ABSTRACT
Cervical cancer (CC) is the second most common malignancy in women worldwide. The mechanism underlying CC development remains unclear. Recently, Circular RNAs (circRNAs)have attracted attention because of its role in tumorigenesis. To investigate circRNAsin CC, RNA sequencing was employed to characterize circRNA expression profile between CC tissues and matched adjacent normal tissues. The expression of hsa_circ_0003204 was examined in CC tissues and cell lines by real-time PCR. Migration assay and invasion assay were used to verify the effect of hsa_circ_0003204 on migration and invasion ability in CC cell lines. Tumor formation assay in nude mice was used to analyze the effect of hsa_circ_0003204 on the tumorigenicity of CC cell lines in vitro. Western blotting analyzes were performed to investigate the role of hsa_circ_0003204 in the regulation of MAPK signaling activation. We found that circRNA hsa_circ_0003204 was significantly upregulated in CC tissues. The function and potential molecular mechanisms of hsa_circ_0003204 were also investigated in vitro and in vivo. Hsa_circ_0003204 knockdown reduced cell growth, migration, and invasion but promoted cells apoptosis. However, the over-expression of hsa_circ_0003204 had the opposite effect. The MAPK pathway was different in hsa_circ_0003204 over-expression or down-expression cells, compared to parental cells. In addition, over-expression of hsa_circ_0003204 significantly increased tumor volume and tumor weight in vivo.Taken together, results indicated hsa_circ_0003204 may serve as a potential therapeutic target for patients with CC.
Background
Cervical cancer (CC) is a cancer arising from the cervix. Approximately 500,000 women worldwide develop cervical cancer each year.Citation1 Human papillomavirus (HPV) is the primary etiologic agent of CC. Although HPV vaccine is a cost-effective approach to protect women from CC, these disease is still the second most frequent cause of cancer-related death among women in developing countries.Citation2–4 Most patients have developed invasive cancer at the time of diagnosis due to insufficient infrastructure, inadequate access to prevent HPV vaccines services and lack of screening test.Citation5 Despite advances in the development of therapies, such as surgery, radiotherapy and chemotherapy. CC patients at advanced stages show very poor prognosis. The reported five-year survival rate remained less than 30% in developing countries and patient outcomes depend on how early the cancer is diagnosed.Citation6 To date, the mechanism underlying CC development remains elusive. Understanding the molecular mechanisms of CC development is essential to discover novel diagnostic and prognostic biomarkers for CC, as well as design effective preventive strategies.
As a class of non-coding RNAs (ncRNAs), circular RNAs (circRNAs) are single-stranded that forms into a covalently closed circular structure rather than a linear structure by special splicing mechanism.Citation7 Accumulated functional evidences showed that circRNAs are involved in various biological processes, such as cell proliferation, survival, migration, senescence, and apoptosis.Citation8,Citation9 In recent years, circRNAs have attracted worldwide attention.Citation10 Advances in high-throughput sequencing technology and novel bioinformatics algorithms have facilitated the systematic detection of circRNAs, which may be potential biomarkers and therapeutic targets of cancer.Citation11 The clinical application of circRNAs as a new diagnostic and prognostic biomarkers for CC have been investigated.Citation12–14 Numerous studies have suggested that circRNAs expression profiles were significantly different between normal cervical epithelial cells and cervical cancer cells,Citation14–17 whereas the functions of most circRNAs in CC are largely unclear. Therefore, it is necessary to further study the role and mechanism of circRNAs in the progression of CC. The aim of this study was to identify differentially expressed circRNAs in CC tissues compared wtih matched adjacent normal tissues and to explore the biological function of hsa_circ_0003204 in CC carcinogenesis.
Methods
Tissue samples
The study was conducted according to the principles in the Declaration of Helsinki and approved by the Medical Ethics Committee of Zhujiang Hospital, Southern Medical University, China. Informed consent was obtained from all patients. All patients had been diagonosed with cervical cancer, according to the Federation International of Gynecology and Obstetrics (FIGO) criteria. All the tumor tissues were assessed by Hematoxylin and Eosin (HE) staining, and the diagnosis of all of the cases were confirmed by two independent pathologists. A total of three pairs of CC tissues and matched adjacent normal tissues were collected from three CC patients at stage I B1, I B2, II A1 during radical hysterectomy at Foshan Women and Children Hospital affiliated to Southern Medical University China. The collected adjacent normal tissues were 2 cm away from the visible CC lesions. All specimens were snap-frozen immediately in liquid nitrogen (−150 °C) and stored at −80 °C until RNA extraction.
