, miR-20b-5p and WWOX were all enriched in the beads containing Ago2 antibody (), unveiling the potential involvement of WWOX-AS1, miR-20b-5p and WWOX in ceRNA network. Subsequently, the binding site between miR-20b-5p and WWOX-AS1 (or WWOX) was, respectively, predicted by DIANA and starBase (). To increase the expression of miR-20b-5p, miR-20b-5p mimics was transfected into HepG2 and Huh7 cells (). Subsequently, results from luciferase reporter assays displayed that miR-20b-5p mimics efficiently decreased the luciferase activity of wild-type WWOX-AS1 or WWOX reporter, but no obvious change was shown in the reporter containing mutant type WWOX-AS1 or WWOX (). Besides, WWOX-AS1 upregulation could reverse the suppressive effect of miR-20b-5p upregulation on the luciferase activity of WWOX-WT reporter (Fig. S2B). Lastly, Pearson’s correlation analysis revealed the negative correlation between miR-20b-5p expression and WWOX-AS1 or WWOX expression (). Conclusively, WWOX-AS1 functions as a ceRNA to sponge miR-20b-5p and upregulate WWOX expression.
Figure 3. WWOX-AS1 acts as a ceRNA to sponge miR-20b-5p and upregulate WWOX expression in HCC. (a) According to subcellular fractionation assay, WWOX-AS1 was mainly distributed in cytoplasm of HepG2 and Huh7 cells. (b) DIANA and starBase predicted seven potential miRNAs that could bind with both WWOX-AS1 and WWOX. (c) The expression levels of potential miRNAs were detected in transfected cells via qRT-PCR after 48 h of transfection. (d) MiR-20b-5p expression in HCC tissues and adjacent non-tumor tissues was detected by qRT-PCR. (e) MiR-20b-5p expression in HCC cell lines and one control cell line was examined through western blot. (f) RIP revealed the significant enrichment of WWOX-AS1, miR-20b-5p and WWOX in anti-Ago2 group after incubation for overnight. (g) The predicted binding sequences between miR-20b-5p and WWOX-AS1 (or WWOX). (h) qRT-PCR was used to detect the overexpression efficiency of miR-20b-5p after 48 h of transfection. (i) Luciferase reporter assay was performed to examine the interaction between miR-20b-5p and WWOX-AS1 (or WWOX) in HepG2 and Huh7 cells after 48 h of transfection. (j) Expression relevance between miR-20b-5p and WWOX-AS1 (or WWOX) in HCC tissues was analyzed via Pearson’s correlation analysis. **P < .01
WWOX-AS1 mediates HCC progression by regulating WWOX expression
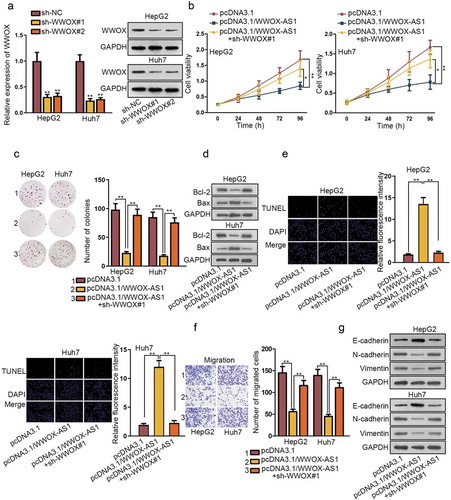
To further demonstrate the regulatory network of WWOX-AS1/miR-20b-5p/WWOX, rescue assays were conducted in HepG2 and Huh7 cells. Firstly, shRNAs targeting WWOX (sh-WWOX#1/2) were transfected into HepG2 and Huh7 cells for reducing the expression of WWOX (). Next, through qRT-PCR and western blot, we observed that sh-WWOX#1 could reverse the promoting effect of pcDNA3.1/WWOX-AS1 on WWOX expression (Fig. S2C). Afterward, CCK-8 and colony formation experiments demonstrated that silenced WWOX reversed the inhibitory effect of overexpressed WWOX-AS1 on cell proliferation (-c). Likewise, knockdown of WWOX reversed the promotive role of WWOX-AS1 upregulation in cell apoptosis (-e). Moreover, cell viability detected after 24 h of cultivation showed no clear changes among different transfected cells (Fig. S2D); meanwhile, the inhibited migration caused by overexpression of WWOX-AS1 was counteracted by WWOX depletion (). In addition, elevated expression of WWOX-AS1 also restrained EMT process, but then depletion of WWOX reversed the effect of WWOX-AS1 upregulation on EMT process (). In conclusion, WWOX-AS1 facilitates HCC progression by downregulating WWOX expression.
