ABSTRACT
Endometrial carcinoma (EC) remains one of the most prevalent forms of cancer to impact the female reproductive system, yet the mechanisms governing its development and progression are incompletely understood. We, therefore, sought to assess the relevance of SOX8 to EC progression and patient prognosis.
Array comparative genomic hybridization (aCGH) was performed using samples from 50 patients with EC. Samples were separated based upon whether patients were positive for lymph node metastasis (LN+ and LN−, respectively). Based on our initial results, the SOX8 gene was selected for further analysis. Immunohistochemical staining of 630 endometrial tissue samples was conducted to understand how SOX8 expression relates to specific EC clinicopathological characteristics. In addition, we explored the impact of SOX8 expression on the growth, invasion, and migration of EC cells through knockdown and overexpression experiments.
In our initial aCGH analysis, SOX family proteins and the Wnt and Notch signaling pathways were significantly associated with EC LN metastasis. SOX8 expression was markedly increased in EC tumor samples relative to normal endometrial tissue (P= .003), and higher SOX8 expression was linked to a high tumor histological grade (P= .032), LN metastasis (P= .027), and shorter patient overall survival (P= .031). When SOX8 was knocked down, this further impaired the proliferative, invasive, and migratory activity of EC cells, whereas overexpressing this gene had the opposite effect.
SOX8 may function in an oncogenic manner to drive EC development and progression, and higher SOX8 expression is associated with a poor EC patient prognosis.
Introduction
Endometrial carcinoma (EC) remains one of the most prevalent forms of gynecological cancer in the world, with 65,620 new diagnoses and 12,590 EC-related deaths predicted to occur in the United States alone in 2020,Citation1,Citation2 and with rising rates of diagnosis in China.Citation3 In patients with early-stage EC, the 5-y survival rate is as high as 90%.Citation4 However, the course of EC is highly variable, and a subset of early-stage patients are at risk for tumor recurrence or metastasis, both of which are associated with an unfavorable prognosis.Citation5 The factors that govern whether EC patients ultimately suffer from a progressive disease, however, are not well understood. The mechanistic basis for EC pathogenesis, therefore, needs to be fully explored in an effort to elucidate how this disease progresses to a metastatic state in a subset of patients as a means of designing appropriate treatments for at-risk individuals.
Sex-determining region Y box (SOX) genes are conserved genes that encode transcription factors which regulate key developmental processes such as embryogenesis, organ development, sex differentiation, and neurogenesis.Citation6,Citation7 These SOX genes contain conserved high-mobility group (HMG) box regions that have been used to classify SOX family proteins into defined subgroups.Citation8 Several studies to date have highlighted the pivotal roles played by certain SOX proteins in the context of tumor cell proliferation, migration, and invasion.Citation9,Citation10 Consistent with these roles, some SOX proteins have been shown to be key markers of cancer patient prognosis in addition to being potentially viable therapeutic targets.Citation9 There is some evidence suggesting that SOX genes can govern EC pathogenesis,Citation11 with SOX3, SOX4, and SOX11 overexpression being associated with the enhanced EC cell proliferative migratory, and invasive activity that was suppressed upon the knockdown of these genes.Citation12–14 In contrast, SOX7, SOX15, and SOX17 were found to have the opposite effect, impairing EC cell proliferation.Citation15–17
SOX8 is a member of the E subgroup of SOX proteins and is expressed at the highest levels in embryonic and adult brain tissues.Citation18 Elevated SOX8 expression has been associated with greater tumor aggression and poorer survival in patients with non-small lung cancer, triple-negative breast cancer, chemo-resistant tongue squamous cell carcinoma, and colorectal cancer.Citation19–23 Whether SOX8 similarly controls EC tumor aggression and progression, however, remains to be determined. Herein, we, therefore, used an immunohistochemistry (IHC) staining approach to analyze SOX8 expression in normal endometrium (NE), hyperplastic endometrium (HE), atypical hyperplastic endometrium (AHE), and EC tissues. We further examined the relationship between SOX8 expression and EC patient clinicopathological features and utilized in vitro approaches to directly examine the role of SOX8 expression in EC cells.