Cell culture and transfection
Human CC cell lines (Hela, SiHa, Caski and MS751) were purchased from the Cellcook (Guangzhou, China). Hela, SiHa and MS751 was maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA) supplemented with 10% (vol/vol) fetal bovine serum (FBS), 100 units/ml of penicillin and 100 μg/ml of streptomycin (Sigma-Aldrich) in 5% COCitation2 at 37°C. Caski was cultured in RPMI-1640 (Invitrogen, NY, U.S.A.) containing 10% FBS, 100 units/ml of penicillin and 100 μg/ml of streptomycin in 5% CO2 at 37°C. Human cervical epithelial cells (HcerEpiC) were cultured in Cervical Epithelial Cell Growth Supplement (CerEpiCGS, Cat #7062), a complete medium designed for optimal growth of normal cervical epithelial cells in vitro. SiRNA against hsa_circ_0003204 and negative controls were synthesized by RIBOBIO (Guangzhou, China). Overexpression of hsa_circ_0003204 were obtained from Invitrogen (Carlsbad, CA, USA). All cell transfections were performed using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA) according to the manufacturer’s protocol. SiRNA used in our study was listed in .
Table 2. Primer sequence of has_circ_0003204
Table 1. SiRNA sequence against has_circ_0003204
RNA extraction from sample and high throughput sequencing
Total RNA of Hela, SiHa, Caski, and MS751 cell lines were extracted using TRIZOL reagent (Invitrogen, USA) following manufacturer’s instructions. The total RNA concentration and purity were measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The RNA integrity and yield were assessed using the Agilent 2100 Bioanalyzer Lab-on-Chip system (Agilent Technologies, Palo Alto, CA, USA) and RNA 6000 Nano LabChip Kit (Agilent Technologies, USA). 10 µg of total RNA was depleted of ribosomal RNA using the RiboMinus Eukaryote Kit (Qiagen, Hilden, Germany). The rRNA-depleted RNA was teated with 10 U/μg RNase R (Epicenter, Madison, WI) and incubated at 37 °C for 1 h to remove linear RNA. The remaining RNA was used as template for the construction of RNA-seq library in accordance with the protocol of NEB Next Ultra Directional RNA Library Prep Kit (Illumina, San Diego, USA). The resulting RNA-seq library was quantified by Agilent 2100 Bioanalyzer and sequenced on HiSeq 2000 platform (Illumina, CA, USA), which generated paired-end reads of 150 nucleotides. A cutoff criterion of |log2(fold-change) | ≥ 1 and FDR<0.05 between two samples were used to identify differentially expressed circRNA and transcripts.
Validation of hsa_circ_0003204 by Sanger sequencing Convergent (for linear RNA) and divergent (for circRNA) primers were designed to validate the existence of hsa_circ_0003204 (http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003204). Details of divergent and convergent primers are listed on . Genomic DNA (gDNA) and cDNA from CC sample were used for PCR reaction to confirm the hsa_circ_0003204 junction. PCR products of divergent and convergent primers from cDNA and gDNA were analyzed by agarose gel electrophoresis. Back-splicing sites of circRNAs were verified by Sanger sequencing at Guangzhou IGE biotechnology Ltd. (IGE, Guangzhou, China).
Quantitative real-time PCR
The hsa_circ_0003204 levels were measured by quantitative real-time PCR (qRT-PCR). Total RNA from cells was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. qRT-PCR was performed using Power SYBR Green PCR Mix from Life Technologies. All experiments were performed in triplicates. GAPDH gene was used as an endogenous control. The primers used in the study were list in .
Construction of stable hsa_circ_0003204 overexpressing cells
The sequences of hsa_circ_0003204 were cloned into an overexpression vector LV003 (Forevergen, Guangzhou, China) containing the green fluorescent protein (GFP) reporter gene and puromycin resistance gene. The overexpression vectors were packaged into lentivirus. SiHa cells were infected with lentivirus according to the manufacturer’s instructions. The cells were then selected with puromycin for 1–2 weeks. The surviving cells were considered as stable hsa_circ_0003204-overexpressing cells and verified using qRT-PCR.
Cell cycle and apoptosis using flow cytometry
Cells apoptosis was evaluated by FITC Annexin V Apoptosis Detection Kit (BD Pharmingen, San Diego, CA, USA) using flow cytometry (Becton Dickinson, San Jose, CA, USA) according to the manufacturer’s guidelines. Twenty-four hours after transfection, 1 × 106 cells MS751 and Caski cells were harvested and stained by FITC Annexin V and propidium iodide (PI). The stained cells were measured by flow cytometry, and data were analyzed using CellQuest software (BD Bioscience, San Diego, CA, USA). For cell cycle analysis, cells were harvested and fixed with 70% ethanol overnight at 4°C. The cell pellet was stained by PI after washing and centrifugation. The cell cycle distribution was determined by flow cytometry.
MTS assay
To determine the effect of hsa_circ_0003204 on MS751 and Caski cell viability, cells were subjected to the MTS assay. Cell proliferation were determined using CellTiter 96® Aqueous One Solution Cell Proliferation Assay (MTS) (Promega, WI, USA) according to manufacturer’s instructions. Cells were seeded in 96-well microtiter plate at 1 × 104 cells/well for 24 h and then transfected with si-circRNA or overexpression of hsa_circ_0003204 vector. About 20 µL of the MTS reagent was added into each well at 24 h after transfection and cells were incubated for 3 h at 37°C under standard culture conditions. The absorbance was evaluated at OD 490 nm using a microplate reader (Multi spectrum, Themo scientific).
