ABSTRACT
Oral squamous cell carcinoma (OSCC) is one of the most common malignancies worldwide. Emerging evidence has suggested that long noncoding RNAs (lncRNAs) play vital roles in various biological processes of cancers, such as cell proliferation, migration, invasion, and apoptosis. As reported previously, long intergenic non-protein coding RNA 284 (LINC00284) is an important regulator in multiple cancers. However, the biological role, as well as regulatory mechanism of LINC00284 in OSCC, has not been investigated. In our study, RT-qPCR results indicated that LINC00284 was significantly upregulated in OSCC tissues and cells. Moreover, loss-of-function experiments demonstrated that LINC00284 downregulation suppressed cell proliferation and migration and facilitated cell apoptosis. Mechanistically, we found that LINC00284 sponged microRNA 211–3p (miR-211-3p) to upregulate MAF bZIP transcription factor G (MAFG) expression in OSCC cells. Additionally, LINC00284 interacted with FUS protein to increase KAZN mRNA stability. Functional assays showed that either MAFG or KAZN overexpression promoted the malignant behaviors of OSCC cells. Through a series of rescue assays, we found that the inhibitory effect of silencing LINC00284 on OSCC cells can be reversed by upregulated MAFG and KAZN. Overall, silencing LINC00284 inhibits the malignant characteristics of OSCC cells by downregulating MAFG and inhibiting the binding of FUS to KAZN mRNA.
Introduction
Oral squamous cell carcinoma (OSCC) is the most common type of oral cancer,Citation1 accounting for over 90% of malignant lesions in the oral cavity,Citation2 with approximately 300,000 new cases reported annually worldwide.Citation3,Citation4 Though great progress in treatment achieved in the past years, 5-year overall survival of patients with OSCC remains less than 50%,Citation5 mainly due to cancer metastasis to lymph nodes.Citation6,Citation7 Therefore, it is urgent to explore new mechanisms of the tumorigenesis and development of OSCC.
Long noncoding RNAs (lncRNAs) are transcripts that are more than 200 nucleotides,Citation8,Citation9 including antisense, intergenic, and intronic transcripts and pseudogenes.Citation10,Citation11 Accumulating evidence has revealed that lncRNAs play important roles in regulating genes at the transcriptional, posttranscriptional, and epigenetic levels and are involved in many biological processes, including cell proliferation, apoptosis, migration, and invasion in cancers.Citation11,Citation12 Moreover, microRNAs (miRNAs) are small noncoding RNAs with the capability of modulating gene expression posttranscriptionally either by inhibiting messenger RNA (mRNA) translation or by promoting mRNA degradation.Citation13–15 As reported, lncRNAs act as sponges that can silence miRNA expression.Citation16,Citation17 Additionally, lncRNAs can interact with RNA-binding proteins (RBP) to stabilize target mRNAs.Citation18,Citation19 According to previous reports, long intergenic non-protein coding RNA 284 (LINC00284) exert carcinogenic functions in ovarian cancer.Citation20 However, the roles of LINC00284 in OSCC have not been investigated yet. MAF bZIP transcription factor G (MAFG), a nearby gene of MAFG-AS1, was previously reported as an oncogene in lung cancer.Citation21 MAFG is upregulated in tissues of human cholangiocarcinoma and hepatocellular carcinoma and its high expression levels promote tumor progression and reduce survival time.Citation22 Kazrin, periplakin interacting protein (KAZN) was reported to be involved in some diseases. A previous study confirmed the association among KAZN and LAMA5 with endometriosis-related infertility.Citation23 KAZN has a high level in active tuberculosis patients as compared with healthy controls.Citation24 Here, we investigated the role of MAFG and KAZN in OSCC.
This study was specially designed for exploring the biological role of LINC00284 and its regulatory mechanism in OSCC. Our findings suggested that LINC00284 may be a new biomarker for the treatment of OSCC.
Results
LINC00284 knockdown suppresses cell proliferation and migration and represses cell apoptosis in OSCC
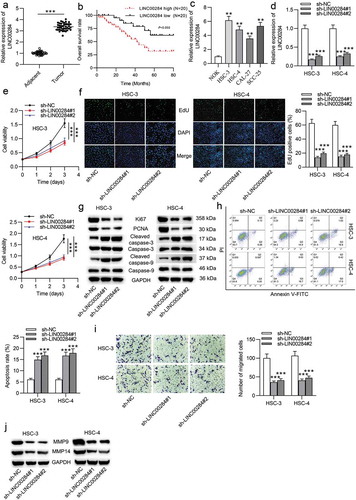
First, we performed RT-qPCR analysis in OSCC tissues and adjacent normal tissues. The results indicated that LINC00284 expression was markedly higher in OSCC tissues than in adjacent normal tissues (). Additionally, we examined the correlation between the LINC00284 level and the prognosis of OSCC patients based on the average of LINC00284 expression in OSCC tissues. Kaplan-Meier survival analysis showed that patients with higher levels of LINC00284 had shorter overall survival than those who had lower levels of LINC00284 (). We further examined the LINC00284 level in OSCC cell lines (HSC-3, HSC-4, CAL-27, and SCC-25) and normal oral keratinocyte cell line (NOK). The results indicated that LINC00284 was significantly upregulated in OSCC cells (). Next, to test the biological role of LINC00284 in OSCC, a series of loss-of-function experiments were conducted. First, we transfected HSC-3 and HSC-4 cells with sh-LINC00284#1/2 to silence LINC00284. The transfection efficacy was examined by RT-qPCR. Compared to the sh-NC group, LINC00284 expression was significantly reduced in HSC-3 and HSC-4 cells in the sh-LINC00284#1/2 groups (). Next, CCK-8 and EdU assays were performed to assess the viability and proliferative ability in OSCC cells. We found that cell viability was inhibited after silencing LINC00284 (). The number of EdU-positive cells was decreased by LINC00284 knockdown (). To further verify the impact of LINC00284 knockdown on cell proliferation and apoptosis, the levels of proliferation-related proteins (Ki67 and PCNA) as well as apoptosis-related proteins (cleaved-caspase-3 and cleaved-caspase-9) were examined by western blot analysis. Both in HSC-3 and HSC-4 cells, the protein expression of Ki67 and PCNA was reduced by LINC00284 downregulation. Additionally, silencing LINC00284 significantly increased the protein expression of cleaved-caspase-3 and cleaved-caspase-9 (). Subsequently, flow cytometry analysis validated the promotive effect of downregulated LINC00284 on OSCC cell apoptosis (). We then explored the impact of silencing LINC00284 on cell migration using wound healing assay and western blot analysis. As shown in , the migratory capability of OSCC cells was significantly suppressed after transfection with sh-LINC00284#1/2. Expression levels of migration-related proteins (MMP-9 and MMP-14) in the sh-LINC00284#1/2 group were decreased (). Overall, these findings suggested that LINC00284 knockdown inhibits the aggressive phenotypes in OSCC cells.