Results
Identification of CNVs in specific genes and pathways associated with EC lymph node metastasis
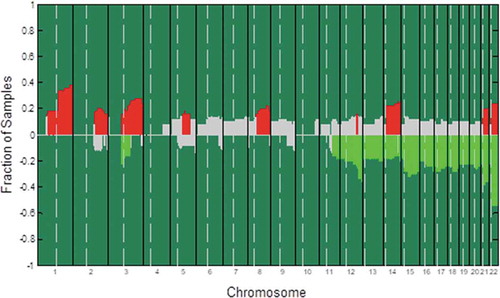
We began by utilizing gDNA extracted for 50 EC patient tumor samples in order to conduct a whole-genome aCGH analysis. The most frequently altered genomic regions in these samples were identified in an effort to identify specific cytoband variations that were significantly associated with LN metastasis in these EC patients ( and Supplementary ). We were able to determine that copy-number variations (CNVs) in certain genes in the SOX, Wnt, and Notch families were significantly more amplified in LN+ samples relative to LN− samples. Specifically, the levels of Wnt1, Wnt10a, Wnt10b, SOX4, SOX8, NCAM1, NOTCH2, NOTCH3, and NRXN2 CNVs were significantly higher in LN+ patient samples relative to those from LN− patientsLN patients. This suggests that these genes may be key regulators of EC metastasis. We have previously highlighted important roles for Wnt10a and Wnt10b in EC, and in the present study, we, therefore, elected to examine the role of SOX8 in this cancer type.
Figure 1. An aCGH-based analysis of DNA copy-number alteration frequencies in 50 endometrial carcinoma samples. Gains or losses for each measured sequence are shown on the y-axis, with sequences being aligned along the x-axis in chromosomal order. The significance threshold is represented by a dashed line. Significant DNA copy-number gain frequencies are marked by red lines, while significant DNA copy-number losses are marked by green lines. Non-significant changes are marked in gray. Vertical bars demarcate different chromosomes, with dashed vertical bars marking the separation between short and long arms of individual chromosomes
Elevated SOX8 expression is linked with EC progression and development
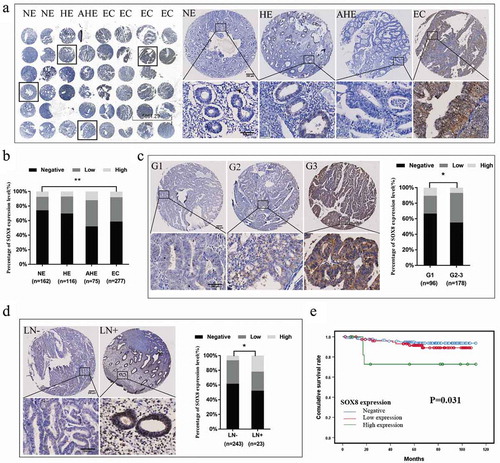
We next measured SOX8 expression levels in TMAs prepared using NE, HE, AHE, and EC tissue samples via IHC, the representative image is shown in . We found that EC tumor samples exhibited significantly higher SOX8 expression relative to NE tissues (χ2 = 11.685, P= .003, ). High SOX8 expression was almost exclusively detected in EC and AHE samples (7.9% and 12%, respectively), while negative SOX8 expression was most frequently observed in NE and HE samples (74.1% and 69.8%, respectively) (, ). In order to further explore how SOX8 expression levels relate to EC patient clinicopathological parameters, we next conducted a correlation analysis of the 277 EC patients included within this TMA sample cohort. Elevated SOX8 expression was found to be significantly associated with higher tumor histological grade (χ2 = 6.907, P= .032) and with LN metastasis (Fisher, P= .027) ( and d, ). There were also trends toward higher SOX8 expression in samples exhibiting deep myometrial invasion, advanced FIGO stage, and 2 type samples (P> .05).
Table 1. Gene alterations in endometrial carcinoma
Table 2. SOX8 expression in NE, HE, AHE, and EC tissues
Table 3. Correlation between SOX8 expression and clinicopathological parameters in EC cases
Figure 2. SOX8 expression levels in human EC tissue samples correlate with aggressive clinicopathological findings. (a) An IHC approach was used to measure SOX8 levels in TMAs. The resultant data revealed that SOX8 levels were higher in EC and AHE samples relative to HE and NE samples. (b) The expression of SOX8 in EC samples was significantly higher than that in NE samples. (c) SOX8 overexpression was linked to high EC histological grade. Left: Representative images of EC tissue SOX8 staining (G1 – G3). Right: Elevated SOX8 expression was confirmed to be significantly associated with high EC histological grade in these patient samples. (d) SOX8 overexpression was associated with EC lymph node metastasis. Left: Representative SOX8 staining in LN− and LN+ EC samples. Right: Elevated SOX8 expression was confirmed to be significantly associated with EC lymph node metastasis in these patient samples. (e) The relationship between SOX8 expression and post-operative EC patient OS was assessed using a Kaplan-Meier curve. (**p< .01, *p< .05)
3. Elevated SOX8 expression correlated with poor prognosis
We next examined the relationship between SOX8 expression and the overall survival (OS) of EC patients from the date of surgery to death or last follow-up. A Kaplan–Meier curve analysis revealed that high SOX8 expression was linked to shorter OS relative to low or no SOX8 expression in these EC patients (χ2 = 6.930, P= .031) ().
