ABSTRACT
Spindle and Kinetochore Associated Complex Subunit 3 (SKA3) is crucial for anaphase mitosis. However, the relationship between SKA3 and lung adenocarcinoma (LUAD) has not been fully clarified. Differentially expressed genes were first identified by analyzing data from TCGA. It was found that miR-133b was significantly lowly expressed in LUAD, while SKA3 was remarkably highly expressed. Cell Counting Kit-8 (CCK8), wound healing assay and Transwell assay uncovered that overexpressing miR-133b could inhibit the proliferation, invasion and migration of LUAD cells. In addition, the targeting relationship between miR-133b and SKA3 was also verified by dual-luciferase analysis. Moreover, it was proved by the rescue assay that the overexpression of miR-133b significantly downregulated SKA3 in LUAD cells. All in all, these findings revealed the role of miR-133b and SKA3 in regulating the proliferation, migration, and invasion of LUAD cells. This study could yield new information about the mechanisms of LUAD.
KEYWORDS:
Introduction
Lung cancer is characterized by the highest incidence among all cancers, with less than 20% of patients having an overall survival of more than 5 years.Citation1 Non-small cell lung cancer (NSCLC) accounts for 80% of all lung cancers and lung adenocarcinoma (LUAD) is the most common histological subtype of NSCLC.Citation2 Since LUAD in early stages is usually asymptomatic, most patients are diagnosed in advanced stages. Patients after surgical resection often experience local or distant tumor recurrence.Citation3 Thus, it is necessary to research the molecular mechanisms for pathogenesis and metastasis of LUAD.
MicroRNAs (miRNAs) are short single-stranded non-coding RNAs in eukaryotes. The expressions of miRNAs vary according to the specific tissue and developmental stage. Additionally, miRNAs play an important regulatory role in cell development and differentiation. By targeting the 3ʹ untranslated region (3ʹUTR) of homologous mRNAs, miRNAs are involved in the translational suppression or direct degradation of mRNAs.Citation4,Citation5 Increasing evidence has indicated that miRNAs regulate the proliferation, metastasis, and apoptosis of tumor cells by post-transcriptional regulation of corresponding genes.Citation6 MiR-133b has been found to play a crucial role in various tumors.Citation7 A study showed that miR-133b suppresses the invasion and migration of gastric cancer via COL1A1/TGF-β axis.Citation8 MiR-133b expression targets SOX9 to modulate the pathogenesis and metastasis of breast cancer.Citation9 MiR-133b modulates epithelial-mesenchymal transition (EMT) of ovarian cancer via CTGF upregulation.Citation10 Currently, the regulatory effect of miR-133b in lung cancer has been preliminarily studied,Citation11 but its regulatory mechanism in the progression of LUAD remains unclear.
SKA3, a subunit located on the outer kinetochore of SKA complex, not only coordinates with NDC80 compounds to control mitosis,Citation12 but also plays an important role in meiosis spindle migration and anaphase spindle stability.Citation13 It was reported that somatic mutations of SKA3 commonly affect breast cancer cell growth.Citation14 It was also reported that SKA3 is related to the prognosis of prostate cancer patients.Citation15 Although current studies have shown that SKA3 disorder is related to the occurrence of LUAD,Citation16 the detailed function and potential mechanism of SKA3 in this lethal cancer are still unknown. The specific functional mechanism of SKA3 in LUAD was explored in this study.
The objective of this study was to investigate the role of miR-133b and SKA3 in LUAD. Based on the predicted result of bioinformatics, it was found that miR-133b and SKA3 were abnormally expressed in LUAD. It was speculated that SKA3 might be a target gene of miR-133b in this cancer. Besides, miR-133b mimics was transfected onto human LUAD cells to study the function of miR-133b. Its effect on the expressions of SKA3 protein and mRNA as well as on cancer cell proliferation, migration and invasion were also explored.
Materials and methods
Bioinformatics analysis
Expression data of mature miRNAs (Normal: 46, Tumor: 521) and mRNAs (Normal: 59, Tumor: 535) of LUAD along with relevant clinical data (2019.09.30) were downloaded from The Cancer Genome Atlas (TCGA) database. “EdgeR” package was used to perform differential expression analysis (|logFC|>2, padj<0.05) on miRNAs and mRNAs in the normal group and the tumor group. Differentially expressed miRNAs and mRNAs were then obtained. Next, “survival” package was applied to perform survival analysis. Afterward, target mRNAs were predicted by miRDB, starBase and miRWalk databases to obtain mRNAs that had binding sites of target miRNA. Lastly, correlation analysis was used to determine the target mRNA.
Cell culture
All cells were cultured in an incubator with 5% CO2 at 37 °C. The cell lines and culture medium used in this study were purchased from BeNa Culture Collection (Shanghai, China), as shown in .