Figure 1. LINC00284 knockdown suppressed cell proliferation and migration but repressed cell apoptosis in OSCC. (a) LINC00284 expression in 40 pairs of OSCC tissues and normal tissues was examined by RT-qPCR. (b) Kaplan-Meier survival analysis of OSCC patients based on the LINC00284 levels. RT-qPCR analysis of LINC00284 expression in OSCC cell lines and normal oral keratinocyte cell line. (d) The transfection efficiency of sh-LINC00284#1/2 in HSC-3 and HSC-4 cells was tested by RT-qPCR analysis. (e) Cell viability in OSCC after silencing LINC00284 was assessed by CCK-8 assay at post-transfection of day 0, 1, 2, 3, and 4. (f) The effect of silenced LINC00284 on cell proliferation was determined by EdU assay. (g) Western blot analysis was used to measure the levels of proliferation and apoptosis-related protein in OSCC cells after silencing LINC00284. (h) Flow cytometry analysis was performed to evaluate the apoptotic rate of OSCC cells after LINC00284 knockdown. (i) The migratory ability of OSCC cells after LINC00284 downregulation was detected by Transwell assay. (j) Western blot analysis was performed to measure the levels of migration-related protein in OSCC cells after silencing LINC00284. **P < .01, ***P < .001
LINC00284 interacts with miR-211-3p
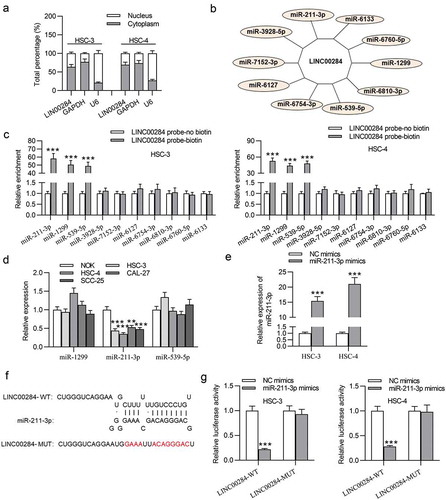
To explore how LINC00284 exerts its function, we tested its subcellular localization and found the preferential distribution of LINC00284 in the cytoplasm of OSCC cells (), which revealed the post-transcriptional regulation of LINC00284 on gene expression. More and more evidence has suggested that lncRNAs act as a competitive endogenous RNA (ceRNA) to sponge miRNAs and exert biological effects in cancers by interacting with miRNAs.Citation25 Therefore, we supposed that LINC00284 may act as a ceRNA in OSCC cells. Ten potential miRNAs of LINC00284 were predicted from the DIANA database (). RNA pull-down experiment demonstrated that three miRNAs (miR-211-3p, miR-1299, and miR-539-5p) might interact with LINC00284 (). RT-qPCR results suggested that only miR-211-3p expression was significantly downregulated in OSCC cells (). Thus, miR-211-3p was selected for further investigation. We further transfected HSC-3 and HSC-4 cells using NC mimics or miR-211-3p mimics. Then, miR-211-3p expression in OSCC cells was significantly upregulated by miR-211-3p mimics (). The potential sequence of miR-211-3p on LINC00284 was predicted at the starBase website (). Luciferase reporter assay indicated that miR-211-3p mimics notably attenuated the luciferase activity of LINC00284-WT, whereas had no impact on the luciferase activity of LINC00284-MUT (). Therefore, LINC00284 can act as a ceRNA by sponging miR-211-3p in OSCC cells.
Figure 2. LINC00284 interacted with miR-211-3p. (a) Subcellular fractionation assay was used to test the localization of LINC00284. (b) Ten possible miRNAs were predicted from online tool DIANA database. (c) RNA pulldown assays were conducted to examine the binding abilities between candidate miRNAs and LINC00284. (d) The expression levels of miRNAs in OSCC cell lines were examined by RT-qPCR analysis. (e) The transfection efficiency of miR-211-3p mimics was verified using RT-qPCR. (f) The binding sequence of miR-211-3p on LINC00284 is predicted from starBase. (g) The interaction between LINC00284 and miR-211-3p was assessed by luciferase reporter assay. ***P < .01
Downregulated miR-211-3p reverses the effect of LINC00284 knockdown on cell phenotypes of OSCC
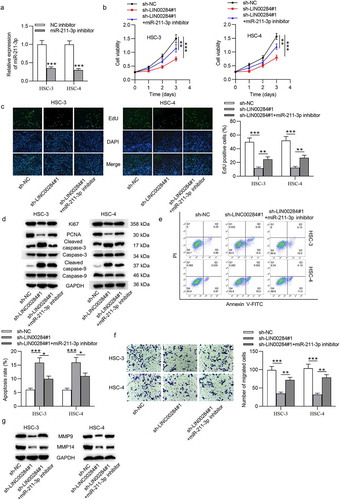
To test whether miR-211-3p is involved in the LINC00284-mediated regulation of OSCC cell functions, rescue experiments were performed. HSC-3 and HSC-4 cells were transfected with miR-211-3p inhibitor to downregulate the expression of mir-211-3p (). As shown in and , the reduced proliferation ability of OSCC cells caused by LINC00284 knockdown was restored after silencing miR-211-3p. Western blot and flow cytometry analyses revealed that miR-211-3p downregulation abrogated the effect of LINC00284 knockdown on cell proliferation and apoptosis ( and ). In addition, miR-211-3p downregulation rescued cell migration inhibited by LINC00284 knockdown ( and ). Overall, miR-211-3p as downstream of LINC00284 is involved in the LINC00284-mediated regulation of cell proliferation, migration, and apoptosis in OSCC.