Cell migration and invasion assays
The CC cell invasion and migration assay in the absence and presence of Matrigel were performed using 24 well transwell chambers (Corning Costar, New York, USA). MS751 and Caski cells were transfected with si-circRNA or overexpression of hsa_circ_0003204 vector for 24 h. A total of 1 × 105 transfeced cells were suspended in 150 µl serum-free DMEM or RPMI-1640 medium, cultured in the upper transwell chamber with 8.0 μm pore polycarbonate membrane insert, the lower chamber contained 600 µl of complete DMEM or RPMI-1640 medium. After 24 h of incubation at 37 °C, the migration cells on the bottom surface of membrane were fixed with 4% paraformaldehyde solution for 30 min, and stained with 0.5% crystal violet for 25 min at room temperature. A total of six random visual fields were selected and calculated. Experiments were independently performed at least three times.
Xenograft assay in vivo
All animal protocols were approved by the Institutional Animal Care and Use Committee. BALB/C mice (4–6-weeks-old) were purchased from the Chinese Academy of Sciences (Shanghai, China), and maintained in a specific pathogen-free facility. Twelve mice were randomly divided into NC group (n = 6) and hsa_circ_0003204 overexpression group (n = 6). 2 × 106 SiHa cells with stably hsa_circ_0003204 overexpressing and SiHa negative control cells were injected into the flanks of nude mice subcutaneously, respectively. Tumor volumes were determined at indicated time points. Tumor volume was calculated based on the formula: Tumor volume (mm3) = (width) × (height)Citation2/2. When the average tumor volume reached 1500 mm3 (study end point), xenograft tumor tissues were collected after sacrifice and weighed.
Western blotting
Cells were collected at 24 h after transfection with si-circRNA and hsa_circ_0003204 overexpression vector. Xenograft tissue were harvest at the termination of tumor xenograft experiment. Cells and xenograft tissue lysates were prepared and subjected to Western blot for the analysis of different proteins of MAPK signaling pathways. Western blot analysis were preformed using standard procedures. Primary antibodies used in this study were: anti-P38(8690s,CST), anti-p-P38(4511S,CST), anti-ERK (9102S,CST), anti-p-ERK(4376S,CST), and anti-GAPDH (A-1310016, Life) antibody. The immune complex were visualized using ECL detection kit (Amersham Biosciences, Buckinghamshire, UK).
Statistical analyzes
All statistical data analyzes were performed using SPSS 17.0 software (IBM, Chicago, IL). The data are expressed as the mean±standard deviation from at least three separate experiments. p < .05 was considered as statistical significance. Student’s t test and Kruskal-Wallis test were utilized to analyze two or multiple groups, respectively.
Result
CircRNAs expression profile in the CC tissue
To identify specific circRNAs that are differentially expressed between CC and adjacent normal tissues, the expression profile of circRNAs was performed by high-throughput sequencing. To obtain consistent biological information, paired samples from three patients with similar baseline characteristics were selected and pooled for circRNA sequencing. The circRNA expression pattern was found to be different between CC and adjacent normal tissues. As shown in , the top 10 upregulated and 10 downregulated circRNAs were identified according to following criteria: FDR <0.05, |log2 Ratio(Tumor/Control)| ≥1, which are included in the circBase database and expressed in both samples. The Scatter-Plot () were produced to visualize the significantly differently expressed circRNAs (FDR <0.05, |log2 Ratio(Tumor/Control)| ≥1) between CC and adjacent normal tissues. To explore potential function and pathways in which the differentially expressed circRNAs were involved, Gene Ontology (GO) term enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed. As shown in , GO terms comprise three classifications: biological process, cellular component, and molecular function. The GO terms of the most differentially expressed circRNAs host genes included cellular process, binding and cell development, regulation of cell proliferation, migration, and apoptosis. As shown in , the host genes of differentially expressed circRNAs were mainly enriched in cancer-related pathways, such as MAPK signaling pathway and PI3K-Akt signaling pathway. As upregulated circRNAs are more suitable to serve as biomarkers than downregulated circRNAs, hsa_circ_0003204 was selected from the upregulation group for further analysis.
