ABSTRACT
Lung squamous cell carcinoma (LUSC) is a prevalent subtype of nonsmall cell lung cancer (NSCLC). Dysregulated long noncoding RNAs (lncRNAs) are increasingly identified as pivotal modulators in cancer progression. NCK1 divergent transcript (NCK1-AS1) is a lncRNA that has been proven to be oncogenic in different types of human cancers. However, whether it exerts similar functions in LUSC remains to be elusive. The present study focused on investigating the influence of NCK1-AS1 on the cellular process in LUSC and exploring its underlying mechanism. Through online bioinformatics analysis, we obtained a high NCK1-AS1 level in LUSC tissues. Meanwhile, we confirmed that NCK1-AS1 was upregulated in LUSC cells. Gain- or loss-of-function assays suggested that NCK1-AS1 prompted cell proliferation and migration, whilst impeded cell apoptosis in LUSC. Mechanistically, we revealed that NCK1-AS1 induced the upregulation of its nearby gene NCK adaptor protein 1 (NCK1) at the transcriptional level by interacting with the transcription factor MYC proto-oncogene (MYC). Rescue assays indicated that NCK1 participated in the regulation of NCK1-AS1 on LUSC progression. In conclusion, we firstly demonstrated the oncogenic role of NCK1-AS1 in LUSC and illustrated its downstream molecular mechanism.
Introduction
Lung cancer is recognized to be the most prevalent malignancy and the main contributor of global cancer-related mortality.Citation1 The commonest histological subtype of lung cancer is nonsmall cell lung cancer (NSCLC), taking up around 84% of all lung cancer cases.Citation2 Despite that therapeutic regimes have been developed greatly in treating LUSC, patients are still burdened with an unsatisfactory 5-year overall survival rate.Citation3 Therefore, new effective therapeutic targets are required to be identified to improve the treatment and prognosis of LUSC.
Long non-coding RNAs (lncRNAs) are defined to be the non-coding transcripts consisting of over 200 nucleotides.Citation4–6 Evidence has proved that lncRNAs are administrators of a myriad of cell functions, such as cell proliferation, cell-cycle, and cell apoptosis.Citation7 In LUSC, it has been reported that three lncRNAs (RP5-821D11.7, APCDD1L-AS1, and RP11-277P12.9) are correlated to the prognosis of LUSC.Citation8 LncRNA SFTA1P promotes apoptosis and induces cisplatin sensitivity via the hnRNP-U-GADD45A axis in LUSC.Citation9 LncRNA NCK1 divergent RNA (NCK1-AS1) has been determined to be upregulated in human cancers and exert oncogenic functions. For example, NCK1-AS1 regulates the miR-6857/CDK1 pathway in cervical cancer, thus facilitating tumorigenesis.Citation10 Knockdown of NCK1-AS1 suppresses cellular processes in cervical cancer through modulating miR-134 expression.Citation11 NCK1-AS1 maintains the stemness of bladder cancer cells through negatively regulating miR-143.Citation12 Recently, NCK1-AS1 has been reported to accelerate tumor growth in glioma through activating the TRIM24/Wnt/β-catenin axis.Citation13 Nevertheless, the functions of NCK1-AS1 in LUSC remains unknown. Importantly, NCK1-AS1 was expressed at a high level in LUSC tissues of the TCGA database. Thus, we tried to unveil the functions of NCK1-AS1 in LUSC in the current study.
LncRNAs can function through regulating their nearby genes.Citation14–16 Here, we explored the regulatory correlation between NCK1-AS1 and NCK1 in LUSC. NCK adaptor protein 1 (NCK1) belongs to the family of Src homology (SH) domain-containing adaptor protein Nck (noncatalytic region of tyrosine kinase).Citation17 NCK1 can be responsible for translation regulation via interacting with the β subunit of eukaryotic initiation factor 2.Citation18 A recent study showed that NCK1 can modulate cap homeostasis through the assembly of cytoplasmic capping complex.Citation19 To date, NCK1 has been delineated to promote cancer development in several cancers, such as colorectal cancer and hepatocellular cancer.Citation20–22 Nevertheless, the role of NCK1 in LUSC has not yet been explored. Here, we also explored the functional role of NCK1 in LUSC.
