ABSTRACT
Background
There is an unmet need to identify novel mechanism-based prognostic genes associated with hepatocellular carcinoma (HCC) recurrence that can predict patient outcomes and provide therapeutic targets. This study aims to identify potential novel driver genes and mutations in HCC.
Methods
Single nucleotide variations (SNVs) contributing to HCC recurrence were identified using whole-exome sequencing of 5 DNA samples extracted from a single HCC patient with HBV-induced cirrhosis. SNVs were verified in primary HCC (n = 87), recurrent HCC (n = 34), and benign liver disease with cirrhosis tissues (n = 43). A candidate gene was identified, and its association and function in HCC development and recurrence were examined.
Results
177 SNVs were identified and 70 SNVs were verified. A MPPE1 missense mutation on chr18_11897016 was the most frequent mutation (16.5%) in primary and recurrent HCC tissues, occurring with a higher frequency in recurrent HCC than primary HCC or benign liver tumor tissues. The MPPE1 mutation was significantly associated with HCC recurrence (P = .003), TNM stage (P = .002), and Child–Pugh classification (P = .039), and was an independent risk factor for HCC recurrence (HR = 1.969; 95%CI = 1.043–3.714, P = .037). Analysis of publically available data deposited in the GEO and TCGA showed MPPE1 expression levels were significantly increased in HCC tumor samples compared to adjacent nontumor tissues. The knockdown of MPPE1 in HCC cell lines significantly inhibited cell proliferation, migration and invasion, induced cell cycle arrest and apoptosis in vitro, and inhibited xenograft tumor growth in nude mice in vivo (P < .05).
Conclusions
MPPE1 is a novel gene associated with HCC malignancy and recurrence.
Introduction
Hepatocellular carcinoma (HCC) accounts for approximately 70% to 85% of primary liver cancersCitation1,Citation2 and is the second leading cause of death from cancer in men worldwide.Citation3 Risk factors for HCC include chronic hepatitis B virus (HBV) or hepatitis C virus infection, consumption of food contaminated with aflatoxin B, and alcoholism.Citation4–6 Despite the progress in diagnosis and treatment, including hepatic resection, liver transplantation, local ablation, and molecular-targeted therapies, the prognosis of HCC patients remains poor due to difficulties in early diagnosis and the high incidence of tumor recurrence and metastasis.Citation4,Citation7,Citation8
HCC is characterized by its propensity for vascular invasion and metastasis, which results in a high incidence of recurrence and poor prognosis after early resection.Citation7–9 Induced gene mutations and/or epigenetic alterations, such as silencing of tumor suppressor genes and/or activation of oncogenes, promote the malignant transformation of hepatocytes, contributing to hepatocarcinogenesis and post-operative HCC recurrence.Citation10 The underlying mechanisms may involve alterations in physiological signaling through the Wnt/β-catenin, RAS-MAPK, JAK-STAT, PI3K-AKT-mTOR, and Notch pathways.Citation11,Citation12 Currently, there is an unmet need to identify novel mechanism-based prognostic genes associated with HCC recurrence that can accurately predict patient outcomes and provide potential therapeutic targets.
The metallophosphoesterase gene MPPE1 encodes a metallophosphoesterase protein that is a member of the calcineurin-like phosphoesterase superfamily.Citation13,Citation14 MPPE1 protein contains metal binding and active sites similar to serine/threonine phosphoprotein phosphatase catalytic subunits.Citation13,Citation14 MPPE1 SNPs have been associated with bipolar disorder,Citation13 and MPPE1 is involved in regulating the transport of glycosylphosphatidylinositol-anchor proteins from the endoplasmic reticulum to the Golgi.Citation14,Citation15 MPPE1 protein expression was detected in the cytoplasm/membranes of the majority of cells in liver cancer tissues (http://www.proteinatlas.org/search/MPPE1); however, the exact function of MPPE1 in cancer remains unclear.
In this study, with whole-exome sequencing and PCR-MassARRAY, we identified MPPE1 as a novel candidate gene associated with HCC recurrence and characterized the role of MPPE1 in HCC development. This study may provide a new clue to develop drug targets for HCC for therapy of HCC patients.
Results
Identification of SNVs and genes potentially associated with recurrent HCC
High-throughput whole-exome sequencing was used to identify SNPs in genomic DNA extracted from one primary HCC sample, one recurrent HCC sample, their adjacent liver tissue samples, and one pre-operative peripheral blood sample from a single HCC patient with HBV-induced cirrhosis (Supplemental materials, Tables S1-S2, Fig S1 and Fig S3). The mutation distribution in all chromosomes is shown in . The distribution of non-synonymous mutations was 10%-18% for each chromosome. One hundred and seventy-seven nonsynonymous SNVs, including 6 nonsense mutations and 171 missense mutations, were potentially associated with HCC recurrence (Fig. S2 and Table S3). Of these, 70 SNVs were verified with PCR-MassARRAY and Sanger sequencing in the additional dataset that included 87 primary HCC, 34 recurrent HCC, and 43 benign liver disease with cirrhosis tissue samples. Among the 70 verified SNVs, C:G->G:C transversions were the predominate mutations in primary HCC tissues () and C:G->T:A transversions were the predominate mutations in recurrent HCC tissues (), suggesting a different gene signature of the recurrent HCC from that of the primary HCC.