Figure 1. Overview of circRNA differential expression. (a) Hierarchical cluster analysis (heat map) for differentially expressed circRNAs between CC tissue and adjacent normal tissue. (b) Scatter plot demonstrating the variation of circRNAs expression in CC tissue (y-axis) versus control (x-axis). (c) GO enrichment of differentially expressed circRNAs d.KEGG pathway enrichment analysis of differentially expressed circRNAs
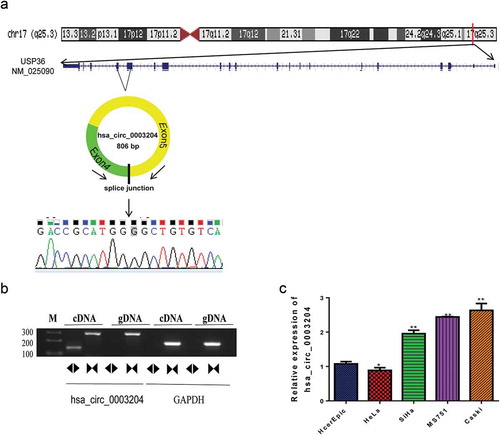
Identification and characterization of differentially expressed circRNAs in CC patients
PCR were employed to validate the selected circRNA expression, and subsequent Sanger sequencing was performed from the PCR product amplified with the divergent primers. PCR results indicate that the circular form was amplified with the divergent primers using cDNA as template. There was no amplification detected at similar sizes using gDNA as template, which suggested the presence of back-site junctions. Back-splice junctions of hsa_circ_0003204 were represented in schematic diagram (). The sequence of hsa_circ_0003204 was shown in . PCR was also performed on gDNA and cDNA using the corresponding convergent primers to confirm that back-spliced junctions were just in RNA transcripts. PCR products were amplified with convergent primers from both the cDNA and the gDNA (). Moreover, the expression of hsa_circ_0003204 from different CC cell lines including HcerEpic, HeLa, SiHa, MS751 and Caski were analyzed using qRT-PCR (). MS751 and Caski with the higher expression of hsa_circ_0003204 were selected for further study.
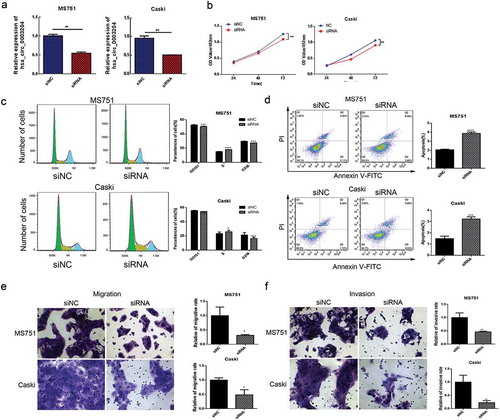
Silencing of hsa_circ_0003204 suppressed CC cell proliferation, migration, invasion, and enhanced cell apoptosis in vitro
In order to investigate the biological functions of hsa_circ_0003204 in CC, the specific siRNA was used to silence the expression of hsa_circ_0003204 in MS751 and Caski CC cell lines. As shown in , the siRNAs significantly decreased hsa_circ_0003204 expression level. Cell proliferation was measured using the MTS assay. Silencing of hsa_circ_0003204 significantly suppressed cell proliferation in both MS751 and Caski cell lines (). Cell cycle phase distribution and apoptosis of CC cells were assessed by flow cytometric analysis. Results showed that knockdown of hsa_circ_0003204 significantly arrested MS751 and Caski cells at S phase (). Cell apoptosis analysis exhibited significantly increased apoptotic rate as compared with negative control (). Moreover, the transwell migration and invasion assays showed knockdown of hsa_circ_0003204 significantly reduced the migration and invasion abilities in MS751 and Caski cells ( and ).
Figure 3. Silencing of hsa_circ_0003204 suppress cervical cancer cell proliferation, migration, invasion, and enhance apoptosis in vitro. (a) Validation of hsa_circ_0003204 in cell by qRT-PCR. (b) Silencing of hsa_circ_0003204 suppressed cell proliferation in both MS751 and Caski cells. (c) Silencing of hsa_circ_0003204 significantly prevent MS751 and Caski cells from entering into S phase. (d) Silencing of hsa_circ_0023404 significantly increased apoptosis of both MS751 and Caski. (e) Silencing of hsa_circ_0003204 significantly decreased migration of both MS751 and Caski. (f) Silencing of hsa_circ_0003204 significantly reduced invasion of both MS751 and Caski
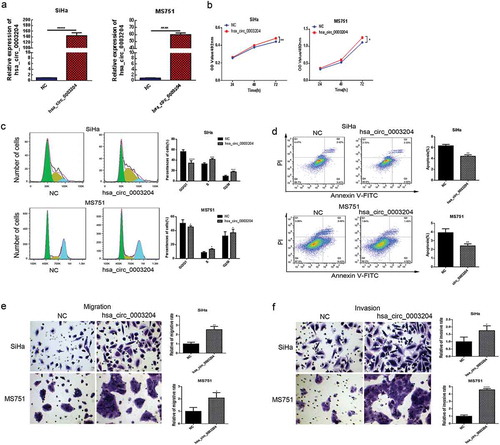
Overexpression of hsa_circ_0003204 promoted CC cell proliferation, migration, invasion, and reduced apoptosis in vitro
Meanwhile, MS751 and SiHa cells were transiently transfected with hsa_circ_0003204 overexpression vector. qRT-PCR analysis indicated that expression of hsa_circ_0003204 was significantly upregulated in both MS751 and SiHa cells after transiently transfection (P < .01, ). Proliferation of MS751 and SiHa cells was markedly increased after overexpression of hsa_circ_0003204, compared with the NC group (). Cell cycle phase distributions results indicated that overexpression hsa_circ_0003204 increased the number of cells in S and G2/M phase compared with control group (). Overexpression of hsa_circ_0003204 also reduced cellular apoptosis in MS751 and SiHa cells (). Compared with the control group, invasion and migration rates were significantly enhanced after overexpression of hsa_circ_0003204 (p < .01; and ). The above in vitro experiments indicated that hsa_circ_0003204 exert biological function in proliferation, migration, invasion, and apoptosis of CC cell.