4. SOX8 promotes the proliferative, migratory, and invasive activity of EC cells
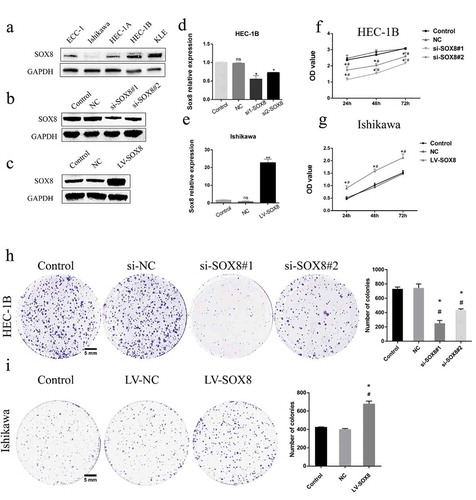
In order to confirm the functional role of SOX8 in the context of EC, we next assessed the expression of this protein in five common EC lines via Western blotting. This analysis revealed that SOX8 levels were higher in HEC-1B cells and lower in Ishikawa cells relative to other tested cell lines (), leading us to select these cells for downstream experimentation. We then generated versions of these cells in which SOX8 had been knocked down (HEC-1B-si-SOX8#1, HEC-1B-si-SOX8#2) or overexpressed (Ishikawa-LV-SOX8), with these changes in SOX8 expression relative to cells transfected with negative control (NC) constructs being confirmed by Western blotting () and RT-PCR (). Using a CCK8 assay, we found that HEC-1B cells in which SOX8 had been knocked down were significantly less proliferative relative to control and NC cells (P< .05, ), whereas SOX8 overexpression had the opposite effect on Ishikawa cell proliferation (P< .01, ). This was in line with the results of a colony-formation assay, wherein SOX8 knockdown decreased colony numbers relative to control and NC groups (251.3 ± 23.25, 436.3 ± 9.615 vs 732 ± 14.57, vs 744.3 ± 32.2, P< .001) while SOX8 overexpression increased colony numbers relative to control and NC groups (681 ± 20 vs 427 ± 1, vs 403 ± 5, P< .01) ().
Figure 3. SOX8 controls EC cell proliferation and growth. (a) Endogenous SOX8 levels in ECC-1, Ishikawa, HEC1A, HEC1B, and KLE cells were assessed by Western blotting. SOX8 knockdown in HEC-1B cells (b,d) and SOX8 overexpression in Ishikawa cells (c,e) was confirmed via Western blotting and qPCR. A CCK8 assay revealed that SOX8 knockdown significantly impaired cell proliferation (f), whereas SOX8 overexpression had the opposite effect (g). A colony-formation assay indicated that SOX8 knockdown impaired EC cell growth (h), while SOX8 overexpression significantly enhanced proliferation (i). Data are means ± SD from triplicate experiments. #p < .05 vs. control; *p < .05 vs. NC control
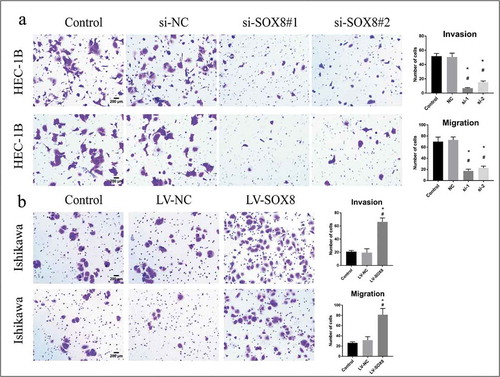
In a Transwell assay system, we further found that SOX8 knockdown in HEC-1B cells impaired their migration relative to control and NC treatment (17.4 ± 1.364, 22.8 ± 1.393 vs 70 ± 3.619, vs 73 ± 2.387, P< .0001), and findings in an invasion assay were comparable (7 ± 0.3162, 15.2 ± 0.860 vs 51.8 ± 1.685, vs 50.6 ± 2.4, P< .0001) (). In contrast, overexpressing SOX8 enhanced Ishikawa cell migration (81.4 ± 5.437 vs 26.2 ± 1.02, vs 31.8 ± 2.99, P< .0001), and invasion (66.2 ± 2.709 vs 21 ± 0.8367, vs 19.4 ± 2.694, P< .0001) relative to control and NC groups ().