LncRNAs can regulate gene expression in epigenetic, transcriptional, and posttranscriptional mechanisms.Citation23–26 Interestingly, studies have shown that nuclear lncRNAs can interact with transcription factors (TFs) to induce the transcription of target genes.Citation27,Citation28 More importantly lncRNAs can epigenetically activate their nearby gene.Citation29 Here, we also investigated whether NCK1-AS1 regulated NCK1 through transcriptional modulation. MYC proto-oncogene (MYC) is a transcription modulator regulating genes and thus is responsible for cell metastasis, survival, and proliferation.Citation30–32 The involvement of MYC as an oncogene in NSCLC has been demonstrated by multiple studies.Citation33,Citation34 Here, we unveiled the interaction of MYC to NCK1-AS1 and NCK1 in LUSC.
To summary, our current study was aimed to monitor the function of NCK1-AS1 and its correlation with NCK1 in LUSC.
Materials and methods
Cell lines and culture
Human bronchial epithelial cell (16HBE) and human LUSC cell lines (H2170, H1703, EBC-1, and SK-MES-1) were all acquired from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). 16HBE and LUSC cells were cultured in DMEM medium (HyClone, Logan, UT, USA) with 10% fetal bovine serum, 1 × 105 U/L penicillin G and 1 × 105 U/L streptomycin (Beyotime, Shanghai, China). Cell culture was conducted at 37℃ in a humidified atmosphere containing 5% CO2.
RNA extraction and RT-qPCR analysis
On the basis of the manufacturer’s instructions, TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was utilized to extract total RNA. Prime Script RT Master Mix (A5001, Promega, Madison, WI, USA) was obtained to reversely transcribe RNA into cDNA. RT-qPCR was performed using the SYBR Green Mix (Kangwei, Beijing, China) to evaluate the expressions of various RNAs. The followings were the PCR conditions: at 95℃ for 5 min, then amplification with 35 cycles at 95℃for 5 s, at 60℃for 20 s, and finally at 70℃for 10 s. All the results of three independent experiments were calculated by 2−ΔΔCt method.
Cell transfection
The knockdown of NCK1-AS1, NCK1, and MYC in SK-MES-1 and H2170 was achieved by transfection with sh-NCK1-AS1#1/2/3, sh-NCK1#1/2/3, sh-MYC#1/2/3 (Genepharma, Shanghai, China) and their corresponding negative control (sh-NC) using Lipofectamine2000 Reagent (Invitrogen, Carlsbad, CA, USA). For the overexpression of NCK1-AS1, NCK1, and MYC, pcDNA3.1 vectors were bought from GeneCopoecia (Guangzhou, China), along with the empty vector as control (termed as pcDNA3.1). The transfected cells were harvested after 48 h transfection.
Cell proliferation assay
In order to investigate the rate of cell proliferation, a CCK-8 assay was conducted on the basis of the manufacturer’s protocol. The transfected SK-MES-1 and H2170 cells were reaped and cultured in 96-well plates for 0, 24, 48, 72, and 96 h. CCK-8 solution (Dojindo, Kyushu Island, Japan) was added for another 4 h’ incubation. Cell viability was assessed via measuring the absorbance values at a wavelength of 450 nm.
Caspase 3 activity test
SK-MES-1 and H2170 cells were cultured in 1 mL PBS. After homogenization, cells were centrifuged at 4℃ for 10 min. Pellets were left alone to incubate in lysis buffer (Beyotime, Shanghai, China). Caspase 3 activity assay was conducted in 96-well plates containing lysates, assay buffer, and Caspase-3 colorimetric substrate Ac-DEVD-pNA at 37℃ for 2 h. The chromospheres pNA was cleaved from the substrate molecule. The absorbance values at a wavelength of 405 nm were acquired from three different replications.
Flow cytometry analysis
Annexin V-PI apoptosis detection kit (BD Biosciences PharMingen) was used to analyze apoptosis. After double staining, apoptotic cells in different stages were measured by flow cytometry.
Cell migration assay
Transwell assay was performed by the use of a transwell chamber (Corning Costar, Cambridge, MA, USA). FBS-free RPMI-1640 medium was applied to suspend cells. And then, cells were plated in the upper chambers. In the lower chambers, RPMI-1640 with 10% FBS was added to be the chemoattractant. The migrating cells were fixed in methanol, staining with 0.1% crystal violet after 48 h incubation. After the top surfaces were softly removed, cells migrated to the lower faces were observed under the microscope.