Figure 4. WWOX-AS1 mediates HCC progression by regulating WWOX expression. (a) The knockdown efficiency of WWOX in HCC cells was detected via qRT-PCR and western blot after 48 h of transfection. (b-c) The proliferative capacity of HepG2 and Huh7 cells was evaluated by CCK-8 and colony formation after transfected with indicated plasmids. Cell viability was detected at indicated time points (0, 24, 48, 72 or 96 h). The number of colonies was detected after 14 days of incubation. (d-e) The effect of WWOX knockdown on WWOX-AS1 overexpression-induced apoptosis was examined via western blot (after 48 h of transfection) and TUNEL (after 24 h of incubation). (f) The effect of silenced WWOX on the inhibitive migration capacity resulted from upregulation of WWOX-AS1 was measured via transwell after 24 h of incubation. (g) The expression levels of EMT-associated proteins were examined in transfected cells through western blot after 48 h of transfection. *P < .05, **P < .01
Discussion
In recent years, an increasing number of studies have uncovered the important role that lncRNAs play in the tumorigenesis and progression of cancers.Citation19,Citation20 Abnormal regulation of lncRNAs is tightly involved in the diagnosis and therapy of cancer patients. For example, upregulated MNX1-AS1 and LINC00346 are reported as a tumor promoter in gastric cancer and cause an unsatisfactory outcome;Citation21,Citation22 lncRNA EGFR-AS1 enhances cell growth in renal cancer by affecting HuR expression.Citation23 LncRNA WWOX-AS1 has been revealed to inhibit the progression of osteosarcoma and serve as an indicator of prognosis.Citation16 Our research identified downregulated WWOX-AS1 in HCC tissues and cell lines. Interestingly, WWOX-AS1 was closely relevant with the proliferation, apoptosis, migration and EMT process of HCC cells and indicated a favorable prognosis. Namely, WWOX-AS1 acted as a tumor suppressor in HCC.
WWOX is the nearby gene of WWOX-AS1 and has been confirmed to exert anti-carcinogenic role in some cancers. For instance, WWOX modulates miR-146a to restrain the metastasis in triple-negative breast cancer.Citation24 WWOX represses the proliferation and neoplasia of hepatocyte by controlling hepatic HIF1α.Citation25 WWOX modulates the response of ER stress to activate the paclitaxel sensibility of ovarian cancer cells.Citation26 In our study, WWOX was found at a low level in HCC tissues and cell lines. Additionally, WWOX-AS1 expression was positively correlated with WWOX expression. Moreover, WWOX overexpression suppressed cell proliferation, migration and EMT process and induced cell apoptosis in HCC. All these data suggested that WWOX was an anti-oncogene in HCC.
MicroRNA (miRNA) is a subtype of ncRNAs with about 25 nucleotides in length.Citation27 LncRNAs have been reported in a ceRNA network by completely sponging miRNAs to upregulate mRNAs expression.Citation28 For example, lncRNA KTN1-AS1 is reported as a tumor facilitator to promote HCC tumor growth via regulating miR-23 c/ERBB2IP axis.Citation29 LINC00518 promotes multidrug resistance of breast cancer via sponging miR-199a and regulating MRP1.Citation30 LncRNA NORAD serves as a sponge of miR-202-5p to promote the progression of HCC by enhancing TGF-β pathway.Citation31 MiR-20b-5p has been studied and revealed to be implicated in the progression of cancers, such as colorectal cancerCitation32 and non-small cell lung cancer.Citation33 In our study, overexpressed miR-20b-5p was discovered in HCC tissues and cells. Moreover, miR-20b-5p was verified to combine with WWOX-AS1 or WWOX in HCC cells. The expression of miR-20b-5p was further validated to be negatively related to that of WWOX-AS1 or WWOX. Rescue assays depicted that WWOX knockdown could reverse the effect of WWOX-AS1 overexpression on HCC progression.