Figure 4. Overexpression of hsa_circ_0003204 promote CC cell proliferation, migration, invasion, and reduce apoptosis in vitro. (a) The expressions of hsa_circ_0003204 in SiHa and MS751 were significantly increased. (b) Overexpression of hsa_circ_0003204 promoted cell proliferation in both MS751 and SiHa cells. (c) Overexpression of hsa_circ_0003204 significantly increased S and G2/M phase of both MS751 and SiHa cells. (d) Overexpression of hsa_circ_0003204 significantly decreased apoptosis of both MS751 and SiHa cells. (e) Overexpression of hsa_circ_0003204 enhance migration of both MS751 and SiHa cells. (f) Overexpression of hsa_circ_0003204 enhance invasion of both MS751 and SiHa cells
Differentially expressed genes and pathways in hsa_circ_0003204 overpression cells
To further investigate the role of hsa_circ_0003204 in CC, SiHa cells stably overexpressing hsa_circ_0003204 was established. The infection efficiency of the lentivirus was assessed by the fluorescence intensity and qRT-PCR ( and ). These results demonstrated the successful establishment of overexpressed cells.
Figure 5. Construction of stable overexpression of hsa_circ_0003204 cell line and mRNA expression profile of hsa_circ_0003204 overpression cell. (a) Establishment of stable overexpression of hsa_circ_0003204 cell line with green fluorescent protein. (b) Validation of overexpression of hsa_circ_0003204 in cell by qRT-PCR. (c) Hierarchical cluster analysis (heat map) for differentially expressed mRNA between overexpression of hsa_circ_0003204 cell and negative control cell. (d) Volcano plot revealed the differentially expressed mRNA in overexpression of hsa_circ_0003204 cell compared to negative control cell
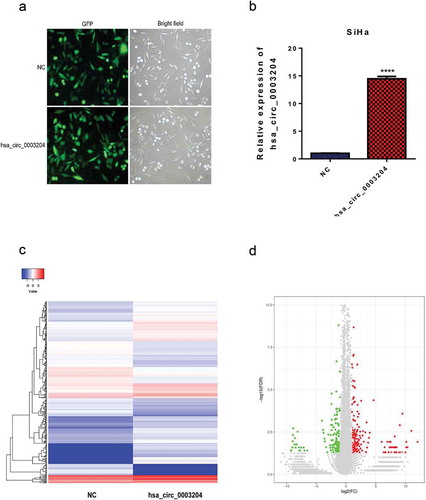
To obtain a comprehensive overview of the transcriptome profile of hsa_circ_0003204, a comparison of mRNA expression profile between hsa_circ_0003204 overexpression cells and negative control (NC) cells was identified by high-throughputsequencing. The heat map and hierarchical clustering showed systematic variations in the expression of mRNA between hsa_circ_0003204 overexpression cells and NC cells (). Differentially expressed mRNAs with statistically significant between hsa_circ_0003204 overexpression cells and NC cells were identified using volcano plot filtering (). The significantly differentially expressed genes were identified according to following criteria: FDR <0.05, |log2 Ratio(Tumor/Control)| ≥1.
GO analysis was applied to analyze the main functions of the differentially expressed genes. The results clearly showed important functions associated with the differentially expressed genes (). The upregulated GO functions (upGOs) and the downregulated GO functions (downGOs) were classified as cellular component, molecular function and biological process. The differentially expressed mRNA mainly enriched in cellular process, binding and apoptotic process, and several protein processing-related terms. KEGG pathway analysis was employed to identify significantly enriched biological pathways of differentially expressed genes. The top 15 enriched pathways were shown in . The mRNAs were mainly enriched in cancer-relative signaling pathway, TLR signaling pathway, TNF signaling pathway and MAPK signaling pathway.