In vivo experiment
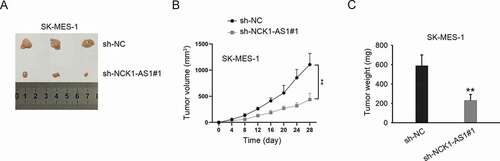
Female athymic nude mice (aged 5 weeks) were purchased from the National Laboratory Animal Center (Beijing, China). SK-MES-1 cells with stable transfection of sh-NC or sh-NCK1-AS1#1 (5 × 106 cells per mouse) suspended in 200 μl of PBS were subcutaneously injected into the right flank of nude mice. Tumor growth was monitored and measured every 4 days. Four weeks later, the nude mice were sacrificed and tumors were resected for subsequent examination.
Western blotting
Western blot was conducted to detect the expression of proteins. The protein was extracted from cells using RIPA lysis buffer and qualified via BCA detecting kit (Beyotime, Shanghai, China). Next, isolation of the proteins was performed using SDS-PAGE. Subsequently, the separated proteins were transferred onto PVDF membranes (Millipore, Bedford, MA, USA). And the membranes were then blocked by bovine serum albumin and incubated with primary antibodies, including anti-NCK1 (1/1000, GTX32738, GeneTex, Southern California, USA), anti-MYC (1/800, ab32, Abcam, Cambridge, USA), anti-Bax (1/1000, ab182734, Abcam), anti-Bcl-2 (1/1000, ab32124, Abcam), anti-Cleaved caspase 3 (1/500, ab32042), anti-caspase 3 (1/5000, ab32351, Abcam) and anti-GAPDH (1/1000, ab8245, Abcam). Afterward, the membrane was incubated with secondary antibodies for 1 h at room temperature. Western blotting was conducted in triplicate.
Luciferase reporter assay
For NCK1 promoter analysis, 293 T cells (AT-1592, ATCC, Manassas, VA, USA) were inoculated into a 24-well plate followed by 24 h of culturing. NCK1 promoter was subjected to PCR amplification and subcloned into the pGL3-Basic vector (Promega, USA). 293 T cells were co-transfected with pRLTK plasmid, NCK1 promoter plasmid and sh-NCK1-AS1#1/2 or sh-MYC#1/2, along with their corresponding control. After transfection for 48 h, the cells were harvested and subjected to the Dual-Luciferase Reporter Assay System (Promega, USA). Biological triplicate was performed independently.
Subcellular fractionation assay
To determine the subcellular localization of NCK1-AS1, the nuclear fraction was isolated from the cytoplasm in accordance with the instruction of NUCLEI EZ PREP NUCLEI ISOLATION KIT (Sigma, St. Louis, MO, USA). SK-MES-1 and H2170 cells were rinsed gently in ice-cold PBS twice. 1 ml of ice-cold Lysis Solution was added to the 25 cm2 flask. Thereafter, 3 × 106 cells were collected using a cell scraper on ice. Lysates were cultured on ice for 5 min. Followed by centrifugation, the nuclear fraction was isolated from the supernatant containing cytoplasmic fraction. At last, the supernatant was removed into a fresh 1.5 ml of EP tube. The precipitate was rinsed in PBS and re-suspended in the Nuclei EZ storage buffer. Experimental result was obtained from three replications.
RNA fluorescence in situ hybridization (FISH)
The RNA FISH probe for NCK1-AS1 was synthesized by Ribobio (Guangzhou, China). Briefly, LUSC cells were fixed in 4% formaldehyde and washed with PBS. After fixation and dehydration, cells were mixed with an NCK1-AS1 probe. Afterward, cells were incubated in a hybridization buffer at 80 ℃for 2 min. After washing and dehydration, the sides were observed and detected with Prolong Gold Antifade Reagent using DAPI.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed in H2170 and SK-MES-1 cells with EZ ChIPTM Chromatin Immunoprecipitation kit (Millipore, Burlington, MA, USA) in line with the user guide. The cross-linked chromatin was sonicated into 200–1,000 base pairs fragments and immunoprecipitated with anti-MYC (Abcam). Normal rabbit immunoglobulin G (IgG) was used as a negative control. RT-qPCR was used to analyze the relative enrichment. Three replications were conducted.
RNA pull-down assay
In vitro biotin-labeled RNAs (NCK1-AS1 and antisense NCK1-AS1) were transcribed using biotin RNA labeling mix (Roche) and T7 RNA polymerase (Roche, Oregon, Hillsboro, USA), followed by treatment with RNase-free DNase I (Promega). Then, RNA was purified with the RNeasy Mini Kit (QIAGEN, New York, USA). Biotinylated RNA was cultured with lysates from H2170 and SK-MES-1 cells. The pull-down proteins were analyzed by RT-qPCR and western blotting.