Discussion
EC is currently among a limited number of cancers for which mortality rates are rising.Citation24 Most patients with EC are diagnosed while the disease is still in its early stages due to obvious symptoms such as postmenopausal vaginal bleeding. While these women typically have favorable disease outcomes, some women are not diagnosed until the disease is in an advanced state, while others present with low-grade, early-stage, well-differentiated endometrioid tumors that recur unexpectedly and that have a poor prognosis. EC LN metastasis is correlated with poorer patient outcomes. However, there is a clear need for the identification of LN metastasis-related biomarkers that can be used to evaluate EC patients in an effort to improve treatment efficacy. In the present study, we compared genomic alterations in LN+ and LN− EC tumor samples via an aCGH approach, revealing SOX8 to be amplified with higher frequency in LN+ samples relative to LN− samples (CNV levels of 33.33% and 0%, respectively), suggesting a role for SOX8 in EC LN metastasis. Recent work has highlighted a role for SOX family proteins in tumor development and progression.Citation9 This, coupled with our aCGH analysis results, suggested that SOX8 may play a yet-to-be-characterized role as a risk factor for EC progression. We, therefore, elected to study the prognostic and functional relevance of SOX8 in EC as a means of understanding how this protein impacts EC development.
This is the first study we are aware of to have demonstrated a difference in SOX8 levels between LN+ and LN− EC patient samples. We further confirmed that SOX8 expression was significantly higher in EC tumor tissue samples relative to NE samples, and we found that elevated SOX8 expression correlated with LN metastasis, high histologic grade, and poor prognosis. When we extended this work in vitro, we confirmed that SOX8 is able to directly drive the migratory and proliferative activity of EC cell lines. Together, these results thus suggest that SOX8 overexpression may be indicative of a higher risk of EC progression or development.
SOX8 dysregulation has previously been documented in multiple human tumor types. For example, SOX8 upregulation has been observed in hepatocellular carcinoma and colorectal cancer (CRC) tumor samples relative to levels in paracancerous tissues.Citation19,Citation21 Chemoresistant tongue squamous cell carcinoma patients were found to express significantly higher levels of SOX8, suggesting that this gene may also influence therapeutic resistance.Citation23 In this analysis, we found that SOX8 expression was significantly upregulated in EC samples relative to NE controls, and we found that this increased expression was correlated with LN metastasis, in line with our aCGH results. This is also consistent with work demonstrating that SOX8 upregulation correlates with LN metastasis in non-small cell lung cancer (NSCLC), chemo-resistant tongue squamous cell carcinoma, and CRC, suggesting that SOX8 is a metastasis-related oncogene.Citation20,Citation21,Citation23 Our Kaplan-Meier survival analyses further revealed that high SOX8 expression was associated with poor EC patient prognosis, consistent with findings in studies of NSCLC, tongue squamous cell carcinoma, CRC, and triple-negative breast cancer.Citation22 SOX8 may thus be a key regulator of EC progression and a valuable prognostic biomarker in patients with this disease.