Figure 1. SNVs potentially associated with recurrent HCC were identified using high-throughput whole-exome sequencing, PCR-MassArray, and Sanger DNA sequencing. (a) Frequency of gene mutations in the genome of a single HCC patient. Data from five exomic SNP profiles were combined. Nucleotide transversions in (b) primary HCC (n = 87) and (c) recurrent HCC (n = 34) tissue samples. (d) Illustration showing the presence of 70 verified SNVs in 69 genes and their chromosomal locations. (e) Mutant genes occurring with the highest frequency in primary HCC, recurrent HCC, and benign liver tumor tissues
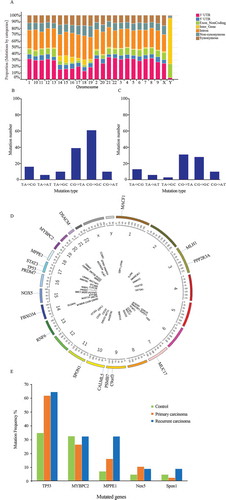
Annotation of the 70 SNVs revealed their presence in 69 genes and all chromosomes, except chromosome 5 (). Some of the 69 mutated genes were enriched in multiple cancer-associated pathways (Table S4). Specifically, TP53, MLH1, STAT3, and LAMA1 were enriched in a cancer pathway (P = .01), CHD8 and TP53 were enriched in the Wnt signaling pathway (P = .02), and CCNB3, ESPL1, and TP53 were enriched in the cell cycle pathway (P = .01). Notably, apart from these known cancer-associated genes that are often involved in KEGG pathways, previously undiscovered (or not previously associated with cancer) frequently mutated genes, such as MPPE1 were identified with the Gene Ontology (GO) and UniProtKB/Swiss-Prot databases in the GeneCard database (https://www.genecards.org/c).
Gene Ontology annotations related to MPPE1 include hydrolase activity and phosphoric diester hydrolase activity (Table S5). The UniProtKB/Swiss-Prot database indicates MPPE1 is required for transport of GPI-anchor proteins from the endoplasmic reticulum to the Golgi apparatus. It acts in the lipid remodeling steps of GPI-anchor maturation by mediating the removal of a side-chain ethanolamine-phosphate (EtNP) from the second Man (Man2) of the GPI intermediate, an essential step for efficient transport of GPI-anchor proteins.
Mutation in the MPPE1 gene is associated with recurrent HCC
Analysis of the frequencies of the candidate gene mutations showed that none of the genotype frequencies deviated significantly from those expected by the HWE for cases or controls. Analysis of the minor allele frequency (MAF) of the 69 mutated genes showed that only mutations in TP53, Mybpc2, and MPPE1 were more abundant in primary HCC or recurrent HCC than in benign liver tumors (MAF >0.05, Table S6).
In the 87 primary HCC, 34 recurrent HCC, and 43 benign liver disease with cirrhosis tissues, TP53 (41.0%) was the most frequently mutated gene, followed by Mybpc2 (26.2%), MPPE1 (16.5%), Nox5 (9.1%), and Spon1 (4.3%). Notably, the frequencies of the TP53, MPPE1, and Nox5 mutations were higher in primary or recurrent HCCs compared to benign liver disease with cirrhosis tissues, and the frequency of the MPPE1 mutation was higher in recurrent HCC than in primary HCC or benign liver disease with cirrhosis tissues (). These results suggest that the mutation in MPPE1 (chr18_11897016, C/T), a missense mutation (NM_001242904:c.A248G:p.E83G) occurring in the exonic (coding) region, is associated with HCC recurrence.
Analysis of the association of mutation frequency with the risk of primary HCC or HCC recurrence showed that the mutation in MPPE1 was significantly associated with recurrent HCC (P = .003) but not primary HCC (P = .16) when compared to benign liver disease with cirrhosis tissue (). Logistic regression analysis indicated that only the MPPE1 mutation was significantly associated with recurrent HCC (P = .009) (). These results suggest that the MPPE1 mutation is significantly associated with the risk of HCC recurrence.