Figure 3. Downregulated miR-211-3p reversed the effect of LINC00284 knockdown on cell phenotypes of OSCC. (a) The transfection efficiency of miR-211-3p inhibitor was tested by RT-qPCR. (b and c) The viability and proliferation of OSCC cells transfected with the indicated plasmids were assessed by CCK-8 and EdU assays. (d) Western blot analysis was performed to measure the levels of proliferation and apoptosis-related protein. (e) Cell apoptosis was detected using flow cytometry analysis. (f) Cell migration was evaluated using Transwell assay. (g) Western blot analysis was performed to measure the levels of migration-related protein. *P < .05, **P < .01, ***P < .001
LINC00284 sponges miR-211-3p to upregulate MAFG expression
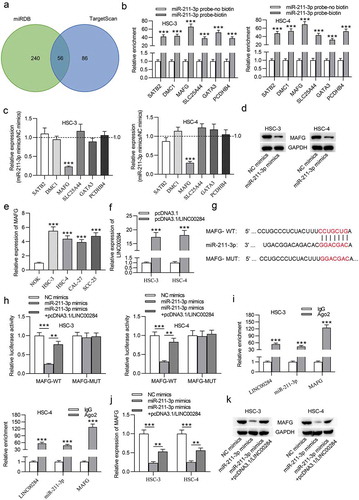
To further investigate the ceRNA pattern, we predicted the target genes of miR-211-3p using bioinformatic analysis. We identified 56 shared mRNAs predicted by two databases (miRDB and TargetScan), as shown in the Venn diagram (). Full lists of predicted target genes by miRDB and TargetScan as well as the list of 56 shared mRNA are shown in Supplementary Data. We used RNA pulldown assay to select the mRNAs interacting with miR-211-3p. The RT-qPCR results revealed that six mRNAs were significantly pulled down by biotinylated miR-211-3p compared to the other 50 mRNA not listed in . Subsequently, we measured the effect of upregulated miR-211-3p on these six mRNAs using RT-qPCR analysis. The results indicated that only MAFG was significantly downregulated in OSCC cells after miR-211-3p upregulation (). Moreover, western blot analysis further verified that the levels of MAFG protein in OSCC cells were decreased by miR-211-3p mimics (). Furthermore, we found that MAFG was upregulated in OSCC cells (). Next, RT-qPCR confirmed the overexpression efficiency of pcDNA3.1/LINC00284 (). The sequence of miR-211-3p on 3ʹ-UTR of MAFG was predicted from TargetScan (). To examine the interaction between miR-211-3p and MAFG, we carried out a luciferase reporter assay in OSCC cells. As indicated in , the luciferase activity of MAFG-WT was notably inhibited by miR-211-3p upregulation, which was restored after cotransfection with pcDNA3.1/LINC00284 (). Furthermore, RIP assay suggested that LINC00284, MAFG, and miR-211-3p were significantly enriched in the Ago2 group, indicating that LINC00284 (MAFG) interacts with miR-211-3p (). We then tested their relationship using RT-qPCR and western blot analyses. As shown in and , overexpression of LINC00284 reversed the miR-211-3p upregulation-mediated inhibition of MAFG mRNA and protein levels. These data demonstrated that LINC00284 upregulates the expression of MAFG by sponging miR-211-3p in OSCC cells.
Figure 4. MAFG was targeted by miR-211-3p. (a) The potential target of miR-211-3p predicted from MiRDB and TargetScan databases. (b) The binding between candidate mRNAs and miR-211-3p was verified by RNA pulldown assay. (c) RT-qPCR analysis of candidate mRNA expression in miR-211-3p overexpressed cells. (d) The impact of miR-211-3p upregulation on MAFG protein level was detected by western blot analysis. (e) MAFG expression in OSCC cell lines was measured by RT-qPCR. (f) RT-qPCR was used to examine the transfection efficiency of pcDNA3.1/LINC00284 in OSCC cells. (g) The binding site of miR-211-3p on 3ʹ-UTR of MAFG was obtained from TargetScan. (h and i) Luciferase reporter and RIP assays were conducted to verify the interaction among, LINC00284, MAFG, and miR-211-3p. (j and k) The levels of MAFG mRNA and protein were measured by RT-qPCR and western blot analyses. **P < .01, ***P < .01
MAFG overexpression promotes cellular processes of OSCC
Next, the biological effect of MAFG in OSCC cells was investigated. We used RT-qPCR and western blot analyses to validate the overexpression efficiency of MAFG in HSC-3 and HSC-4 cells (Fig. S1A). Functional assays showed that upregulated MAFG significantly facilitated cell proliferation (Fig. S1C and S1D), cell migration (Fig. S1F), as well as suppressed cell apoptosis (Fig. S1E). Western blot analysis demonstrated that overexpression of MAFG decreased the levels of pro-apoptotic proteins (Fig. S1G). Additionally, we assayed the effects of different concentrations of pcDNA3.1/MAFG (5 µg, 10 µg, 15 µg, and 20 µg) on proliferation/migration protein levels and discovered that their concentration above a threshold would not result in a corresponding increase in proliferation/migration (Fig. S1H). The above findings suggested that MAFG overexpression contributes to the malignant phenotypes in OSCC cells.
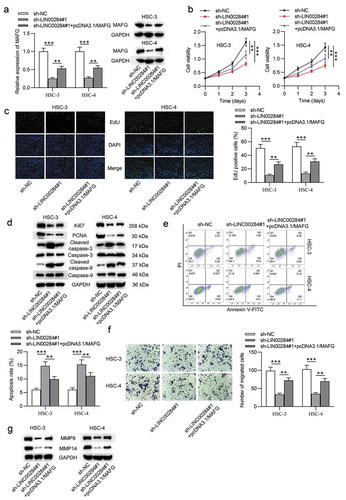
Upregulated MAFG reverses the effect of LINC00284 knockdown on cell phenotypes of OSCC
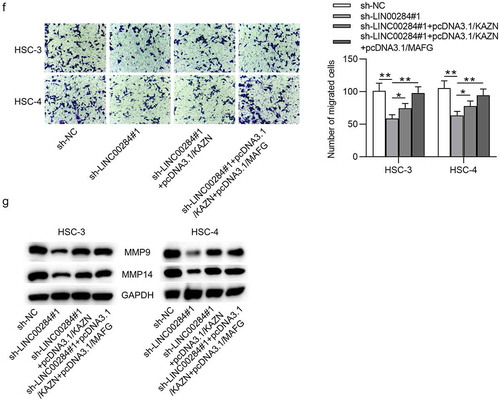
According to the aforesaid results, we supposed that the LINC00284/miR-211-3p/MAFG axis may be associated with the cellular progression of OSCC. To verify this supposition, we performed a series of rescue experiments. First, HSC-3 and HSC-4 cells were transfected with pcDNA3.1/MAFG after LINC00284 was knocked down and RT-qPCR and western blot analyses showed that the reduced MAFG levels by sh-LINC00284#1 were rescued after cotransfection with pcDNA3.1/MAFG (). Next, we assessed cell viability, proliferation, apoptosis, and migration. As shown in and , upregulated MAFG partially rescued the inhibitory function of LINC00284 knockdown on the viability and proliferation of OSCC cells. Furthermore, overexpressed MAFG can partially counteract the effect of silencing LINC00284 on the proliferation and apoptosis of OSCC cells ( and ). Furthermore, Transwell assays revealed that the migratory ability of OSCC cells inhibited by downregulated LINC00284 was restored by upregulated MAFG (). And partial rescue of MAFG overexpression on the migratory capability of OSCC cells was further demonstrated by western blot results (). Overall, MAFG is involved in the LINC00284-mediated regulation of cell phenotypes in OSCC.