Statistical analyses
Statistical analyses were conducted by student’s t test and one-way ANOVA using GraphPad Prism 6.0 (GraphPad, San Diego, CA, USA) and SPSS 19.0 statistical software (SPSS, Chicago, IL, USA). The data were expressed as mean ± SD. P < .05 was considered as statistical significance.
Results
NCK1-AS1 was upregulated in LUSC cells and promotes proliferation and migration in vitro
By analyzing the TCGA data through using GEPIA (http://gepia.cancer-pku.cn/), an online bioinformatics website, we found that NCK1-AS1 level was elevated in LUSC samples ()). Consistently, NCK1-AS1 presented high expression in LUSC cell lines, among which SK-MES-1 expressed the highest level of NCK1-AS1, whereas H2170 expressed the lowest level ()).
Figure 1. NCK1-AS1 was upregulated in LUSC cells and promotes proliferation and migration in vitro.
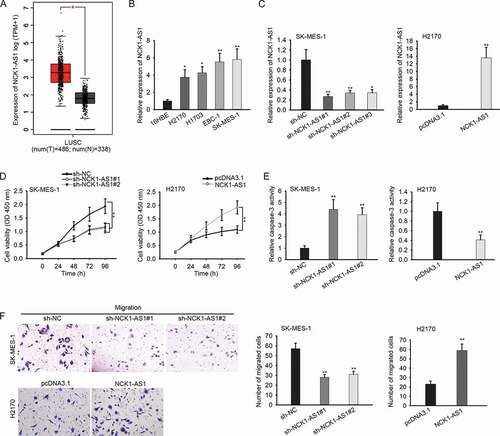
To figure out the biological function of NCK1-AS1 in LUSC, we silenced the expression of NCK1-AS1 in SK-MES-1 cells but overexpressed NCK1-AS1 in H2170 cells for loss- and gain-of-function assays. Since RT-qPCR data confirmed that sh-NCK1-AS1#1/2 knocked down NCK1-AS1 in SK-MES-1 cells more efficiently than sh-NCK1-AS1#3, and pcDNA3.1/NCK1-AS1 overexpressed NCK1-AS1 overtly in H2170 cells ()). To avoid the off-target effect, we used sh-NCK1-AS1#1/2 (with relative highest knockdown efficiency) for loss-of-function assays. Proliferation of SK-MES-1 cells was retarded by NCK1-AS1 silence, whereas that of H2170 cells was facilitated by NCK1-AS1 overexpression ()). NCK1-AS1 depletion prompted apoptosis in SK-MES-1 cells, whereas NCK1-AS1 overexpression had opposite effects ()), which was further demonstrated by flow cytometry analysis (Figure S1). Apoptsis-related proteins were also mesaured by indicating the effect of NCK1-AS1 on apoptosis. As a result, the levels of Bax and Cleaved caspase 3 (pro-apoptosis proteins) were enhanced by the knockdown of NCK1-AS1 but were decreased by the overexpression of NCK1-AS1 (Figure S1B). However, the level of Bcl-2 (anti-apoptosis protein) was positively regulated by NCK1-AS1. Additionally, we observed that inhibiting NCK1-AS1 expression weakened the migration of SK-MES-1 cells, while ectopic expression of NCK1-AS1 led to opposite results in H2170 cells ()). Together, NCK1-AS1 was upregulated in LUSC and accelerates cell growth and migration in vitro.