Table 1. Associations of mutation frequency with the risk of primary HCC or HCC recurrence
Table 2. Logistic regression analysis of the associations of gene mutations with the risk of primary HCC or HCC recurrence
Mutation in the MPPE1 gene is associated with TNM stage, Child–Pugh classification, and post-operative HCC recurrence
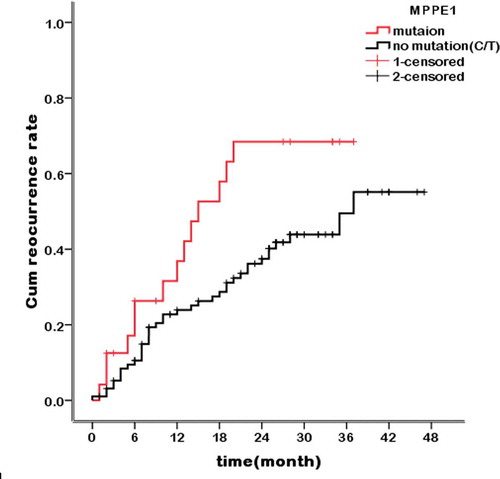
The association between the MPPE1 mutation and the clinicopathological features of disease and prognosis in 121 HCC patients was analyzed. The baseline characteristics of the HCC patients are summarized in . The MPPE1 mutation was associated with TNM stage (P = .002) and Child–Pugh classification (P = .04). To determine the association between the MPPE1 mutation and HCC recurrence, a Kaplan–Meier curve and the log-rank test were used to compare postoperative HCC recurrence in patients with and without the MPPE1 mutation. Among HCC patients without the MPPE1 mutation, 1-, 2-, and 3-y postoperative tumor recurrence rates were 28.0%, 45.0%, and 56.0%, respectively. Among HCC patients with the MPPE1 mutation 1-, 2-, and 3-y postoperative tumor recurrence rates were 53.0%, 69.0%, and 69.0%, respectively. HCC patients with the MPPE1 mutation showed a higher tumor recurrence rate than those without the MPPE1 mutation (P = .02; ).
Table 3. Associations between the MPPE1 mutation and the clinicopathological features of HCC disease and prognosis
Figure 2. Kaplan–Meier curves showing the association between tumor recurrence rate and the MPPE1 mutation. n = 121 HCC patients stratified by presence/absence of the MPPE1 mutation
In multivariate analysis using a Cox proportional hazards model for tumor recurrence, including age, number of tumor lesions, tumor size, venous invasion, lymph node invasion, tumor differentiation, serum alpha-fetoprotein (AFP) level, HBV DNA, Child–Pugh score, pre-operative antitumor therapy, and the MPPE1 mutation as variables, the MPPE1 mutation was an independent risk factor for HCC recurrence (P = .04, HR = 1.969; CI: 1.043–3.714, Table S7). These results suggest that the MPPE1 mutation is a predictor of HCC recurrence.
To determine whether the MPPE1 mutation exists in HCC cell lines, we examined the MPPE1 sequence in two HCC cell lines (Huh7 and HepG2) and found that both cell lines contained wild-type MPPE1 (base TT) (Fig. S5). The TCGA database showed that no mutation was found in the MPPE1 gene in 27 HCC cell lines in the sequencing database.
MPPE1 is overexpressed in HCC
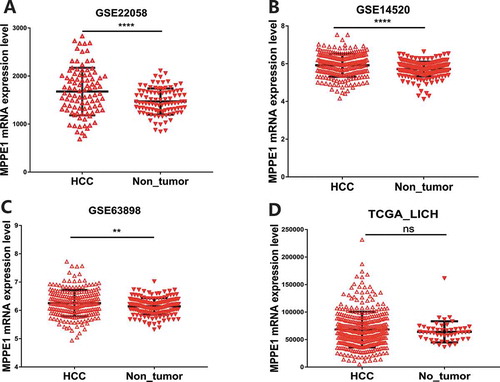
As the role of MPPE1 in tumors remains unknown, we used a data mining strategy to examine the expression of MPPE1 in HCC tissues. Data extracted from the GEO showed that MPPE1 expression was significantly increased in HCC tumor samples compared to adjacent nontumor tissues (). This finding was verified using data extracted from the TCGA, although the difference in MPPE1 expression in HCC tumor samples and adjacent nontumor tissues was not significant (), possibly because of the limited number of nontumor samples included in the TCGA analysis.