Figure 5. Upregulated MAFG reversed the effect of LINC00284 knockdown on cell phenotypes of OSCC. (a) The levels of MAFG mRNA and protein were measured by RT-qPCR and western blot analyses. (b and c) The viability and proliferation of OSCC cells transfected with the indicated plasmids were assessed by CCK-8 and EdU assays. (d) Western blot analysis was performed to measure the levels of proliferation and apoptosis-related protein. (e) Cell apoptosis was detected using flow cytometry analysis. (f) Cell migration was evaluated using Transwell assay. (g) Western blot analysis was performed to measure the levels of migration-related protein. **P < .01, ***P < .01
LINC00284 increases KAZN mRNA stability by interacting with FUS protein
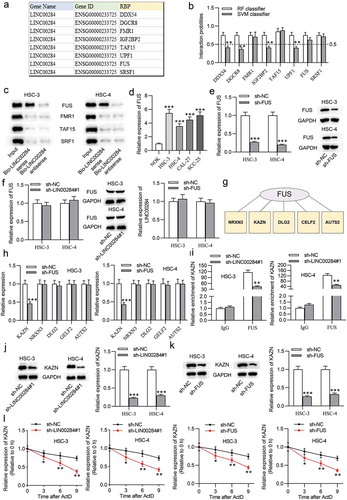
As reported, cytoplasmic lncRNAs could interact with RBPs in cancers.Citation18 We thus hypothesized that LINC00284 can interact with RBP to exert its regulatory function in OSCC. First, we predicted eight potential RBPs by bioinformatics analysis (). Next, we predicted the binding ability between LINC00284 and RBPs at the RPISeq website. Using Random Forest Classifier (RF) and Support Vector Machines (SVM), we identified that LINC00284 may have binding potential with FUS, FMR1, TAF15, or SRF1 (). To test the binding abundance between LINC00284 and these RBPs, western blot analysis on RNA pull-down products was carried out. As revealed in , only FUS protein was pulled down by biotinylated LINC00284, demonstrating the binding between LINC00284 and FUS protein. Therefore, FUS was identified as an RBP recruited by LINC00284. Additionally, we found that FUS expression showed a marked upregulation in OSCC cells (). Subsequently, the knockdown efficiency of sh-FUS was confirmed by RT-qPCR and western blot analyses, shown by the reduced levels of FUS mRNA and protein (). We further found that FUS expression was unchanged after LINC00284 knockdown, and LINC00284 expression showed no change after FUS knockdown (), which suggested that FUS and LINC00284 were not downstream or regulator of each other. It has been reported that RBPs have characteristics of interacting with target mRNAs and enhancing mRNA stability.Citation26 We investigated whether FUS regulated its target mRNA in OSCC. Five putative target genes were predicted from starBase (). RT-qPCR results showed that only KAZN was markedly downregulated in FUS knockdown cells (). Moreover, we found that the binding of “FUS-KAZN” was attenuated after LINC00284 knockdown by RIP analysis in OSCC cells (). In addition, KAZN showed a notable reduction in mRNA and protein levels after LINC00284 or FUS silencing (-). These results demonstrated the “LINC00284-FUS” coregulation of KAZN expression. OSCC cells were treated with actinomycin D to further investigate whether LINC00284/FUS had a regulatory effect on the stability of KAZN mRNA, and the decay of preexisting mRNA was examined. The results indicated that downregulated either LINC00284 or FUS shortened the half-life of KAZN mRNA (). These findings suggested that LINC00284 can increase KAZN mRNA stability by recruiting FUS. We further explored the role of KAZN in OSCC cells. As revealed in Fig. S1B, the levels of KAZN mRNA and protein were increased after transfection with pcDNA3.1/KAZN. The subsequent assays suggested that KAZN overexpression exerted the same effect on OSCC cell phenotypes as MAFG overexpression (Fig. S1C-S1G), which proved the oncogenic role of KAZN in OSCC. Additionally, we found that the concentration of pcDNA3.1/KAZN above a threshold would not lead to a corresponding increase in proliferation/migration (Fig. S1I).
Figure 6. LINC00284 increased KAZN mRNA stability by interacting with FUS protein. (a) Eight putative RBPs was predicted from starBase database. (b) RPISeq website was searched to assess the binding possibilities between LINC00284 and RBPs. (c) The binding between LINC00284 and RBPs was tested by biotinylated LINC00284 pulldown assay and the products were measured by western blot. (d) RT-qPCR analysis of FUS expression in OSCC cells. (e) The transfection efficiency of sh-FUS was analyzed by RT-qPCR and western blot analyses. (f) The expression of FUS after LINC00284 silencing and the expression of LINC00284 after FUS knockdown were measured by RT-qPCR and western blot analyses. (g) Five potential target of FUS was predicted from starBase. (h) RT-qPCR analysis was performed to measure the expression level of targets in FUS knockdown cells. (i) RIP assay was performed to examine the enrichment of targets in FUS RIP after silencing LINC00284 in OSCC cells. (j-k) RT-qPCR and western blot analyses respectively determined KAZN mRNA and protein levels after downregulating LINC00284 or FUS in OSCC cells. (l) Half-life of KAZN in OSCC cells after LINC00284 or FUS silencing was tested by RNA stability assay. **P < .01, ***P < .01
The interaction between FUS and LIN00284 or KAZN mainly occurs in the nucleus of OSCC cells
FUS is a predominantly nuclear RBP widely considered to be involved in splicing regulation.Citation27 We thus examined the subcellular localization of FUS in OSCC cells. The results revealed that FUS retained its nuclear localization in OSCC cells (Fig. S2A). Given the preferential localization of FUS and LINC00284 in the nucleus and cytoplasm, respectively, the compartment where the interaction between them occurred was determined by performing LINC00284 pulldown and FUS RIP assays from nuclear and cytoplasmic fractions independently. As indicated in Fig. S2B, the enrichment of FUS protein was significantly higher in the biotinylated sense-strand LINC00284 probe in the nucleus than in the cytoplasm. Moreover, RIP assays with an antibody against FUS also confirmed a stronger interaction between FUS and LINC00284 in the nucleus than in the cytoplasm (Fig. S2C). The same results occurred in the binding between FUS and KAZN (Fig. S2D and S2E), suggesting that FUS affected KAZN mRNA stability through alternative splicing in the nucleus rather than interacting with KAZN in the cytoplasm. Therefore, we concluded that the interaction between FUS and LIN00284 or KAZN mainly occurs in the nucleus of OSCC cells.