NCK1 was upregulated in LUSC cells and was positively regulated by NCK1-AS1
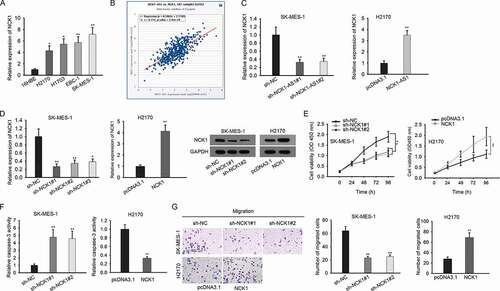
Thereafter, we investigated the mechanism of NCK1-AS1 in LUSC. It has been reported that lncRNAs potentially regulate their nearby genes in cancer development.Citation35,Citation36 Through searching UCSC (http://genome.ucsc.edu/), we found that NCK1 was a gene neighboring to NCK1-AS1. Here, we speculated that NCK1-AS1 might regulate NCK1 in LUSC. At first, we identified the upregulation of NCK1 in LUSC cell lines ()). We analyzed the correlation between NCK1-AS1 and NCK1 through the Starbase pan-cancer (http://starbase.sysu.edu.cn/panCancer.php). Intriguingly, NCK1-AS1 had a positive expression correlation with NCK1 in LUSC tissues ()). Then, we evaluated the influence of NCK1-AS1 on NCK1 expression. Silence of NCK1-AS1 reduced NCK1 mRNA and protein levels in SK-MES-1 cells, and overexpression of NCK1-AS1 exerted opposite influences in H2170 cells ()). NCK1 has been reported to drive tumor progression in several cancers, such as in colorectal cancer and hepatocellular cancer.Citation21,Citation22 Then, we also probed the function of NCK1 in LUSC cells. We silenced NCK1 in SK-MES-1 cells but overexpressed it in H2170 cells ()). Proliferaion of SK-MES-1 cells was hampered by NCK1 silence, whereas the proliferation of H2170 cells was strengthened by NCK1 overexpression ()). Next, the activity of caspase 3 was increased in SK-MES-1 cells upon the knockdown of NCK1, while the opposite results were observed in NCK1-overexpressed H2170 cells ()). Besides, the depletion of NCK1 resulted in less migrating SK-MES-1 cells ()). Therefore, the data above suggested that NCK1 was positively regulated by NCK1-AS1 and exerted an oncogenic role in LUSC.
Figure 2. NCK1 was upregulated in LUSC cells and was positively regulated by NCK1-AS1
NCK1-AS1 upregulated NCK1 at transcriptional level by interacting with MYC
Subsequently, we explored the mechanism by which NCK1-AS1 regulated NCK1 expression. Luciferase reporter assay showed that the silencing of NCK1-AS1 expression reduced the luciferase activity of NCK1 promoter reporter, whereas overexpression of NCK1-AS1 led to the opposite results ()), indicating that NCK1-AS1 could regulate the transcription of NCK1. Besides, we identified that NCK1-AS1 expressed higher in the nucleus of LUSC cells () and Figure S1C). Studies have shown that nealer lncRNAs could regulate the transcription of target genes through interacting with transcriptional factors.Citation27,Citation28 Therefore, we hypothesized that NCK1-AS1 regulated NCK1 transcription in this manner.
Figure 3. NCK1-AS1 upregulated NCK1 at transcriptional level by interacting with MYC
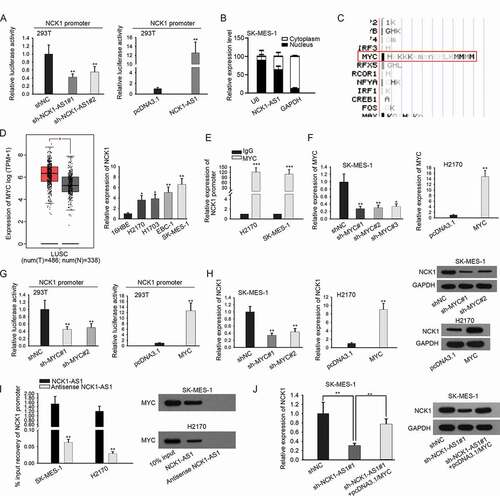
By searching UCSC, we found that the NCK1 promoter contained potential MYC sites ()). MYC is known to be a proto-cancer transcription factor involved in the progression of cancers including NSCLC.Citation30–34 Therefore, we focused on the investigation of MYC. We confirmed that MYC was upregulated in LUSC through GEPIA, and we then confirmed the elevation of MYC in LUSC cell lines ()). ChIP assay validated the enrichment of the NCK1 promoter in the precipitates of the MYC antibody ()). Then, we evaluated the effect of MYC on NCK1. MYC was silenced in SK-MES-1 cells and overexpressed in H2170 cells as confirmed by RT-qPCR ()). Luciferase reporter assay demonstrated that the silenced expression of MYC weakened the transcription of NCK1, whereas induced expression of MYC led to opposite results ()). Moreover, the levels of NCK1 mRNA and protein were decreased responding to MYC silence in SK-MES-1 cells, whereas opposite results were observed in the presence of MYC overexpression vector in H2170 cells ()).