Figure 6. GO and KEGG analysis of differentially expressed mRNA function and pathways in hsa_circ_0003204 overpression cell. (a) GO enrichment of differentially expressed mRNA. (b) KEGG pathway enrichment analysis of differentially expressed mRNA. (c) Western blot analysis verified MAPK pathway regulated by hsa_circ_0003204
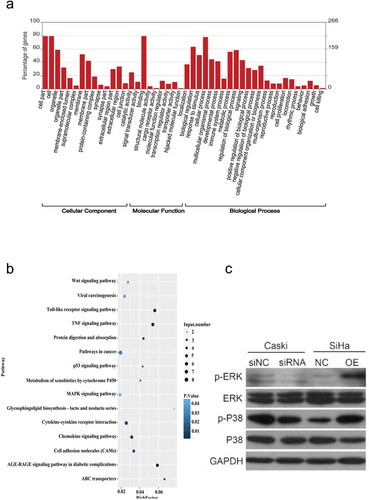
We also examined the regulatory effect of hsa_circ_0003204 on the MAPK signaling pathway. Key proteins of MAPK signaling pathway related to tumor proliferation were analyzed using Western blotting. The results indicated that p-ERK and p-P38 was markedly decreased in hsa_circ_0003204 knockdown cells, while increased in the hsa_circ_0003204 overexpression cells. No alteration of ERK and P38 was found in knockdown and overexpression cell ().
The effect of hsa_circ_0003204 on CC progression in vivo
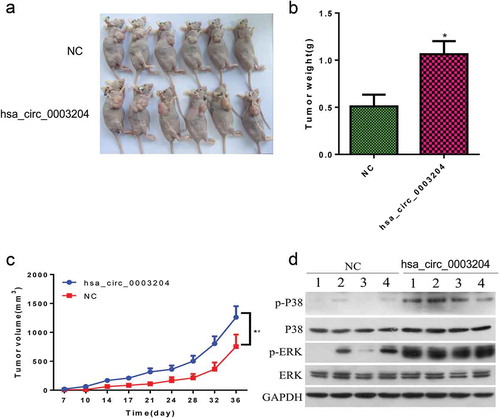
To further confirm the effect of hsa_circ_0003204 on CC progression, xenograft assay was conducted on nude recipient mice. Two million hsa_circ_0003204 overexpression SiHa cells and NC cells were subcutaneously injected into the flank of each nude recipient mouse, respectively. Results showed that hsa_circ_0003204 overexpression significantly increased tumor volumes ( and ) and tumor weight (). Western blot result in vivo were consistent with in vitro (). Taken together, these results indicated that hsa_circ_0003204 acts as a tumor inducer in CC via activation of the MAPK signaling pathway.
Figure 7. The effect of hsa_circ_0003204 on CC progression in vivo. (a) Subcutaneous xenografts excised from nude mice. (b) Bar graph shows tumor weight at the endpoint of the experiment. (c) The tumor growth curves. (e) Western blot analysis verified MAPK pathway regulated by hsa_circ_0003204. All data were expressed as mean ± standard deviation for three replicate determination. Student’s t-test was used for data analysis, p < .05 was considered statistically significant difference
Discussion
In the present study, we characterized the expression profile of circRNAs between CC tissues and matched adjacent normal tissues by high-throughput sequencing, and identified the most elevated circRNA hsa_circ_0003204 in CC tissues. We also investigated the effects of hsa_circ_0003204 on CC in vitro and in vivo, and explored the potential underlying molecular mechanisms. The knock-down expression of hsa_circ_0003204 reduced cell growth, migration and invasion, and increased the apoptosis of CC cells. The overexpression of hsa_circ_0003204 enhanced cell growth, migration and invasion, while it suppressed the apoptosis of CC cells. Results revealed that hsa_circ_0003204 could promote CC cell proliferation, migration and invasion by regulating MAPK pathway. We established hsa_circ_0003204 overexpression in SiHa cell line and identified the mRNA expression profile in hsa_circ_0003204 overexpression cell, compared with parental cells. Moreover, we found that overexpression of hsa_circ_0003204 significantly increased tumor volumes and tumor weight in vivo. Therefore, these data indicated hsa_circ_0003204 may serve as a potential therapeutic target for patients with CC.
Circular RNAs (circRNAs) are an novel class of endogenous noncoding RNAs characterized by a covalently closed circularc structure, lacking poly-adenylated tails, high stability, and are implicated in gene regulation.Citation18,Citation19 In recent years, accumulating studies focus on various aspects of circRNA biology have revealed the crucial role of these molecules in normal cellular differentiation, tissue homeostasis, and in disease development.Citation20 CircRNAs were characterized by tissue-specific expression patternCitation21, and stable in saliva, blood and exosomes.Citation22,Citation23 Due to highly conserved sequences and stability, circRNAs may serve as effective diagnostic biomarkers or promising therapeutic targets for treatment of cancer. Though many researches have been conducted on tumor-associated circRNAs, while relevance with CC has yet to be discovered.Citation20 Serval circRNAs participated in CC development have been found and identified, such as hsa_circ_0023404, hsa_circRNA_101996, hsa_circ_0000263, hsa_circ_0018289, circRNA8924, and circRNA-000284.Citation12–Citation15,17,Citation24 CircRNAs are considered as a competitive endogenous RNA (ceRNA) to regulate mRNA by acting as ‘sponge’ involved in cancer initiation and progression.Citation25,Citation26 In addition, circRNAs might alter gene expression by regulating alternative splicingCitation27 or transcription via interacting with RNA-binding proteins (RBPs).Citation28 To our knowledge, there has been no effort to define the effects of hsa_circ_0003204 on CC. This study identified the circRNA expression profile in human CC tissues and performed functional assays based on computational analysis. We found that the hsa_circ_0003204 specifically over-expressed in CC tissues and cell lines compared with that in adjacent normal tissues and HcerEpiC cells. Our data from knockdown and overexpression experiments confirmed that hsa_circ_0003204 plays an important role in the proliferation, migration and invasion process of CC cells. We also showed that overexpression of hsa_circ_0003204 induced CC cell tumorigenesis in vivo.