LINC00284 mediates cell behaviors in OSCC by regulating KAZN and MAFG
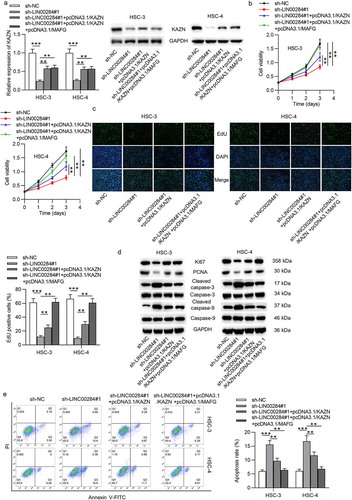
To explore whether KAZN expression was responsible for cell progression mediated by LINC00284, we performed the following rescue experiments. First, HSC-3 and HSC-4 cells were cotransfected with sh-LINC00284#1, pcDNA3.1/KAZN and pcDNA3.1/MAFG. Using RT-qPCR and western blot analyses, we found that KAZN overexpression in combination with MAFG overexpression can completely rescue the expression of KAZN reduced by downregulated LINC00284 (). KAZN overexpression partially rescued the inhibitory effect of LINC00284 downregulation on cell viability and proliferation, while KAZN and MAFG overexpression can completely abolish the inhibitory effect of LINC00284 downregulation on cell viability and proliferation ( and ). Furthermore, the levels of proliferation-related proteins and apoptosis-related proteins in OSCC were partially reversed by overexpressed KAZN but can be completely reversed by upregulation of KAZN and MAFG (). The apoptotic rate of OSCC cells showed a partial restoration under KAZN upregulation but showed a full recovery under KAZN and MAFG upregulation (). Additionally, the suppressive effect of silenced LINC00284 on OSCC cell migration was completely eliminated after upregulating KAZN and MAFG (). Overall, LINC00284 regulates cell proliferation, migration, and apoptosis in OSCC by regulating KAZN and MAFG simultaneously.
Figure 7. LINC00284-mediated cell behaviors in OSCC by regulating KAZN and MAFG. (a) The levels of KAZN mRNA and protein were measured by RT-qPCR and western blot analyses. (b and c) CCK-8 and EdU assays were used to assess cell viability and proliferation in OSCC cells after indicated transfection. (d) Western blot analysis was performed to measure the levels of proliferation and apoptosis-related protein. (e) Cell apoptosis was detected using flow cytometry analysis. (f) Cell migration was evaluated using Transwell assay. (g) Western blot analysis was performed to measure the levels of migration-related protein. **P < .01, ***P < .01
Discussion
LncRNAs have been identified to act as miRNA sponges to interact with miRNAs and play crucial roles in human carcinogenesis by regulating the expression of miRNA target genes.Citation28 As reported, LINC00284 has an oncogenic effect on some cancers, such as ovarian cancer and.Citation20,Citation29 In our research, we first demonstrated that LINC00284 expression was upregulated in OSCC tissues and cells. We performed a series of loss-of-function experiments and investigated the biological role of LINC00284 in OSCC cells. The results indicated that silencing of LINC00284 suppressed OSCC cell viability, proliferation, and migration, as well as promoted cell apoptosis. Therefore, we concluded that LINC00284 may act as an oncogene in OSCC.
Next, we investigated the potential mechanism of LINC00284 in OSCC cells. The localization of lncRNAs in cells primarily determines its molecular function.Citation30 It is well known that lncRNAs exert their influences through diverse ways among which acting as ceRNAs is a very general and important path for lncRNAs. Cytoplasmic lncRNAs can regulate mRNAs by sponging miRNAs through the ceRNA network.Citation31,Citation32 According to the cytoplasmic localization of LINC00284 in OSCC cells, we hypothesized that LINC00284 acted as a ceRNA to sponge miRNA to modulate target gene expression in OSCC cells. Through bioinformatics analysis, we predicted that miR-211-3p was a potential downstream of LINC00284. As reported, miR-211-3p exert an inhibitory function in some cancers, such as lung cancerCitation33 and breast cancer.Citation34 In this study, we validated that LINC00284 interacted with miR-211-3p in OSCC cells. Additionally, miR-211-3p downregulation rescued the inhibitory effect caused by silencing LINC00284 on the cellular progression of OSCC. To further support the ceRNA theory, MAFG was identified to be a potential target of miR-211-3p. Through a series of mechanistic experiments, LINC00284 upregulated MAFG expression by competitively binding to miR-211-3p in OSCC. Moreover, MAFG was involved in the LINC00284-mediated regulation of cellular processes in OSCC. All findings showed the ceRNA role of the LINC00284/miR-211-3p/MAFG axis in OSCC.
Gene expression can be regulated at the posttranscriptional level by RNA-binding proteins (RBPs), which plays a key role in the occurrence and progression of tumors.Citation35,Citation36 The interaction with one or more RBPs is an important way for lncRNAs to exert their effects.Citation18 Thus, we hypothesized that LINC0084 might interact with RBP to regulate target mRNA. The RNA-binding protein FUS, an RBP that regulates RNA metabolism including alternative splicing, miRNA processing, mRNA transport, and local translation.Citation37 In the current study, FUS was predicted as a potential RBP recruited by LINC00284. We further confirmed its abundant binding to LINC00284. Furthermore, we identified that KAZN, a novel cancer-specific transcript detected in thyroid tumorsCitation38 was a putative target of FUS. The results of mechanistic assays revealed that “LINC00284-FUS” targeted KAZN and LINC00284 increased KAZN mRNA stability by abundantly binding to FUS. Functional assays revealed that KAZN promoted OSCC cells proliferation and migration and inhibited cell apoptosis. Additionally, rescue assays indicated that overexpression of KAZN reversed the LINC00284-mediated suppressive effect on the malignant behaviors of OSCC cells.