Using in vitro knockdown and overexpression techniques, we further sought to confirm the role of SOX8 in regulating EC progression and proliferation. The results of these experiments indicated that knocking down this transcription factor markedly impaired EC cell proliferative, migratory, and invasive activity, whereas its overexpression had the opposite effect. This is in line with findings in several other cancer types. For example, SOX8 knockdown inhibited chemoresistance, tumorsphere formation, and EMT via Wnt/β-catenin signaling in cisplatin-resistant tongue squamous cell carcinoma cells, whereas its overexpression in triple-negative breast cancer cells enhanced their migratory and proliferative activity. SOX8 overexpression in hepatocellular carcinoma cell lines also enhanced their proliferation via a mechanism associated with Wnt/β-catenin signaling. Our current study did not explore the mechanistic basis whereby SOX8 drives the progression and development of EC. However, our aCGH analysis revealed that in addition to SOX8, amplifications were also common in several Wnt and Notch signaling pathway proteins in LN+ EC patient samples. Wnt and Notch signaling are well known to be linked to EC activation and development,Citation25–29 and SOX genes have been shown to interact with the Wnt/β-catenin signaling pathway in many cancers.Citation10,Citation30 We also found that SOX8 is predicted to interact with the Wnt/β-catenin signaling pathway via CTNNB1 (β-catenin) in the STRING database (https://string-db.org/). Future studies exploring whether SOX8 influences EC cell proliferation and metastasis via the Wnt/β-catenin signaling pathway are therefore warranted.
In conclusion, in the present study, we determined that EC samples exhibit significantly elevated SOX8 expression relative to NE tissues. We additionally provided novel evidence indicating that high expression of SOX8 is correlated with LN metastasis, a high histologic grade, and poor patient prognosis. Using cell lines, we further confirmed that SOX8 can directly promote EC cell proliferative, migratory, and invasive activity, confirming the oncogenic relevance of this protein. Together, these findings suggest that SOX8 may be a viable prognostic biomarker and therapeutic target in EC, although further analyses will be necessary in order to understand the molecular mechanisms whereby SOX8 influences the development and progression of this form of cancer.
Materials and methods
Patient sample collection
For this study, we analyzed previously collected samples from 50 EC patients that had been archived by Tianjin Medical University General Hospital (TMUGH, Tianjin, China) between 2000 and 2009. These samples were utilized for aCGH and were separated into two groups based upon whether they were positive for lymph node metastasis (LN+, 18 cases) or negative for lymph node metastasis (LN−, 32 cases). These tissues had been snap-frozen within 30 minutes of isolation and stored at −80°C. In addition, we utilized a tissue microarray that had been constructed from 630 endometrial tissue samples collected at TMUGH from 2008 to 2016. This latter cohort included samples from 277 EC patients, 75 patients with AHE, 116 patients with HE, and 162 patients with NE tissue collected during hysterectomy. Carcinosarcoma was not included in EC. Two pathologists had independently graded the histological typing of each sample based on the WHO classification criteria, and EC tumor staging was conducted using the 2014 FIGO staging system. Patients in this study that did not have EC had been admitted to the hospital for the treatment of uterine prolapse, cystocele, or urethrocele. No patients had undergone radio- or chemotherapy prior to tissue collection. The Ethics Committee of TMUGH approved these sample collection procedures.
Array comparative genomic hybridization (aCGH)
A DNA/RNA Prep Kit (Qiagen, CA, USA) was used to isolate gDNA from the 50 EC patient samples, after which labeled DNA was hybridized to a human whole-genome CGH microarray (4 × 44 k; Agilent Technologies, CA, USA). This array covered 43,000 coding and non-coding genomic sequences, with an average spatial resolution of 35kbp per oligonucleotide probe. The array contained a minimum of one target sequence per well-characterized gene and a minimum of two probes per cancer-related gene. The University of California Santa Cruz hg17 human genome (National Center for Biotechnology Information, NCBI Build 35) was used to guide probe design.
Immunohistochemistry
IHC staining of TMA samples was conducted as in prior studies,Citation31 using anti-SOX8 from Proteintech (20627-1-AP, China) and an appropriate secondary from ZSGH-Bio (PV9001, China). Hematoxylin and eosin (H&E) were used to counterstain slides based on standard protocols. Both staining frequency and intensity were then used to assess SOX8 staining levels, the scoring system was derived from the previous report.Citation21 Scores for the percentage of positively stained cells were as follows: 0 = 0–5%; 1 = 6%-25%; 2 = 26%-50%; 3 = 51–75%; 4, 76%-100%. Intensity scoring was conducted as follows: 0 = no staining; 1 = weak; 2 = moderate; 3 = strong. These two scores were then multiplied together to yield a final score, which was graded as follows: 0 [0] = negative expression; 1 [1–3] = low expression; 2 [4–12] = high expression. Two pathologists independently scored each sample.