Based on in vitro data, we attempt to elucidate the molecular mechanisms of hsa_circ_0003204 that may play a critical role in the malignant transformation process. Therefore, we compared the expression profile between of hsa_circ_0003204 overexpression cells and negative control cells. Overexpression of hsa_circ_0003204 caused alteration of many genes associated with oncogenesis in cells. Most of the differentially expressed genes were mianly enriched in cancer-related signaling pathway, TLR signaling pathway, TNF signaling pathway and MAPK signaling pathway.
Mitogen-activated protein kinases (MAPKs) are important signaling molecules which regulate diverse cellular processes, such as proliferation, differentiation, migration, and apoptosis.Citation29–31 MAPK pathways comprise multiple molecules including the extracellular signal-regulated kinases 1 and 2 (ERK1/2), p38 (α, β, γ, and δ), c-Jun amino-terminal kinases 1–3 (JNK1 to −3), and ERK5 families.Citation32 Activated MAPK pathways can transduce stimuli into inside cells. Activation of each of MAPKs is part of a signaling cascade that depends on the phosphorylation of the kinases, such as JNK, ERK, or p38 within the cells.Citation33,Citation34 In our study, in order to identify the MAPK signaling pathway regulated by hsa_circ_0003204, we analyzed key proteins of MAPK signaling pathway (ERK and P38) using Western blotting. The results indicated that p-ERK and p-P38 was markedly decreased in hsa_circ_0003204 knockdown cells, the opposite was observed in the hsa_circ_0003204 overexpression cells. No alteration of ERK and P38 were found between hsa_circ_0003204 knockdown and hsa_circ_0003204 overexpression cells. These results revealed that hsa_circ_0003204 promotes CC cells proliferation, migration, and invasion by regulating MAPK pathway.
Conclusion
In summary, our data suggested that hsa_circ_0003204 acts as an oncogenic circRNA which could induce CC tumorigenesis through regulating MAPK signaling pathway. Our study supported that hsa_circ_0003204 may serve as a potential therapeutic target for patients with CC.
Abbreviations
| Cervical cancer | = | (CC) |
| Human papillomavirus | = | (HPV) |
| Non-coding RNAs | = | (ncRNAs) |
| Circular RNA | = | (circRNAs) |
| Federation International of Gynecology and Obstetrics | = | (FIGO) |
| Hematoxylin and Eosin | = | (HE) |
| Genomic DNA | = | (gDNA) |
| Quantitative real-time PCR | = | (qRT-PCR) |
| Dulbecco’s Modified Eagle’s Medium | = | (DMEM) |
| Cervical Epithelial Cell Growth Supplement | = | (CerEpiCGS) |
| Propidium iodide | = | (PI) |
| Gene Ontology | = | (GO) |
| Kyoto Encyclopedia of Genes and Genomes | = | (KEGG) |
| Negative control | = | (NC) |
| RNA-binding proteins | = | (RBPs) |
| Mitogen-activated protein kinases | = | (MAPKs) |
| c-Jun amino-terminal kinases 1–3 | = | (JNK1 to −3) |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ contributions
Yuan-li He conceived and designed the study, Xiao-bin Huang, Kai-jing Song, and Rui Liu performed bioinformatics analysis and writing of the manuscript. Xiao-bin Huang, Kai-jing Song, Guo-bin Chen, and Zhuo-fei Jiang completed the experiments. Yuan-li He were involved in drafting and revising the manuscript. All authors read and approved the final manuscript.
Competing interests
All authors declare no conflict of interest.
Acknowledgments
None.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Ezem B. Awareness and uptake of cervical cancer screening in Owerri, South-Eastern Nigeria. Ann Afr Med. 2007;6(3):94. doi:10.4103/1596-3519.55727.
- Haie-Meder C, Morice P, Castiglione M. Group EGW. Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21(suppl_5):v37–v40. doi:10.1093/annonc/mdq162.
- Arbyn M, Castellsagué X, de Sanjosé S, Bruni L, Saraiya M, Bray F, Ferlay J. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22(12):2675–2686. doi:10.1093/annonc/mdr015.