Down-regulation of MPPE1 inhibits HCC cell proliferation and affects cell apoptosis
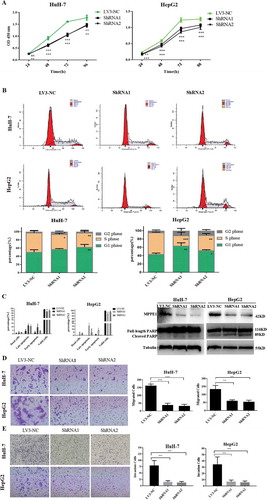
To investigate the function of MPPE1 in tumor cells, MPPE1 was knocked down in two human HCC cell lines, HuH-7 and HepG2, using MPPE1-shRNA (Fig. S4A, 4B). Knockdown of MPPE1 expression in HuH-7 and HepG2 cells significantly inhibited cell proliferation (P < .001) () and induced cell-cycle arrest. The proportion of HuH-7 cells and HepG2 in the G0/G1 phase was significantly increased and the proportion of HuH-7 cells and HepG2 cells in the S phase was substantially reduced at 72 h after MPPE1 knockdown (). These results suggest that MPPE1 plays an essential role in the proliferation of HCC cells by regulating the cell cycle transition from the G0/G1 to the S phase. Apoptosis assay indicated that knockdown of MPPE1 significantly increased the percentage of early and late apoptotic HuH-7 and HepG2 cells compared to control (), and obviously increased the cleavage of PARP (), the hallmark of apoptosis. These results suggest that MPPE1 may help HCC cells resist apoptosis.
Figure 4. Functional analysis of MPPE1 knockdown in Huh-7 and HepG2 cells. (a) Knockdown of MPPE1 expression significantly inhibited cell growth in a CCK-8 assay (n = 5). (b) Knockdown of MPPE1 expression significantly increased the proportion of HuH-7 cells and HepG2 in the G0/G1 phase and significantly decreased the proportion of HuH-7 cells and HepG2 cells in the S phase. The DNA content of HCC cells was analyzed using PI staining and flow cytometry (n = 4). (c) Knockdown of MPPE1 expression significantly increased the percentage of apoptotic HCC cells, detected by flow cytometry, and increased the level of cleaved PARP, identified with Western blotting analysis. Tubulin was used as an internal control. (d) Knockdown of MPPE1 expression inhibited cell invasion in a Transwell assay. Invasive cells were counted under a microscope (200×) as the mean of ten randomly selected fields of view. (e) Knockdown of MPPE1 expression inhibited cell migration in a Transwell assay. Migrated cells were counted under a microscope (200×) as the mean of ten randomly selected fields of view. Error bars represent the SD of the mean value. Statistical analysis was performed by the two-tailed least-significant difference (LSD) t-test. *, P < .05, **P < .01, ***P < .001
Down-regulation of MPPE1 inhibits HCC cell invasion and migration
Invasion and migration contribute to the malignant phenotype of HCC cells. Knockdown of MPPE1 expression in HuH-7 cells and HepG2 cells significantly reduced cell invasion () and migration abilities compared to controls ().
The invasion and migration potential of cancer cells is closely related to epithelial–mesenchymal transition (EMT) status. We examined the EMT markers, E-cadherin, and N-cadherin, in HepG2 cells with MPPE1 knockdown (shRNA2) or without (LV3-NC). Results showed that the expression of E-cadherin was significantly up-regulated (P= .002), while the expression of N-cadherin was significantly down-regulated in HepG2 cells (P< .001) after transcriptional silencing of MPPE1 (Fig S6), indicating that MPPE1 may influence invasion and migration by regulating EMT in HCC cells.
Down-regulation of MPPE1 inhibits tumor growth in vivo
A xenograft tumor model in nude mice was used to detect the effect of MPPE1 knockdown on tumor growth in vivo. Transcriptional silencing of MPPE1 (shRNA2) in HepG2 cells significantly reduced tumor weight and tumor volume in vivo, compared to the LV3-NC control group (P = .049) (Fig S7A-C). The results suggest that transcriptional silencing of MPPE1 inhibited tumor growth in vivo.
Discussion
In the present study, we found that an MPPE1 missense mutation on chr18_11897016 was among the most frequent mutations in primary or recurrent HCC. The frequency of the MPPE1 mutation was higher in recurrent HCC than in primary HCC or benign liver disease with cirrhosis tissues and was significantly associated with HCC recurrence. The MPPE1 mutation was significantly associated with TNM stage and Child–Pugh classification. In multivariable analysis of data from 121 HCC patients, the MPPE1 mutation was an independent risk factor for HCC recurrence. HCC patients with the MPPE1 mutation had a higher post-operative tumor recurrence rate. Further, knockdown of MPPE1 in the HCC cell lines, HepG2 or HuH-7, significantly inhibited cell proliferation, migration and invasion, induced cell cycle arrest and apoptosis, and inhibited tumor growth in nude mice in vivo. These data indicate that the MPPE1 mutation is a risk factor for HCC recurrence and that MPPE1 has potential as a novel therapeutic target for HCC.