In summary, our study first demonstrated that silencing LINC00284 inhibits the malignant biological behaviors of OSCC cells by the miR-211-3p/MAFG axis and inhibiting the binding of FUS to KAZN mRNA. This work suggested that LINC00284 may be a promising biomarker for the treatment of OSCC.
Materials and methods
Bioinformatics analysis
The interaction between LINC00284 and miR-211-3p was predicted by “starBase” (http://starbase.sysu.edu.cn/rbpClipRNA.php?source). The interaction between MAFG and miR-211-3p was obtained from “TargetScan” (http://www.targetscan.org/vert_72/). Additionally, the binding between LINC00284 and FUS was predicted at the RNA–Protein interaction prediction (RPISeq) website (http://pridb.gdcb.iastate.edu/RPISeq/).
OSCC tissue samples
A total of 40 paired OSCC tissues and adjacent noncancerous tissues (within 3 cm around tumors) were collected from patients (age range 35 to 62 years old, mean age 49.5 ± 3.1 years old) at the Dezhou People’s Hospital (Shandong, China). Inclusion criteria were as follows: (1) OSCC diagnosed by histopathological examinations; (2) no history of previous malignancies; (3) newly diagnosed and no therapy received before admission. Exclusion criteria: (1) patients with multiple clinical disorders besides OSCC; (2) patients who transferred from other hospitals. These tissues were immediately snap-frozen in liquid nitrogen after collection and stored at −80°C until the extraction of RNA and protein. According to the AJCC criteria, there were 7 cases of stage I, 10 cases of stage II, 8 cases of stage III, and 15 cases of stage IV. This study was approved by the ethics committee of Dezhou People’s Hospital (Shandong, China) and informed consent was signed by all patients.
Cell lines and culture conditions
Human OSCC cell lines (SCC-25 and CAL-27) were supplied by the American Type Culture Collection (ATCC; Rockville, MD, USA). Human OSCC cell lines (HSC-3 and HSC-4) were purchased from Suershengwu (Shanghai, China). A normal oral keratinocyte cell line (NOK) was purchased from Shanghai Zishi Biotechnology Co., Ltd. HSC-3, HSC-4, and CAL-27 cells were maintained in DMEM (Gibco, Rockville, MD, USA) with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA). SCC-25 cells were cultured in DMEM/F12 (1:1) (Gibco, USA) containing 10% FBS. NOK was incubated in K-SFM (Gibco, USA). All cells were cultured at 37°C in a 5% CO2 atmosphere.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from frozen tissues or cell lines with RNAiso Plus (Takara, Dalian, China) based on the manufacturer’s protocols. RNA was reverse transcribed to cDNA using a Reverse Transcription Kit (Takara). A SYBR Premix Ex Taq miRNA kit (Takara) was used to examine the expression of miR-211-3p, and U6 was used as an internal control. A SYBR Premix Ex TaqTM II kit (Takara) was used to analyze the expression of LINC00284, MAFG, FUS and KAZN, and GAPDH acted as an internal control. The expression of miR-211-3p, MAFG, FUS, and KAZN were calculated using the 2−ΔΔCt method. All primer sequences are listed in Supplementary Table 1.
Construction of vectors and cell transfection
Short harpin RNAs (sh-RNAs) for LINC00284 (sh-LINC00284#1:5ʹ-GGTTATGTGGTAACCAGTTTATTCAAGAGATAAACTGGTTACCACATAACCTTTTTT-3ʹ; sh-LINC00284#2: 5ʹ-GGTAACCAGTTTATGACTTGATTCAAGAGATCAAGTCATAAACTGGTTACCTTTTTT-3ʹ), FUS (sh-FUS) and the negative control (sh-NC; 5ʹ-GGTGGAGGTGGAGGTAACTATTTCAAGAGAATAGTTACCTCCACCTCCACCTTTTTT-3ʹ) were constructed by Genepharma Company (Shanghai, China). The miR-211-3p mimics (5ʹ-GCAGGGACAGCAAAGGGGUGC-3ʹ) and NC mimics (5ʹ-UUCUCCGAACGUGUCACG-3ʹ), miR-211-3p inhibitor (5ʹ-UACAAAAGGGUAGUGAAAGUA-3ʹ) and NC inhibitor (5ʹ-UAGAACUUGCAUUGCAACCGG-3ʹ) were synthesized by RiboBio Co., Ltd. (Guangzhou, China). Sequence of LINC00284, KAZN (NM_015209.2), or MAFG (NM_002359.4) was synthesized and subcloned into the pcDNA3.1 vector (V79520; Invitrogen). All plasmid transfection was performed for 48 h using Lipofectamine 2000 (Invitrogen). RT-qPCR was performed to test the transfection efficiency.
CCK-8 assay
Cell viability was using a cell counting kit 8 (CCK8) assay (Dojindo, Kumamoto, Japan). Briefly, the cells were seeded into 96-well plates and transfected with the indicated plasmids. CCK-8 reagent (10 μL) was added to each well at day 0, 1, 2, and 3 followed by incubation for 4 h at 37°C. The absorbance was measured at 450 nm using a spectrophotometer (Perkin Elmer, MA, USA).
5-Ethynyl-2′-deoxyuridine (EdU) assay
The Edu assay kit (RiboBio, China) was used to assess cell proliferation. In brief, the cells from different groups were treated with EdU for 2 h. After washing 3 times with 0.5 g/ml phosphate-buffered saline (PBS), the cells were stained with 4′,6-diamid-ino-2-phenylindole (DAPI; Invitrogen) for 10 minutes in the dark room at room temperature. DAPI-marked cells were washed 3 times with PBS. Finally, the EdU-positive cells were observed using a flow cytometer FACSCalibur DXP (BD Biosciences, Franklin Lakes, NJ, USA).
Western blot
Total protein was extracted from OSCC cells using a RIPA lysis buffer (ProMab Biotechnology, USA). The concentration of total protein was examined using a BCA protein assay kit. Equal amounts of protein were separated by 10% SDS polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes (Millipore, Bedford, MA, USA). The membranes were blocked with 5% bovine serum albumin for 1 h at room temperature and then were incubated with specific primary antibodies overnight at 4°C. After washing, the membranes were incubated with HRP-conjugated anti-IgG at room temperature for 1 h, followed by detection and visualization with an enhanced chemiluminescence (ECL) system (Beyotime, Shanghai, China). Primary antibodies include anti-Ki67 (ab1667; Abcam, Cambridge, UK), anti-PCNA (ab92552; Abcam), anti-cleaved-caspase-3 (ab32042; Abcam), anti-cleaved-caspase-9 (ab2324; Abcam), anti-MMP-9 (ab76003; Abcam), anti-MMP-14 (ab168726; Abcam), anti-KAZN (ab74114; Abcam), anti-FUS (ab124923; Abcam), and anti-MAFG (ab154318; Abcam). Anti-GAPDH (ab181602; Abcam) was USED as an internal control.