- LaVigne AW, Triedman SA, Randall TC, Trimble EL, Viswanathan AN. Cervical cancer in low and middle income countries: addressing barriers to radiotherapy delivery. Gynecologic Oncol Rep. 2017;22:16–20. doi:10.1016/j.gore.2017.08.004.
- Pimple S, Mishra G, Shastri S. Global strategies for cervical cancer prevention. Curr Opin Obstet Gynecol. 2016;28(1):4–10. doi:10.1097/GCO.0000000000000241.
- Jiang L-H, Sun D-W, Hou J-C, Ji Z-L. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7.
- Beermann J, Piccoli M-T, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325.
- Maiese K. Disease onset and aging in the world of circular RNAs. J Transl Sci. 2016;2(6):327. doi:10.15761/JTS.1000158.
- Chen L, Zhang S, Wu J, Cui J, Zhong L, Zeng L, Ge S. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551. doi:10.1038/onc.2017.89.
- Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu Y-M, Dhanasekaran SM, Engelke CG, Cao X, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881. e813. doi:10.1016/j.cell.2018.12.021.
- Zhang J, Zhao X, Zhang J, Zheng X, Circular LF. RNA hsa_circ_0023404 exerts an oncogenic role in cervical cancer through regulating miR-136/TFCP2/YAP pathway. Biochem Biophys Res Commun. 2018;501(2):428–433. doi:10.1016/j.bbrc.2018.05.006.
- Song T, Xu A, Zhang Z, Gao F, Zhao L, Chen X, Gao J, Kong X. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR‐8075. J Cell Physiol. 2019;234(8):14296–14305. doi:10.1002/jcp.28128.
- Liu J, Wang D, Long Z, Liu J. CircRNA8924 promotes cervical Cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p/519-5p family and modulating the expression of CBX8. Cell Physiol Biochem. 2018;48(1):173–184. doi:10.1159/000491716.
- Ma H-B, Yao Y-N, Yu -J-J, Chen -X-X, Li H-F. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Transl Res. 2018;10(2):592.
- Gao YL, Zhang MY, Xu B, Han LJ, Lan SF, Chen J, Dong YJ, Cao LL. 2017. Circular RNA expression profiles reveal that hsa_circ_0018289 is up-regulated in cervical cancer and promotes the tumorigenesis. Oncotarget. 8(49):86625–86633. DOI:10.18632/oncotarget.21257
- Cai H, Zhang P, Xu M, Yan L, Liu N, Wu X. RNA hsa_circ_0000263 participates in cervical cancer development by regulating target gene of miR‐150‐5p. J Cell Physiol. 2019;234(7):11391–11400. doi:10.1002/jcp.27796.
- Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi:10.1186/s13059-014-0409-z.
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci. 1976;73(11):3852–3856. doi:10.1073/pnas.73.11.3852.
- Kristensen L, Hansen T, Venø M, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555. doi:10.1038/onc.2017.361.
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber A, Gregory P, Goodall G, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi:10.1016/j.cell.2015.02.014.
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981. doi:10.1038/cr.2015.82.
- Bahn JH, Zhang Q, Li F, Chan T-M, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61(1):221–230. doi:10.1373/clinchem.2014.230433.
- Gao Y-L, Zhang M-Y, Xu B, Han L-J, Lan S-F, Chen J, Dong Y-J, Cao -L-L. Circular RNA expression profiles reveal that hsa_circ_0018289 is up-regulated in cervical cancer and promotes the tumorigenesis. Oncotarget. 2017;8(49):86625. doi:10.18632/oncotarget.21257.
- Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster K, Weiss D, Perez-Mancera P, et al. In Vivo Identification of Tumor- Suppressive PTEN ceRNAs in an Oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147(2):382–395. doi:10.1016/j.cell.2011.09.032.
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384. doi:10.1038/nature11993.
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi:10.1016/j.molcel.2014.08.019.
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. 2015. The RNA binding protein quaking regulates formation of circRNAs. Cell. 160(6):1125–1134. DOI:10.1016/j.cell.2015.02.014
- Osaki L, Gama P. MAPKs and signal transduction in the control of gastrointestinal epithelial cell proliferation and differentiation. Int J Mol Sci. 2013;14(5):10143–10161.
- Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75(1):50–83.
- Shaul YD, Seger R. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim Biophys Acta Mol Cell Res. 2007;1773(8):1213–1226.
- Plotnikov A, Zehorai E, Procaccia S, Seger R. The MAPK cascades: signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim Biophys Acta Mol Cell Res. 2011;1813(9):1619–1633. doi:10.1016/j.bbamcr.2010.12.012.
- Manna PR, Stocco DM. The role of specific mitogen-activated protein kinase signaling cascades in the regulation of steroidogenesis. J Signal Transduct. 2011;2011:1–13. doi:10.1155/2011/821615.
- Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68(2):320–344. doi:10.1128/MMBR.68.2.320-344.2004.