In conclusion, the present study demonstrated the tumor suppressive effect of WWOX-AS1 on HCC progression. WWOX-AS1 could function as a ceRNA to sequester miR-20b-5p and upregulate WWOX expression. These findings indicated a new WWOX-AS1/miR-20b-5p/WWOX network in HCC, providing a new revelation for the treatment of patients diagnosed with HCC.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download Zip (650 KB)Acknowledgments
We appreciate all the participants who provide supports for the study.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi:10.1053/j.gastro.2007.04.061.
- Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi:10.1038/nrgastro.2010.100.
- Befeler AS, Di Bisceglie AM. Hepatocellular carcinoma: diagnosis and treatment. Gastroenterology. 2002;122:1609–1619. doi:10.1053/gast.2002.33411.
- Forner A, Hessheimer AJ, Isabel Real M, Bruix J. Treatment of hepatocellular carcinoma. Crit Rev Oncol Hematol. 2006;60:89–98. doi:10.1016/j.critrevonc.2006.06.001.
- Xia L, Huang W, Tian D, Zhu H, Qi X, Chen Z, Zhang Y, Hu H, Fan D, Nie Y, et al. Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology. 2013;57:610–624. doi:10.1002/hep.26029.
- Hammerle M, Gutschner T, Uckelmann H, Ozgur S, Fiskin E, Gross M, Skawran B, Geffers R, Longerich T, Breuhahn K, et al. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatology. 2013;58:1703–1712. doi:10.1002/hep.26537.
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi:10.1158/2159-8290.CD-11-0209.
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi:10.1126/science.1138341.
- Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–61. doi:10.1093/hmg/ddq353.
- Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, Wu F, Mo YY. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi:10.1093/nar/gkt182.
- Sun M, Nie FQ, Zang C, Wang Y, Hou J, Wei C, Li W, He X, Lu KH. The pseudogene DUXAP8 promotes non-small-cell lung cancer cell proliferation and invasion by epigenetically silencing EGR1 and RHOB. Mol Ther. 2017;25:739–751. doi:10.1016/j.ymthe.2016.12.018.
- Sheng N, Li Y, Qian R, Li Y. The clinical significance and biological function of lncRNA RGMB-AS1 in hepatocellular carcinoma. Biomed Pharmacother. 2018;98:577–584. doi:10.1016/j.biopha.2017.12.067.
- Zhang L, He X, Jin T, Gang L, Jin Z. Long non-coding RNA DLX6-AS1 aggravates hepatocellular carcinoma carcinogenesis by modulating miR-203a/MMP-2 pathway. Biomed Pharmacother. 2017;96:884–891. doi:10.1016/j.biopha.2017.10.056.
- Ma J, Li T, Han X, Yuan H. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144:205–214. doi:10.1007/s00432-017-2543-y.
- Hu D, Shen D, Zhang M, Jiang N, Sun F, Yuan S, Wan K. MiR-488 suppresses cell proliferation and invasion by targeting ADAM9 and lncRNA HULC in hepatocellular carcinoma. Am J Cancer Res. 2017;7:2070–2080.
- Qu G, Ma Z, Tong W, Yang J. LncRNA WWOXAS1 inhibits the proliferation, migration and invasion of osteosarcoma cells. Mol Med Rep. 2018;18:779–788. doi:10.3892/mmr.2018.9058.
- Yan X, Zhang D, Wu W, Wu S, Qian J, Hao Y, Yan F, Zhu P, Wu J, Huang G, et al. Mesenchymal stem cells promote hepatocarcinogenesis via lncRNA-MUF Interaction with ANXA2 and miR-34a. Cancer Res. 2017;77:6704–6716. doi:10.1158/0008-5472.CAN-17-1915.
- Chen DL, Lu YX, Zhang JX, Wei XL, Wang F, Zeng ZL, Pan ZZ, Yuan YF, Wang FH, Pelicano H, et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7:4836–4849. doi:10.7150/thno.20942.
- DeOcesano-Pereira C, Amaral MS, Parreira KS, Ayupe AC, Jacysyn JF, Amarante-Mendes GP, Reis EM, Verjovski-Almeida S. Long non-coding RNA INXS is a critical mediator of BCL-XS induced apoptosis. Nucleic Acids Res. 2014;42:8343–8355. doi:10.1093/nar/gku561.