Furthermore, pulldown assay depicted the abundance of NCK1 promoter and MYC protein in the pulldown of NCK1-AS1 rather than antisense NCK1-AS1 ()), indicating that NCK1-AS1 interacted with MYC to target NCK1. Furtherly, we observed that the downregulation of NCK1 induced by NCK1-AS1 silencing was recovered after forced expression of MYC ()). Altogether, these results suggested that NCK1-AS1 upregulated NCK1 by interacting with MYC to activate NCK1 transcription.
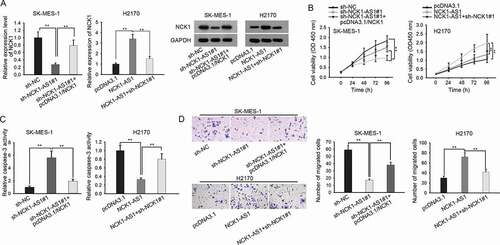
NCK1-AS1 facilitated LUSC progression through regulating NCK1
To detect whether NCK1-AS1 regulated LUSC progression through NCK1, we designed rescue experiments. The mRNA and protein levels of NCK1 decreased by NCK1-AS1 knockdown were recovered by transfection of pcDNA3.1/NCK1 in SK-MES-1 cells (), left). In addition, the enhanced levels of NCK1 caused by NCK1-AS1 overexpression were reduced after the silencing of NCK1 (), right). Functionally, NCK1 overexpression or knockdown abolished the effects of NCK1-AS1 silencing or upregulation on cell proliferation ()). Meanwhile, the apoptosis of SK-MES-1 cells facilitated by NCK1-AS1 silencing or overexpression was reversed by overexpressing or downregulating NCK1 ()). Finally, the migratory ability of SK-MES-1 cells hampered by NCK1-AS1 knockdown was recovered by NCK1 overexpression (), left). In H2170 cells, the migration induced by strengthened NCK1-AS1 was attenuated after the depletion of NCK1 (), right). In sum, these data implied that NCK1-AS1 facilitated proliferation, migration, and inhibited apoptosis through regulating NCK1 in LUSC.
Figure 4. NCK1-AS1 facilitated LUSC progression through regulating NCK1. SK-MES-1 cells were transfected with sh-NC, sh-NCK1-AS1#1, and sh-NCK1-AS1#1+ pcDNA3.1/NCK1, respectively. H2170 cells were transfected with pcDNA3.1, pcDNA3.1/NCK1-AS1, and pcDNA3.1/NCK1-AS1+ sh-NCK1#1, respectively. Cells were harvested after 48 hours for all subsequent rescue assays
NCK1-AS1 silencing led to the inhibition of in vivo tumor growth
Finally, in vivo animal model was used to verify the effect of NCK1-AS1 silencing on tumor growth. As shown in ), tumors in sh-NCK1-AS1#1 group had a smaller size compared to those in the sh-NC group. After calculating, we determined both the volume and weight of tumors in sh-NCK1-AS1#1 were smaller than those in the sh-NC group (). Therefore, we confirmed that NCK1-AS1 accelerated LUSC cell growth in vivo.
Discussion
Mounting reports have revealed that lncRNAs exert crucial impacts on NSCLC progression.Citation8,Citation9 Identification of new lncRNAs in LUSC may be helpful to provide a novel treatment target in LUSC. The present study was aimed at investigating the role of NCK1-AS1 in LUSC. Herein, we firstly revealed that NCK1-AS1 level was upregulated in LUSC samples according to TCGA data, and confirmed the upregulation of NCK1-AS1 in LUSC cell lines, indicating the association of NCK1-AS1 with LUSC. Results of loss- and gain-of-function assays suggested that NCK1-AS1 promoted proliferation, migration, and reduced apoptosis in LUSC cells.
Previous studies have unveiled that antisense RNAs could regulate their neighbor genes.Citation35,Citation36 Accordingly, our study firstly confirmed that NCK1 neighbor to NCK1-AS1, and that NCK1-AS1 positively regulated NCK1 expression in LUSC. Also, we confirmed the positive correlation between NCK1-AS1 and NCK1 in LUSC samples. Previously, several studies have illustrated that NCK1 exhibited oncogenic functions in cancer progression.Citation21,Citation22 In concordance, we firstly validated that NCK1 was upregulated in LUSC cells and its silence-retarded proliferation, migration, and induced apoptosis of LUSC cells, indicating that NCK1 was a positive regulator of LUSC progression.