Evidence suggests that HCC is characterized by the accumulation of genetic mutations and epigenetic changes during oncogenesis and progression.Citation10,Citation12,Citation16–18 In this study, our data identified unique genetic signatures for recurrent HCC and primary HCC (). The C:G-> G:C transversion may be a characteristic mutational signature of primary HCC. In recurrent HCC, the preferential C:G > T:A transversion may result from the high frequency of CpG methylation in the genome sequence, and represents a mutation commonly found in solid tumors.Citation19 Annotation of the 70 SNVs revealed their presence in 69 genes, some of which, such as TP53, MLH1 and STAT3, contribute to HCC development.Citation17,Citation18,Citation20,Citation21 Notably, we identified MPPE1 as a novel gene whose mutation is potentially associated with recurrent HCC.
Several genes, such as TP53, β-catenin (CTNNB1), telomerase reverse transcriptase (TERT), ARID1A, ARID2, and AXIN1 are frequently mutated and contribute to HCC development.Citation17–19,Citation22–25 Although mutations in the TP53 and β-catenin genes are the most common aberrations in HCC progression,Citation26,Citation27 interventions that reverse the effect of these mutations may not be beneficial.Citation28 Reactivation of proteins such as p53 in tumors holds great promise as anticancer therapy; however, some studies suggest that chronic activation of these molecules may be deleterious.Citation27
p53 is a well-known tumor suppressor. p53 protein maintains genomic stability and mediates DNA repair, the induction of apoptosis, cell cycle regulation, and interaction with other genes.Citation13,Citation29 p53 is frequently mutated in human HCC as HBV and HCV can damage DNA and mutate cancer-related genes, such as p53. The X gene of HBV (HBx) is commonly integrated into the host genome and binds to p53, attenuating DNA repair and p53-mediated apoptosis.Citation30,Citation31 The present study identified p53 among the mutated genes in primary and recurrent HCC tissue samples. According to KEGG pathway analysis and consistent with the findings in previous reports, the genes identified in this study were found to participate in several signaling pathways, including cell cycle control, p53 signaling, pathways in cancer, and the Wnt signaling pathway.
MPPE1 encodes a metallophosphoesterase protein belonging to the calcineurin-like phosphoesterase superfamily.Citation13,Citation14 The phosphoesterases of this family are involved in a variety of biological processes, including gene expression, cell growth, and cell differentiation.Citation14 MPPE1 is required to transport glycosylphosphatidylinositol-anchor proteins from the endoplasmic reticulum to the Golgi.Citation15 Gene Ontology annotations include manganese ion and GPI-anchor binding. However, the role of MPPE1 in cancer remains to be elucidated. The mechanisms and pathways underlying the role of MPPE1 mutation in cancer are unknown.
While analyzing the sequence of MPPE1, we found the mutation site (p. E83G) to be close to the putative active sites (D77, H79, D119), so we speculate that the mutation may influence the activity of MPPE1. Furthermore, according to data extracted from the TCGA and COSMIC, MPPE1 expression was significantly increased in HCC tumor samples compared to adjacent nontumor tissues. Therefore, we knocked down MPPE1 expression to investigate phenotypic changes in HCC cells. To the authors’ knowledge, this is the first study to demonstrate that down-regulation of MPPE1 expression significantly inhibits cell proliferation, disrupts cell cycle progression by inhibiting G1 to S phase transition, induces cell apoptosis, inhibits invasion and migration, and inhibits HepG2 tumor growth in vivo. The impact of the MPPE1 mutation on gene function will be explored in future research.
In conclusion, we identified MPPE1 as a novel candidate gene whose mutation is potentially associated with HCC recurrence. In addition, this study provides novel information regarding the function of MPPE1 in cancer cells. Enzymes are generally accepted as ideal drug targets, and developing drugs (such as small molecular inhibitors) that target enzyme activity is warranted. Findings from this study identified MPPE1 as a potential therapeutic target for HCC and indicate that investigating MPPE1 will further understanding of the molecular mechanisms underlying HCC recurrence.
Materials and methods
Patients and tissue samples
Five specimens (a primary HCC tissue, a recurrent HCC tissue, their adjacent non-tumor tissues, and peripheral blood mononuclear cells) were obtained from a single patient who had been suffering from HBV-induced cirrhosis for 23 y and had undergone surgical resection for primary HCC and recurrent HCC in February 2013 and November 2013, respectively, at the General Hospital of the Chinese People’s Armed Police Force. Histological characteristics of the primary and recurrent HCC samples, as well as their adjacent non-tumor tissues, are shown in Fig. S1A and S1B. Genomic DNA was extracted from the five samples and subjected to whole-exome sequencing analysis.
In addition, genomic DNA was extracted from freshly frozen hepatitis B surface antigen (HBsAg)-positive primary HCC (n = 87), recurrent HCC (n = 34) and benign liver disease with cirrhosis tissues (n = 43). Specimens were collected between June 2012 and December 2016 following surgical resection at the General Hospital of the Chinese People’s Armed Police Force. All cases were histologically confirmed.