- Kong Y, Zhang L, Huang Y, He T, Zhang L, Zhao X, Zhou X, Zhou D, Yan Y, Zhou J, et al. Pseudogene PDIA3P1 promotes cell proliferation, migration and invasion, and suppresses apoptosis in hepatocellular carcinoma by regulating the p53 pathway. Cancer Lett. 2017;407:76–83. doi:10.1016/j.canlet.2017.07.031.
- Zhang W, Huang L, Lu X, Wang K, Ning X, Liu Z. Upregulated expression of MNX1-AS1 long noncoding RNA predicts poor prognosis in gastric cancer. Bosn J Basic Med Sci. 2019;19:164–171. doi:10.17305/bjbms.2019.3713.
- Xu TP, Ma P, Wang WY, Shuai Y, Wang YF, Yu T, Xia R, Shu YQ. KLF5 and MYC modulated LINC00346 contributes to gastric cancer progression through acting as a competing endogenous RNA and indicates poor outcome. Cell Death Differ. 2019;26(11):2179–2193.
- Wang A, Bao Y, Wu Z, Zhao T, Wang D, Shi J, Liu B, Sun S, Yang F, Wang L, et al. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis. 2019;10:154. doi:10.1038/s41419-019-1331-9.
- Khawaled S, Suh SS, Abdeen SK, Monin J, Distefano R, Nigita G, Croce CM, Aqeilan RI. WWOX inhibits metastasis of triple-negative breast cancer cells via modulation of miRNAs. Cancer Res. 2019;79:1784–1798. doi:10.1158/0008-5472.CAN-18-0614.
- Abu-Remaileh M, Khalaileh A, Pikarsky E, Aqeilan RI. Author correction: WWOX controls hepatic HIF1alpha to suppress hepatocyte proliferation and neoplasia. Cell Death Dis. 2018;9:1159. doi:10.1038/s41419-018-1158-9.
- Janczar S, Nautiyal J, Xiao Y, Curry E, Sun M, Zanini E, Paige AJ, Gabra H. WWOX sensitises ovarian cancer cells to paclitaxel via modulation of the ER stress response. Cell Death Dis. 2017;8:e2955. doi:10.1038/cddis.2017.346.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi:10.1016/S0092-8674(04)00045-5.
- Qian K, Liu G, Tang Z, Hu Y, Fang Y, Chen Z, Xu X. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Arch Biochem Biophys. 2017;615:1–9. doi:10.1016/j.abb.2016.12.011.
- Zhang L, Wang L, Wang Y, Chen T, Liu R, Yang W, Liu Q, Tu K. LncRNA KTN1-AS1 promotes tumor growth of hepatocellular carcinoma by targeting miR-23c/ERBB2IP axis. Biomed Pharmacother. 2019;109:1140–1147. doi:10.1016/j.biopha.2018.10.105.
- Chang L, Hu Z, Zhou Z, Zhang H. Linc00518 contributes to multidrug resistance through regulating the MiR-199a/MRP1 axis in breast cancer. Cell Physiol Biochem. 2018;48:16–28. doi:10.1159/000491659.
- Yang X, Cai JB, Peng R, Wei CY, Lu JC, Gao C, Shen ZZ, Zhang PF, Huang XY, Ke AW, et al. The long noncoding RNA NORAD enhances the TGF-beta pathway to promote hepatocellular carcinoma progression by targeting miR-202-5p. J Cell Physiol. 2019;234:12051–12060. doi:10.1002/jcp.27869.
- Tang D, Yang Z, Long F, Luo L, Yang B, Zhu R, Sang X, Cao G, Wang K. Long noncoding RNA MALAT1 mediates stem cell-like properties in human colorectal cancer cells by regulating miR-20b-5p/Oct4 axis. J Cell Physiol. 2019;234:20816–20828. doi:10.1002/jcp.28687.
- Peng L, Li S, Li Y, Wan M, Fang X, Zhao Y, Zuo W, Long XY. Regulation of BTG3 by microRNA-20b-5p in non-small cell lung cancer. Oncol Lett. 2019;18:137–144. doi:10.3892/ol.2019.10333.