Flow cytometry analysis
The cells were seeded into 6-well plates and transfected with the indicated plasmids for 48 h. Then, the cells were collected and washed twice with PBS. Next, cells were stained with an Annexin V-FITC/PI Apoptosis Detection Kit (Multisciences, China) in the dark for 15 min and measured by a flow cytometry (BD Biosciences).
Transwell assay
Transwell assay was conducted in transwell chambers (8 μm diameter; Corning Inc., Corning, NY, USA). The transfected cells (1 × 105 cells/well) in serum-free DMEM were added to the upper chambers. And DMEM containing 10% FBS was added to lower chambers. After 48 h, the cells were harvested, washed with PBS, and resuspended in FBS-free DMEM. After 24 h, nonmigratory cells were removed. And the migratory cells were fixed with 100% methanol, stained with 0.5% crystal violet, washed with PBS, and photographed using an inverted microscope (Olympus Corporation, Tokyo, Japan).
Subcellular fractionation assay
The cytoplasmic and nuclear extracts from HSC-3 and HSC-4 cells were obtained using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Waltham, MA, USA) following the manufacturer’s protocol. The levels of cytoplasmic control (GAPDH), nuclear control (U6), and LINC00284 or FUS were measured by RT-qPCR analysis.
RNA pulldown assay
Biotinylated LINC00284 or negative control was respectively transfected into HSC-3 and HSC-4 cells. Biotinylated miR-211-3p or scramble control were transfected into cells. Next, cells were lysed and collected. After incubation with Dynabeads M-280 Streptavidin (Invitrogen) for 10 minutes, the bound RNA was analyzed by RT-qPCR. This assay was used to verify the binding between LINC00284 and predicted miRNAs or the binding between miR-211-3p and predicted mRNAs.
T7 RNA polymerase (Ambio Life) was used to in vitro transcribe LINC00284, RNeasy Plus Mini Kit (Qiagen) was used to purify LINC00284, and RNase-free DNase I (Qiagen) was used to treat LINC00284. Next, the transcribed LINC00284 was biotin labeled with the Biotin RNA Labeling Mix (Ambio Life). Then RNA pulldown assays were performed using a PierceTM Magnetic RNA-Protein Pull-Down Kit (Pierce, Thermo Fisher Scientific, USA). This method was used to assess the binding between LINC00284 and predicted RBPs.
Dual luciferase reporter assay
The cells were seeded into 96-well plates and a wide type full-length sequence of LINC00284 (MAFG) and the mutant-type LINC00284 (MAFG) were cloned into the firefly luciferase gene reporter vector pmirGLO (Promega, Madison, WI, USA). The pmirGLO-LINC00284 (MAFG)-WT or pmirGLO-LINC00284 (MAFG)-MUT was cotransfected with indicated vectors into OSCC cells. After 48 h, the luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega).
RNA immunoprecipitation (RIP) assay
RIP assay was performed using an EZ-Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore). The cells in different groups were lysed in lysis buffer containing protease inhibitor cocktail and RNase inhibitor. The cells were incubated with RIP buffer containing magnetic beads coated with human anti-FUS (ab243880, Abcam) or with negative control normal rabbit IgG (ab172730, Abcam). After incubation for 2 h at 4°C, the coprecipitated RNA was eluted from the beads and examined by RT-qPCR.
RNA stability assay
After transfection with the indicated plasmids, the cells were treated with actinomycin D (1 µg/ml). Next, cells were collected at different time points and the RNA was extracted with TRIzol reagent (Invitrogen). Finally, RT-PCR analysis was used to measure mRNA levels.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.0. Statistical significance was analyzed by Student’s t-test (two groups) or ANOVA (three or four groups) All results are shown as the means ± standard error of the mean (SEM) of at least three independent experiments. Differences were significant when P < .05.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download Zip (2.3 MB)Acknowledgments
The authors thank all the members participating in this study.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- Lindemann A, Takahashi H, Patel AA, Osman AA, Myers JN. Targeting the DNA damage response in OSCC with TP53 mutations. J Dent Res. 2018 Jun;97(6):635–644. doi:10.1177/0022034518759068.
- Bhatia N, Lalla Y, Vu AN, Farah CS. Advances in optical adjunctive AIDS for visualisation and detection of oral malignant and potentially malignant lesions. Int J Dent. 2013;2013:194029. doi:10.1155/2013/194029.
- Salazar C, Nagadia R, Pandit P, Cooper-White J, Banerjee N, Dimitrova N, Coman WB, Punyadeera C. A novel saliva-based microRNA biomarker panel to detect head and neck cancers. Cell Oncol (Dordrecht). 2014 Oct;37(5):331–338. doi:10.1007/s13402-014-0188-2.
- Durr ML, Li D, Wang SJ. Oral cavity squamous cell carcinoma in never smokers: analysis of clinicopathologic characteristics and survival. Am J Otolaryngol. 2013 Sep-Oct;34(5):388–393. doi:10.1016/j.amjoto.2013.01.017.
- Taghavi N, Yazdi I. Prognostic factors of survival rate in oral squamous cell carcinoma: clinical, histologic, genetic and molecular concepts. Arch Iran Med. 2015 May;18(5):314–319. doi:0151805/AIM.0010.
- Hosni A, McMullen C, Huang SH, Xu W, Su J, Bayley A, Bratman SV, Cho J, Giuliani M, Kim J, et al. Lymph node ratio relationship to regional failure and distant metastases in oral cavity cancer. Radiother Oncol. 2017 Aug;124(2):225–231. doi:10.1016/j.radonc.2017.06.018.
- Liu X, Lao X, Liang L, Zhang S, Li K, Liao G, Liang Y. Neck observation versus elective neck dissection in management of clinical T1/2N0 oral squamous cell carcinoma: a retrospective study of 232 patients. Chin J Cancer Res= Chung-kuo Yen Cheng Yen Chiu. 2017 Jun;29(3):179–188. doi:10.21147/j..1000-9604.2017.03.03.
- Kumar MM, Goyal R. LncRNA as a therapeutic target for angiogenesis. Curr Top Med Chem. 2017;17(15):1750–1757. doi:10.2174/1568026617666161116144744.