Moreover, we interrogated the mechanism by which NCK1-AS1 regulated NCK1 expression. We found that NCK1-AS1 positively regulated promoter activity of NCK1, and that NCK1-AS1 could be expressed in the nucleus of LUSC cells, indicating that NCK1-AS1 might regulate NCK1 expression at the transcriptional level. Previously, studies have proved that nuclear lncRNAs can interact with transcription factors (TFs) to induce the transcription of target genes.Citation27,Citation28 MYC is a TF that can modulate the expression of genes responsible for cell metastasis, survival, and proliferation.Citation30–32 Here, we determined that MYC could bind to the promoter of NCK1 to induce its upregulation in LUSC cells. Furthermore, we disclosed that NCK1-AS1 interacted with MYC to target NCK1. Finally, rescue assays indicated the involvement of NCK1 in NCK1-AS1-modulated progression of LUSC.
In conclusion, our study first revealed that NCK1-AS1 promoted the progression of LUSC through transcriptionally activating NCK1 via interacting with MYC, which was a novel molecular pathway in LUSC, indicating NCK1-AS1 as a promising target for LUSC treatment. However, clinical studies are required in the future to further validate the therapeutic significance of NCK1-AS1 in LUSC.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download Zip (717.4 KB)Acknowledgments
Thank you to all participants involved in this research.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29.
- Foss KM, Sima C, Ugolini D, Neri M, Allen KE, Weiss GJ. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol. 2011;6:482–488. doi:10.1097/JTO.0b013e318208c785.
- Cheng T-YD, Cramb SM, Baade PD, Youlden DR, Nwogu C, Reid ME. The international epidemiology of lung cancer: latest trends, disparities, and tumor characteristics. J Thorac Oncol. 2016;11:1653–1671. doi:10.1016/j.jtho.2016.05.021.
- Flintoft L. Structure and function for lncRNAs. Nat Rev Genet. 2013;14:598. doi:10.1038/nrg3561.
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2013;15:7. doi:10.1038/nrg3606.
- Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi:10.1038/bjc.2013.233.
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi:10.4161/rna.20481.
- Luo Y, Xuan Z, Zhu X, Zhan P, Wang Z. Long non-coding RNAs RP5-821D11.7, APCDD1L-AS1 and RP11-277P12.9 were associated with the prognosis of lung squamous cell carcinoma. Mol Med Rep. 2018;17:7238–7248.
- Chen DL, Lu YX, Zhang JX, Wei XL, Wang F, Zeng ZL, Pan -Z-Z, Yuan Y-F, Wang F-H, Pelicano H, et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7:4836–4849. doi:10.7150/thno.20942.
- Guo F, Tang C, Li Y, Liu Y, Lv P, Wang W, Mu Y. The interplay of LncRNA ANRIL and miR-181b on the inflammation-relevant coronary artery disease through mediating NF-κB signalling pathway. J Cell Mol Med. 2018;22(10):5062–5075. doi:10.1111/jcmm.13790.
- Huang L, Gan X, He L, Wang L, Yu J. Silencing of long non-coding RNA NCK1-AS1 inhibits cell proliferation and migration via inhibition of microRNA-134 in cervical cancer. Exp Ther Med. 2019;18:2314–2322.
- Qiao Z, Dai H, Zhang Y, Li Q, Zhao M, Yue T. LncRNA NCK1-AS1 promotes cancer cell proliferation and increase cell stemness in urinary bladder cancer patients by downregulating miR-143. Cancer Manag Res. 2020;12:1661–1668. doi:10.2147/CMAR.S223172.
- Huang L, Li X, Ye H, Liu Y, Liang X, Yang C, Hua L, Yan Z, Zhang X. Long non-coding RNA NCK1-AS1 promotes the tumorigenesis of glioma through sponging microRNA-138-2-3p and activating the TRIM24/Wnt/β-catenin axis. J Exp Clin Cancer Res. 2020;39(1):63. doi:10.1186/s13046-020-01567-1.
- Zhao Y, Wang N, Zhang X, Liu H. LncRNA ZEB1-AS1 down-regulation suppresses the proliferation and invasion by inhibiting ZEB1 expression in oesophageal squamous cell carcinoma. J Cell Mol Med. 2019;23:8206–8218. doi:10.1111/jcmm.14692.
- Wu X, Xiao Y, Zhou Y, Zhou Z, Yan W. LncRNA FOXP4-AS1 is activated by PAX5 and promotes the growth of prostate cancer by sequestering miR-3184-5p to upregulate FOXP4. Cell Death Dis. 2019;10:472. doi:10.1038/s41419-019-1699-6.