This study was approved by the ethics committee of the General Hospital of the Chinese People’s Armed Police Force. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from each patient.
Whole-exome sequencing and identification of SNVs and their genes potentially associated with HCC recurrence
Procedures for DNA extraction and high-throughput whole-exome sequencing are described in the Supplemental File. Single nucleotide polymorphisms (SNPs) were identified from corrected whole-exome sequencing data using SOAPsnp software (BGI Tech Solutions Co., Ltd. Shenzhen, China), as previously described.Citation32–35 Single nucleotide variants (SNVs) potentially associated with HCC recurrence were identified using the dbSNP (version 135) pipeline (Fig. S2). SNVs were annotated and classified using ANNOVAR, as previously described.Citation36
The initial list of candidate nonsynonymous SNVs potentially associated with HCC recurrence was verified using polymerase chain reaction (PCR)-MassARRAY analysis in the dataset that included the HBsAg-positive primary HCC (n = 87), recurrent HCC (n = 34) and benign liver disease with cirrhosis tissues (n = 43). After the PCR products were modified using shrimp alkaline phosphatase (SAP) and single-base extension, the purified products were analyzed on a MassARRAY platform (Sequenom, Inc., San Diego, CA, USA), according to the manufacturer’s instructions. Candidate SNVs were further verified using standard procedures for Sanger sequencing.Citation37 Primers with high amplification efficiency were designed to localize 80–120 bp upstream and downstream of the target mutation. The frequency of each SNV was recorded.
Function and pathway enrichment analyses of mutated genes with candidate SNVs potentially associated with HCC recurrence were performed using WebGestaltCitation38 based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.kegg.jp/) and the Gene Ontology (GO) and UniProtKB/Swiss-Prot databases in the GeneCard database (https://www.genecards.org/c). Significance of the enrichment data was determined by the hypergeometric test and adjusted for multiple testing using the Benjamini-Hochberg false discovery rate (FDR). KEGG terms were considered enriched at P ≤ 0.05.Citation38
Cell lines and RNA interference
Two human HCC cell lines, HuH-7 and HepG2, and human embryonic kidney 293 T cells were purchased from the Chinese Academy of Sciences.Citation39 Two shRNA sequences targeting human MPPE1 mRNA were designed using an online tool (http://rnaidesigner.thermofisher.com/) and is shown in the Supplemental File. A scrambled sequence that shared no homology with the human genome was used as a negative control. Successful cloning of these sequences into the pLV-H1-Puro-GFP lentivector was confirmed by Sanger sequencing. Procedures for amplification of shRNA-expressing recombinant lentiviruses and stable transfection of LV-shRNA-MPPE1 in HepG2 and HuH-7 cells are described in the Supplemental File.
Western blotting
Western blotting was performed as previously describedCitation28 using antibodies for MPPE1 (ab177092, Abcam, USA), poly (ADP-ribose) polymerase (PARP) (9542s, Parp CST, USA) and tubulin (ca-8035, Santa Cruz, USA), and horseradish peroxidize (HRP)-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA).
Cell proliferation assay
Proliferation of HuH-7 and HepG2 cells after transfection with lentiviral ShRNA1, ShRNA2, or LV3-NC was determined using the Cell Counting Kit-8 (CCK-8, Dojindo, Tokyo, Japan), according to the manufacturer’s instructions.
Cell cycle analysis and apoptosis assay
The cell cycle was assessed in 70% ethanol-fixed cells by propidium iodide (Sigma, USA) staining and a FACS flow cytometer (BD Biosciences, San Jose, CA). Apoptosis was assessed in floating and attached cells by annexin V/propidium iodide staining using the Annexin V-APC/7-AAD Apoptosis Detection Kit (BioVision, Palo Alto, CA) according to the manufacturer’s instructions, and the flow cytometer.
Cell invasion and migration assays
Cell invasion and migration assays were performed in chambers, as previously described.Citation40 The number of cells was counted under a microscope (200×) in ten randomly selected fields of view.
HCC datasets from GEO and TCGA
MPPE1 expression in HCC tumor samples and adjacent nontumor tissues was investigated in publically available data deposited in the Gene Expression Omnibus (GEO) (GSE22058, GSE14520 and GSE63898) and The Cancer Genome Atlas (TCGA) (TCGA _LIHC dataset: n = 371 HCC tumor samples, n = 50 adjacent nontumor samples) (as of Jan 28, 2016).