- Lin CL, Lee CH, Chen CM, Cheng CW, Chen PN, Ying TH, Hsieh YH. Protodioscin Induces apoptosis through ROS-mediated endoplasmic reticulum stress via the JNK/p38 activation pathways in human cervical cancer cells. Cell Physiol Biochem. 2018;46(1):322–334. doi:10.1159/000488433.
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013 Mar 14;152(6):1298–1307. doi:10.1016/j.cell.2013.02.012.
- Cao W, Liu JN, Liu Z, Wang X, Han ZG, Ji T, Chen WT, Zou X. A three-lncRNA signature derived from the Atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017 Feb;65:94–101. doi:10.1016/j.oraloncology.2016.12.017.
- Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinf. 2017 Jun;15(3):177–186. doi:10.1016/j.gpb.2016.12.005.
- Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs’ action through miRNA editing. Int J Mol Sci. 2019 Dec 11;20(24):6249. doi:10.3390/ijms20246249.
- Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y, Ren J. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019 Feb;76(3):441–451. doi:10.1007/s00018-018-2940-7.
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010 Sep;11(9):597–610. doi:10.1038/nrg2843.
- Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Current Biol. 2010 Oct 12;20(19):R858–61. doi:10.1016/j.cub.2010.08.052.
- Ballantyne MD, McDonald RA, Baker AH. lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol Ther. 2016 May;99(5):494–501. doi:10.1002/cpt.355.
- Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Briefings Bioinf. 2016 Jan;17(1):106–116. doi:10.1093/bib/bbv031.
- Panda AC, Martindale JL, Gorospe M. Affinity pulldown of biotinylated RNA for detection of protein-RNA complexes. Bio-protocol. 2016 Dec 20;6:24. doi:10.21769/BioProtoc.2062.
- Ruan Z, Zhao D. Long intergenic noncoding RNA LINC00284 knockdown reduces angiogenesis in ovarian cancer cells via up-regulation of MEST through NF-κB1. FASEB J. 2019 Nov;33(11):12047–12059. doi:10.1096/fj.201900101RR.
- Sui Y, Lin G, Zheng Y, LncRNA HW. MAFG-AS1 boosts the proliferation of lung adenocarcinoma cells via regulating miR-744-5p/MAFG axis. Eur J Pharmacol. 2019 Sep 15;859:172465. doi:10.1016/j.ejphar.2019.172465.
- Liu T, Yang H, Fan W, Tu J, Li TWH, Wang J, Shen H, Yang J, Xiong T, Steggerda J, et al. Mechanisms of MAFG dysregulation in cholestatic liver injury and development of liver cancer. Gastroenterology. 2018 Aug;155(2):557–571.e14. doi:10.1053/j.gastro.2018.04.032.
- Christofolini DM, Mafra FA, Catto MC, Bianco B, Barbosa CP. New candidate genes associated to endometriosis. Gynecol Endocrinol. 2019 Jan;35(1):62–65. doi:10.1080/09513590.2018.1499090.
- Satproedprai N, Wichukchinda N, Suphankong S, Inunchot W, Kuntima T, Kumpeerasart S, Wattanapokayakit S, Nedsuwan S, Yanai H, Higuchi K, et al. Diagnostic value of blood gene expression signatures in active tuberculosis in Thais: a pilot study. Genes Immun. 2015 Jun;16(4):253–260. doi:10.1038/gene.2015.4.
- Chan JJ, Noncoding TY. RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018 Apr 27;19:5. doi:10.3390/ijms19051310.
- Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Xiao R, Blue SM, Chen J-Y, Cody NAL, Dominguez D, et al. A large-scale binding and functional map of human RNA-binding proteins. Nature. 2020 Jul;583(7818):711–719. doi:10.1038/s41586-020-2077-3.
- Yamaguchi A, Takanashi K. FUS interacts with nuclear matrix-associated protein SAFB1 as well as Matrin3 to regulate splicing and ligand-mediated transcription. Sci Rep. 2016 Oct 12;6:35195. doi:10.1038/srep35195.
- Chang YN, Zhang K, Hu ZM, Qi HX, Shi ZM, Han XH, Han YW, Hong W. Hypoxia-regulated lncRNAs in cancer. Gene. 2016 Jan 1;575(1):1–8.
- Vidovic D, Huynh TT, Konda P, Dean C, Cruickshank BM, Sultan M, Coyle KM, Gujar S, Marcato P. ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ. 2020 Jan;27(1):363–378. doi:10.1038/s41418-019-0362-1.
- Carlevaro-Fita J, Johnson R. Global positioning system: understanding long noncoding RNAs through subcellular localization. Mol Cell. 2019 Mar 7;73(5):869–883. doi:10.1016/j.molcel.2019.02.008.
- Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015 Oct;52(10):710–718. doi:10.1136/jmedgenet-2015-103334.
- Smillie CL, Sirey T, Ponting CP. Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit Rev Biochem Mol Biol. 2018 Jun;53(3):231–245. doi:10.1080/10409238.2018.1447542.
- Zaimoku R, Miyashita T, Tajima H, Takamura H, Harashima AI, Munesue S, Yamamoto Y, Ninomiya I, Fushida S, Harada K, et al. Monitoring of heat shock response and phenotypic changes in hepatocellular carcinoma after heat treatment. Anticancer Res. 2019 Oct;39(10):5393–5401. doi:10.21873/anticanres.13733.
- Kong Q, Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem Biophys Res Comm. 2018 Jan 8;495(2):1594–1600. doi:10.1016/j.bbrc.2017.12.013.
- Anders G, Mackowiak SD, Jens M, Maaskola J, Kuntzagk A, Rajewsky N, Landthaler M, Dieterich C. doRiNA: a database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res. 2012 Jan;40(Database issue):D180–6. doi:10.1093/nar/gkr1007.
- Blin K, Dieterich C, Wurmus R, Rajewsky N, Landthaler M, Akalin A. DoRiNA 2.0–upgrading the doRiNA database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res. 2015 Jan;43(Database issue):D160–7. doi:10.1093/nar/gku1180.
- Ishigaki S, Sobue G. Importance of functional loss of FUS in FTLD/ALS. Front Mol Biosci. 2018;5:44. doi:10.3389/fmolb.2018.00044.
- Le Pennec S, Konopka T, Gacquer D, Fimereli D, Tarabichi M, Tomás G, Savagner F, Decaussin-Petrucci M, Trésallet C, Andry G, et al. Intratumor heterogeneity and clonal evolution in an aggressive papillary thyroid cancer and matched metastases. Endocr-relat Cancer. 2015 Apr;22(2):205–216. doi:10.1530/ERC-14-0351.