Cell culture
The Ishikawa and HEC-1B cell lines were obtained from the China Infrastructure of Cell Line Resource. The ECC-1, HEC-1A, and KLE EC cell lines were provided by the TMUGH obstetrics and gynecology lab. All cells were cultured using standard protocols, as detailed previously.Citation32 No mycoplasma contamination in all cells during cell culture.
RT-PCR
Total RNA was isolated from cells according to TRIzol protocol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction. Total RNA was converted to cDNA using a Reverse Transcriptase Kit. Then, real-time PCR analyses were carried out in triplicate for each sample. The PCR primers were listed as follows: SOX8: forward: CCGTGTCGCAGGTGCTCA, reverse: CGCCGTGGCTGGTACTTGTAG; GAPDH: forward: TGACTTCAACAGCGACACCCA, reverse: CACCCTGTTGCTGTAGCCAAA.
Western blotting
Standard western blotting procedures were used in this study.Citation33 Using the following primary antibodies: anti-SOX8 (ab125858, 1:1000), anti-GAPDH (ZSGH-Bio, 1:5000). In addition, an appropriate secondary antibody was used (1:8000; ZSGH-Bio), and Immobilon Western HRP (Millipore, USA) was used for protein visualization.
Modulation of SOX8 expression
The Ubi-SOX8-3 FLAG-CBh-gcGFP-IRES-puromycin lentiviral vectors were constructed by GeneChem (Shanghai, China). These viruses were used to transduce appropriate target cells, and stably transduced cells were selected using puromycin. Western blotting was used to confirm SOX8 overexpression. SOX8 knockdown was conducted by transducing cells with siRNA constructs specific for SOX8 from Integrated Biotech Solutions (Shanghai, China). Sequences for these constructs were as follows: 5ʹ-AGAACAUCGACUUCAGCAACG-3ʹ and 5ʹ-UCCACGAGUUCGACCAGUATT-3ʹ. In addition, a negative control siRNA with the following sequence was used: 5‘-UUCUCCGAACGUGUCACGUdTdT-3ʹ. Manufacturer’s protocols were used to conduct SOX8 knockdown and overexpression experiments.
Cell proliferation assay
Proliferation was measured with a Cell Counting Kit-8 (CCK8) (Dojindo, Japan) assay. Briefly, 6 × 103 Ishikawa cells or 5 × 103 HEC-1B cells were added into each well of a 96-well plate in a 100 μL volume. At appropriate time points (24, 48, or 72 h), 10 μL of CCK-8 solution was added per well and absorbance at 450 nm was assessed after a 4 h incubation at 37°C using a microplate reader (EnSpire, China).
Colony-formation assay
A total of 1000 Ishikawa or HEC-1B cells were added per well of a 6-well plate, and colony formation was assessed via microscopy each day. Following a 10-d incubation, 4% paraformaldehyde was used to fix colonies for 20 minutes, after which they were stained with 2.5% crystal violet and counted.
Cell migration/invasion assays
A Transwell system (8.0 μm pore size; Corning, USA) with or without a Matrigel coating (Corning) was used based on provided directions, as in prior studies.Citation34 Briefly, appropriately transfected cells in serum-free media were added to the upper chamber of this system (8 × 104/100 μl for Ishikawa; 10 × 104/100 μl for HEC-1B), while a 500 μl volume of media containing 20% FBS was added to the lower well. After 24 or 48 h, cells on the upper surface were gently removed with a swab, with cells that had undergone migration or invasion were fixed with 4% paraformaldehyde and stained using 2.5% crystal violet. A ZEISS HB100 (×100) was then used to count cells in five random fields of view, with data being compiled from three experimental replicates.
Statistical analysis
SPSS v25.0 (IBM, USA) was used for statistical testing, while GraphPad Prism 7.0 (GraphPad, USA) was used to prepare figures. The relationships between SOX8 expression levels and clinicopathological variables were assessed via χ2 or Fisher’s exact tests, as appropriate, with the former being used when values had an expected frequency of ≤5. Survival analyses were conducted using the Kaplan–Meier method, and the relationship between SOX8 expression and postoperative survival in EC patients was also assessed via univariate analyses. Data generated in vitro were compared using ANOVAs and Student’s t-tests as appropriate. P < .05 was the significance threshold.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Authorship
Yingmei Wang and Fengxia Xue: study design, supervision of the study, review, and editing the manuscript.
Wenyan Tian: collection of tissue specimens and scoring IHC results.