Xenograft tumor model in nude mice
BALB/c nude mice (6–8 weeks) were supplied by the Liver Cancer Institute of Fudan University (Shanghai, China). Animal studies were approved by the local regulatory agency (Institutional Animal Care and Use Committee of Laboratory Animal Center of Academy of The Fourth Military Medical University). Nude mice were randomly divided into two groups: shRNA2- and LV-3NC- (control) infected HepG2 cells (n = 6 mice per group). After the cells were digested and centrifuged (5 min, 1000 R/min), they were resuspended in serum-free DMEM and stored at 4°C. Then, 150 μ L of cells (1.5 × 106/mL) were injected into the back of nude mice.
Tumor volume was measured on Day 14, 21, 28, 32, 37, 42 and 49 after implantation. Tumor volume was calculated using the formula: V (volume) = (π × length × width2)/6. Tumor tissues were removed, weighed, and immediately fixed in formalin on Day 49 after implantation.
Statistical analysis
Statistical analysis was performed using SPSS for Windows version 16.0 (SPSS, Chicago, IL, USA). A case–control model (dichotomous) was applied to identify associations between candidate mutant genes and primary and recurrent HCC. The Hardy–Weinberg Equilibrium (HWE), the Fisher’s exact test, chi-square test, and allele logistic regression was used to investigate associations between mutant genes SNV and disease of primary or recurrent HCC. The Kaplan–Meier method and the log-rank test were conducted to evaluate time to recurrence, defined as the duration from surgery to recurrence diagnosis. Cox proportional hazards model was used to identify independent risk factors for tumor recurrence. Data are presented as the mean ± standard deviation. P-≤0.05 was considered statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Authors’ contributions
QZ: Conception, design, and supervision of the study, technical support, and writing the manuscript.
YKS: Conception, design, and supervision of the study, data analysis and interpretation, and writing the manuscript.
XC, JX, PXW, LS and JG: Study design, technical support, statistical analysis and interpretation of data.
JG, LHH and YAJ: Patient enrollment, technical support, ethics-related work.
JHC and HJC: Study supervision, technical support, data collection and follow-up.
Ethics approval
This study was approved by the ethics committee of the General Hospital of the Chinese People’s Armed Police Force. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from all individual participants included in the study.
Supplemental Material
Download MS Word (15.6 MB)Acknowledgments
We thank BGI Tech Solutions Co., Ltd, Cancer Research Department, and FOREVERGEN Biomedical Technology (Guangdong) Co., Ltd, Dr. Minghui He, Tang Tang Biomedical Technology (Beijing) Co., Ltd, Beijing Zhong Zheng DNA Evidence Institute, and Premier Scientific Partner (Beijing, China) for their technical support.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Data availability
The datasets generated and analyzed during the present study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. 2011. Global cancer statistics. CA Cancer J Clin. 61:69–90. doi:10.3322/caac.20107.
- Altekruse SF, McGlynn KA, Reichman ME. 2009. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 27:1485–1491. doi:10.1200/JCO.2008.20.7753.
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. 2015. Global cancer statistics, 2012. CA Cancer J Clin. 65:87–108. doi:10.3322/caac.21262.
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390.
- Bosch FX, Ribes J, Borras J. 1999. Epidemiology of primary liver cancer. Semin Liver Dis. 19:271–285. doi:10.1055/s-2007-1007117.
- Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. 2006. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 45:529–538. doi:10.1016/j.jhep.2006.05.013.
- Galle PR, Tovoli F, Foerster F, Worns MA, Cucchetti A, Bolondi L. 2017. The treatment of intermediate stage tumours beyond TACE: from surgery to systemic therapy. J Hepatol. 67:173–183. doi:10.1016/j.jhep.2017.03.007.
- Sapisochin G, Bruix J. 2017. Liver transplantation for hepatocellular carcinoma: outcomes and novel surgical approaches. Nat Rev Gastroenterol Hepatol. 14:203–217. doi:10.1038/nrgastro.2016.193.
- Choi JH, Kim MJ, Park YK, Im JY, Kwon SM, Kim HC, Woo HG, Wang HJ. 2017. Mutations acquired by hepatocellular carcinoma recurrence give rise to an aggressive phenotype. Oncotarget. 8:22903–22916. doi:10.18632/oncotarget.14248.
- Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346.
- Aravalli RN, Cressman EN, Steer CJ. 2013. Cellular and molecular mechanisms of hepatocellular carcinoma: an update. Arch Toxicol. 87:227–247. doi:10.1007/s00204-012-0931-2.
- Nguyen DX, Bos PD, Massague J. 2009. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 9:274–284. doi:10.1038/nrc2622.
- Lohoff FW, Ferraro TN, Brodkin ES, Weller AE, Bloch PJ. 2010. Association between polymorphisms in the metallophosphoesterase (MPPE1) gene and bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 153B:830–836. doi:10.1002/ajmg.b.31042.
- Vuoristo JT, Ala-Kokko L. cDNA cloning, genomic organization and expression of the novel human metallophosphoesterase gene MPPE1 on chromosome 18p11.2. Cytogenet Cell Genet. 2001;95:60–63.