- Wu F, Zhong Y, Lang XB, Tu YL, Sun SF. MNX1-AS1 accelerates the epithelial-mesenchymal transition in osteosarcoma cells by activating MNX1 as a functional oncogene. Eur Rev Med Pharmacol Sci. 2019;23:8194–8202.
- Lettau M, Pieper J, Janssen O. Nck adapter proteins: functional versatility in T cells. Cell Commun Signal. 2009;7:1. doi:10.1186/1478-811X-7-1.
- Kebache S, Zuo D, Chevet E, Larose L. Modulation of protein translation by Nck-1. Proc Natl Acad Sci U S A. 2002;99:5406–5411. doi:10.1073/pnas.082483399.
- Mukherjee C, Bakthavachalu B, Schoenberg DR. The cytoplasmic capping complex assembles on adapter protein nck1 bound to the proline-rich C-terminus of Mammalian capping enzyme. PLoS Biol. 2014;12:e1001933–e. doi:10.1371/journal.pbio.1001933.
- Lane C, Qi J, Fawcett JP. NCK is critical for the development of deleted in colorectal cancer (DCC) sensitive spinal circuits. J Neurochem. 2015;134:1008–1014. doi:10.1111/jnc.13137.
- Zhang F, Lu YX, Chen Q, Zou HM, Zhang JM, Hu YH, Li X-M, Zhang W-J, Zhang W, Lin C, et al. Identification of NCK1 as a novel downstream effector of STAT3 in colorectal cancer metastasis and angiogenesis. Cell Signal. 2017;36:67–78. doi:10.1016/j.cellsig.2017.04.020.
- Shen S-L, Qiu F-H, Dayarathna TK, Wu J, Kuang M, Li SSC, Peng B-G, Nie J. Identification of Dermcidin as a novel binding protein of Nck1 and characterization of its role in promoting cell migration. Biochim Biophys Acta Mol Basis Dis. 2011;1812(6):703–710. doi:10.1016/j.bbadis.2011.03.004.
- Yoon J-H, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723–3730. doi:10.1016/j.jmb.2012.11.024.
- Li G, Zhang H, Wan X, Yang X, Zhu C, Wang A, He L, Miao R, Chen S, Zhao H. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014;2014:780521.
- Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta Mol Basis Dis. 2013;1832:1613–1623.
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi:10.1101/gad.1800909.
- Lei JJ, Li HQ, Mo ZH, Liu KJ, Zhu LJ, Li CY, Chen WL, Zhang L. Long noncoding RNA CDKN2B-AS1 interacts with transcription factor BCL11A to regulate progression of cerebral infarction through mediating MAP4K1 transcription. Faseb J. 2019;33:fj201802252R.
- Luo G, Liu D, Huang C, Wang M, Xiao X, Zeng F, Wang L, Jiang G. LncRNA GAS5 inhibits cellular proliferation by targeting P27 Kip1. Mol Cancer Res. 2017;15:789–799. doi:10.1158/1541-7786.MCR-16-0331.
- Su W, Xu M, Chen X, Chen N, Gong J, Nie L, Li L, Li X, Zhang M, Zhou Q, et al. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol Cancer. 2017;16(1):142. doi:10.1186/s12943-017-0711-y.
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12(3):247–256. doi:10.1038/ncb2024.
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi:10.1016/j.cell.2012.03.003.
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976.
- Mei Y, Liu Y-B, Hu D-L, Zhou -H-H. Effect of RIF1 on response of non-small-cell lung cancer patients to platinum-based chemotherapy by regulating MYC signaling pathway. Int J Biol Sci. 2018;14:1859–1872. doi:10.7150/ijbs.27710.
- Kim EY, Kim A, Kim SK, Chang YS. MYC expression correlates with PD-L1 expression in non-small cell lung cancer. Lung Cancer. 2017;110:63–67. doi:10.1016/j.lungcan.2017.06.006.
- Meng W, Cui W, Zhao L, Chi W, Cao H, Wang B. Aberrant methylation and downregulation of ZNF667-AS1 and ZNF667 promote the malignant progression of laryngeal squamous cell carcinoma. J Biomed Sci. 2019;26:13. doi:10.1186/s12929-019-0506-0.
- Wang A, Bao Y, Wu Z, Zhao T, Wang D, Shi J, Liu B, Sun S, Yang F, Wang L, et al. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis. 2019;10:154. doi:10.1038/s41419-019-1331-9.