Zhanghuan Li: conducting the cell biological experiment, statistical integration, writing, review, and editing the manuscript.
Lu Bai: conductingthe aCGH experiment.
Lingli Chen: conducting the qPCR experiment.
Ye Yan: database maintenance and statistical analysis.
Huihui Li: conducting the IHC experiment and collection of patient information
Yanyan Han: scoring IHC results.
Fei Teng and Chao Gao: collection of tissue specimens and follow-up of the patient.
Supplemental Material
Download MS Word (15.8 KB)Acknowledgments
The authors thank Tianjin key laboratory of female reproductive health and eugenics for the facilities.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. 2020. Cancer statistics, 2020. CA Cancer J Clin. doi:10.3322/caac.21590
- Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet (London, England). 2016;387:1094–1108. doi:10.1016/S0140-6736(15)00130-0.
- Chen W, Zheng R, PD B, Zhang S, Zeng H, Bray F, Jemal A, XQ Y, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132.
- McDonald ME, Bender DP. Endometrial cancer: obesity, genetics, and targeted agents. Obstet Gynecol Clin North Am. 2019;46(1):89–105. doi:10.1016/j.ogc.2018.09.006.
- Njølstad TS, Trovik J, Hveem TS, Kjæreng ML, Kildal W, Pradhan M, Marcickiewicz J, Tingulstad S, Staff AC, Haugland HK, et al. DNA ploidy in curettage specimens identifies high-risk patients and lymph node metastasis in endometrial cancer. Br J Cancer. 2015;112(10):1656–1664. doi:10.1038/bjc.2015.123.
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf A-M, Lovell-Badge R, Goodfellow PN, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346(6281):240–244. doi:10.1038/346240a0.
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346(6281):245–250. doi:10.1038/346245a0.
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227(2):239–255. doi:10.1006/dbio.2000.9883.
- Grimm D, Bauer J, Wise P, Kruger M, Simonsen U, Wehland M, Infanger M, Corydon TJ. The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol. 2019. doi:10.1016/j.semcancer.2019.03.004.
- Xu YR, Yang WX. SOX-mediated molecular crosstalk during the progression of tumorigenesis. Semin Cell Dev Biol. 2017;63:23–34. doi:10.1016/j.semcdb.2016.07.028.
- Hu J, Li K, Li Z, Gao C, Guo F, Wang Y, Xue F. Sex-determining region Y box-containing genes: regulators and biomarkers in gynecological cancers. Cancer Biol Med. 2019;16:462–474.
- Gong B, Yue Y, Wang R, Zhang Y, Jin Q, Zhou X. Overexpression of microRNA-194 suppresses the epithelial-mesenchymal transition in targeting stem cell transcription factor Sox3 in endometrial carcinoma stem cells. Tumour Biol. 2017;39:1010428317706217. doi:10.1177/1010428317706217.
- Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, Miller DS, Huang TH. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–9046. doi:10.1158/0008-5472.CAN-09-1499.
- Chang L, Yuan Z, Shi H, Bian Y, Guo R. miR-145 targets the SOX11 3ʹUTR to suppress endometrial cancer growth. Am J Cancer Res. 2017;7:2305–2317.
- Chan DW, Mak CS, Leung TH, Chan KK, Ngan HY. Down-regulation of Sox7 is associated with aberrant activation of Wnt/b-catenin signaling in endometrial cancer. Oncotarget. 2012;3:1546–1556. doi:10.18632/oncotarget.667.
- Rui X, Xu Y, Jiang X, Guo C, Jiang J. 2017. SOX15 regulates proliferation and migration of endometrial cancer cells. Biosci Rep. 37. DOI:10.1042/BSR20171045
- Zhang Y, Bao W, Wang K, Lu W, Wang H, Tong H, Wan X. SOX17 is a tumor suppressor in endometrial cancer. Oncotarget. 2016;7(46):76036–76046. doi:10.18632/oncotarget.12582.
- Pfeifer D, Poulat F, Holinski-Feder E, Kooy F, Scherer G. The SOX8 gene is located within 700 kb of the tip of chromosome 16p and is deleted in a patient with ATR-16 syndrome. Genomics. 2000;63(1):108–116. doi:10.1006/geno.1999.6060.
- Zhang S, Zhu C, Zhu L, Liu H, Liu S, Zhao N, Wu J, Huang X, Zhang Y, Jin J, et al. Oncogenicity of the transcription factor SOX8 in hepatocellular carcinoma. Med Oncol (Northwood, London, England). 2014;31(4):918. doi:10.1007/s12032-014-0918-3.