- Fujita M, Maeda Y, Ra M, Yamaguchi Y, Taguchi R, Kinoshita T. 2009. GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell. 139:352–365. doi:10.1016/j.cell.2009.08.040.
- Liu M, Jiang L, Guan XY. 2014. The genetic and epigenetic alterations in human hepatocellular carcinoma: a recent update. Protein Cell. 5:673–691. doi:10.1007/s13238-014-0065-9.
- Lee JS. 2015. The mutational landscape of hepatocellular carcinoma. Clin Mol Hepatol. 21:220–229. doi:10.3350/cmh.2015.21.3.220.
- Garcia-Fernandez N, Macher HC, Rubio A, Jimenez-Arriscado P, Bernal-Bellido C, Bellido-Diaz ML, Suarez-Artacho G, Guerrero JM, Gomez-Bravo MA, Molinero P. 2016. Detection of p53 mutations in circulating DNA of transplanted hepatocellular carcinoma patients as a biomarker of tumor recurrence. Adv Exp Med Biol. 924:25–28. doi:10.1007/978-3-319-42044-8_5.
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature. 446:153–158. doi: 10.1038/nature05610.
- Zhu X, Liu W, Qiu X, Wang Z, Tan C, Bei C, Qin L, Ren Y, Tan S. 2017. Single nucleotide polymorphisms in MLH1 predict poor prognosis of hepatocellular carcinoma in a Chinese population. Oncotarget. 8:80039–80049. doi:10.18632/oncotarget.16899.
- Liu B, Yu S. 2018. Amentoflavone suppresses hepatocellular carcinoma by repressing hexokinase 2 expression through inhibiting JAK2/STAT3 signaling. Biomed Pharmacother. 107:243–253. doi:10.1016/j.biopha.2018.07.177.
- Huang J, Deng Q, Wang Q, Li KY, Dai JH, Li N, Zhu ZD, Zhou B, Liu XY, Liu RF, et al. 2012. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 44:1117–1121. doi: 10.1038/ng.2391.
- Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJ, et al. 2011. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 43:828–829. doi: 10.1038/ng.903.
- Takai A, Dang HT, Wang XW. 2014. Identification of drivers from cancer genome diversity in hepatocellular carcinoma. Int J Mol Sci. 15:11142–11160. doi:10.3390/ijms150611142.
- Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, et al. 2012. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 44:694–698. doi: 10.1038/ng.2256.
- Sheen IS, Jeng KS, Wu JY. Is p53 gene mutation an indication of the biological behaviors of recurrence of hepatocellular carcinoma? World J Gastroenterol. 2003;9:1202–1207.
- Panera N, Crudele A, Romito I, Gnani D, Alisi A. 2017. Focal adhesion kinase: insight into molecular roles and functions in hepatocellular carcinoma. Int J Mol Sci. 18:E99. doi:10.3390/ijms18010099.
- Gross-Goupil M, Riou P, Emile JF, Saffroy R, Azoulay D, Lacherade I, Receveur A, Piatier-Tonneau D, Castaing D, Debuire B, et al. 2003. Analysis of chromosomal instability in pulmonary or liver metastases and matched primary hepatocellular carcinoma after orthotopic liver transplantation. Int J Cancer. 104:745–751. doi: 10.1002/ijc.11017.
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. 2007. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 28:622–629. doi:10.1002/humu.20495.
- Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. 2007. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 26:2166–2176. doi:10.1038/sj.onc.1210279.
- Kew MC. 2011. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol. 26(Suppl 1):144–152. doi: 10.1111/j.1440-1746.2010.06546.x.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079.
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760.
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics (Oxford, England). 2010;25:1754–1760.
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22:568–576. doi:10.1101/gr.129684.111.
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164. doi:10.1093/nar/gkq603.
- Murphy KM, Berg KD, Eshleman JR. 2005. Sequencing of genomic DNA by combined amplification and cycle sequencing reaction. Clin Chem. 51:35–39. doi:10.1373/clinchem.2004.039164.
- Zhang B, Kirov S, Snoddy J. 2005. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33:W741–748. doi:10.1093/nar/gki475.
- Kim BJ, Ryu SW, Song BJ. 2006. JNK- and p38 kinase-mediated phosphorylation of Bax leads to its activation and mitochondrial translocation and to apoptosis of human hepatoma HepG2 cells. J Biol Chem. 281:21256–21265. doi:10.1074/jbc.M510644200.
- Zhan XH, Xu QY, Tian R, Yan H, Zhang M, Wu J, Wang W, He J. MicroRNA16 regulates glioma cell proliferation, apoptosis and invasion by targeting Wip1-ATM-p53 feedback loop. Oncotarget. 2017;8:54788–54798.