- Xie C, Han Y, Liu Y, Han L, Liu J. miRNA-124 down-regulates SOX8 expression and suppresses cell proliferation in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:7518–7526.
- Wang Y, Yang W, Liu T, Bai G, Liu M, Wang W. Over-expression of SOX8 predicts poor prognosis in colorectal cancer: A retrospective study. Medicine. 2019;98:e16237. doi:10.1097/MD.0000000000016237.
- Tang H, Chen B, Liu P, Xie X, He R, Zhang L, Huang X, Xiao X, Xie X. SOX8 acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Carcinogenesis. 2019. doi:10.1093/carcin/bgz034.
- Xie SL, Fan S, Zhang SY, Chen WX, Li QX, Pan GK, Zhang H-Q, Wang -W-W, Weng B, Zhang Z, et al. SOX8 regulates cancer stem-like properties and cisplatin-induced EMT in tongue squamous cell carcinoma by acting on the Wnt/β-catenin pathway. Int J Cancer. 2018;142:1252–1265. doi:10.1002/ijc.31134.
- Dou Y, Kawaler EA, Cui Zhou D, Gritsenko MA, Huang C, Blumenberg L, Karpova A, Petyuk VA, Savage SR, Satpathy S, et al. Proteogenomic characterization of endometrial carcinoma. Cell. 2020;180(4):729–48.e26. doi:10.1016/j.cell.2020.01.026.
- Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi:10.1038/nature12113 7447 497 4164-73
- Chen H, Wang Y, Xue F. Expression and the clinical significance of Wnt10a and Wnt10b in endometrial cancer are associated with the Wnt/beta-catenin pathway. Oncol Rep. 2013;29:507–514. doi:10.3892/or.2012.2126.
- Gao Y, Liu T, Huang Y. MicroRNA-134 suppresses endometrial cancer stem cells by targeting POGLUT1 and Notch pathway proteins. FEBS Lett. 2015;589:207–214. doi:10.1016/j.febslet.2014.12.002.
- Jonusiene V, Sasnauskiene A, Lachej N, Kanopiene D, Dabkeviciene D, Sasnauskiene S, Kazbariene B, Didziapetriene J. Down-regulated expression of Notch signaling molecules in human endometrial cancer. Med Oncol. 2013;30(1):438. doi:10.1007/s12032-012-0438-y.
- Mitsuhashi Y, Horiuchi A, Miyamoto T, Kashima H, Suzuki A, Shiozawa T. Prognostic significance of Notch signalling molecules and their involvement in the invasiveness of endometrial carcinoma cells. Histopathology. 2012;60(5):826–837. doi:10.1111/j.1365-2559.2011.04158.x.
- Kormish JD, Sinner D, Zorn AM. Interactions between SOX factors and Wnt/beta-catenin signaling in development and disease. Dev Dyn. 2010;239:56–68.
- Tian W, Zhu Y, Wang Y, Teng F, Zhang H, Liu G, Ma X, Sun D, Rohan T, Xue F, et al. Visfatin, a potential biomarker and prognostic factor for endometrial cancer. Gynecol Oncol. 2013;129(3):505–512. doi:10.1016/j.ygyno.2013.02.022.
- Wang Y, Hu L, Ji P, Teng F, Tian W, Liu Y, Cogdell D, Liu J, Sood AK, Broaddus R, et al. MIIP remodels Rac1-mediated cytoskeleton structure in suppression of endometrial cancer metastasis. J Hematol Oncol. 2016;9(1):112. doi:10.1186/s13045-016-0342-6.
- Wang Y, Gao C, Zhang Y, Gao J, Teng F, Tian W, Yang W, Yan Y, Xue F. Visfatin stimulates endometrial cancer cell proliferation via activation of PI3K/Akt and MAPK/ERK1/2 signalling pathways. Gynecol Oncol. 2016;143(1):168–178. doi:10.1016/j.ygyno.2016.07.109.
- Wang H, Wang H, Shen W, Huang H, Hu L, Ramdas L, Zhou YH, Liao WS, Fuller GN, Zhang W. Insulin-like growth factor binding protein 2 enhances glioblastoma invasion by activating invasion-enhancing genes. Cancer Res. 2003;63:4315–4321.