Table 1. Cell lines and culture medium used in this study
Cell transfection
LUAD cells were inoculated in 6-well plates at a density of 5 × 105 cells/well. After cell number reached 60–70% confluency, cell transfection was performed. According to the manufacturer’s instruction, Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used to transfect 50 nM miR-133b mimics (miR-mimics), 50 nM negative control mimics (miR-NC), 1 µg pcDNA3.1-SKA3 plasmids (oe-SKA3) and 1 µg empty pcDNA3.1 plasmid carriers (oe-NC) with LUAD cell line A549/H1299. All the mimics and plasmids were ordered from RiboBio (China). All cells were cultured for no less than 24 h after transfection.
Real-time fluorescence quantitative PCR detection (qRT-PCR)
Following the manufacturer’s instruction, TRIzol reagent (TaKaRa, Dalian, China) was used to extract total RNA. Prime Script RT Kit (TaKaRa) was employed to reversely transcribe the total RNA to cDNA. qRT-PCR detection was performed on CFX96™ real-time system (Bio-Rad Hercules, California, USA) using SYBR Green (SYBR Premix Ex Taq™ II; TaKaRa). Conditions were as follows: pre-denaturation at 95 °C for 30 s; denaturation at 95 °C for 35 s; annealing at 55–60 °C for 30 s; repeating for 35 cycles; extension at 72 °C for 1 min; extension at 72 °C for 10 min. Differences in relative expressions of miR-133b and SKA3 mRNA between the control group and experimental group were calculated by 2−ΔΔCt method. SKA3 mRNA and miR-133b took β-actin and U6 as internal references, respectively. Primer sequences were provided in .
Table 2. qRT-PCR primer sequences
Western blot assay
Cells were washed 3 times with cold phosphate buffered saline (PBS). Total proteins were extracted on ice using modified radioimmunoprecipitation assay (RIPA) lysis buffer. Protein samples (40 µg) were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Then, protein samples were transferred onto a 0.22 mm polyvinylidene fluoride membrane (PVDF) (Millipore, Burlington, MA, USA) using Trans-Blot SD (Bio-Rad). Then, the membrane was blocked with 5% skim milk for 2 h and incubated with primary antibodies at 4 °C overnight. Next, the membrane was washed with TBST 3 times and incubated for 1 h after being added with secondary antibody. Electrochemiluminescence (ECL) detection kit (Tanon, China, Shanghai) was used to visualize protein bands. ImageJ software (Wikimedia Foundation, San Francisco, CA, USA) was applied to analyze the imprints. The experiment was repeated 3 times. The primary antibodies rabbit anti-SKA3 (ab118560), rabbit anti-β-actin (ab8227, 1:1000–1:5000) and the secondary antibody goat anti-rabbit IgG (ab205718, 1:2000–1:50000) were purchased from Abcam (Shanghai, China).
Cell proliferation assay
Cell Counting Kit-8 (CCK-8) (CK04, Dojindo Kumamoto, Japan) was used to detect cell proliferation. Stable A549 and H1299 cell lines were inoculated in 96-well plates (5 × 103 cells per well). After 0 h, 24 h, 48 h, 72 h, and 96 h of culture, the plates were added with 10 μl CCK-8 reagent per well, followed by 1 h of incubation. A microplate reader was applied to measure the optical density (OD) at 490 nm. The experiment was repeated 3 times.
Cell invasion and migration assays
Transwell chamber (8 μm aperture, BD Biosciences) was used to detect the invasive and migratory abilities of LUAD cells. In Transwell invasion assay, the upper chamber was coated with 50 mg/L Matrigel (1:8; BD Biosciences, Franklin Lakes, NJ, USA) and the lower chamber was filled with Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS). Two-hundred μL cell suspension was added into the serum-free medium-supplemented upper chamber (1 × 105 cells per well) for incubation at 37 °C. After 24 h, cells in the lower chamber were fixed and stained with 0.1% crystal violet (Beyotime, Wuhan, China). To image the invading cells, 5 random fields were selected in each well and the number of cells was counted under an inverted microscope (Olympus, Wuhan, China) (100×). All procedures were repeated 3 times. In the Transwell migration assay, the upper chamber did not require Matrigel coating. The other steps were the same as invasion assay.
Wound healing assay
A549 and H1299 cells were inoculated in 6-well plates (3 × 106 cells each well) to grow to nearly 100% Confluency. Sterile pipette tips were used to scratch cell monolayer, and PBS was used to remove separated cells. Then, cells were cultured in serum-free medium for 24 ~ 36 h. An inverted microscope (40×) was applied to capture images of corresponding wound areas at 0 h and 24 h/36 h. Image-pro plus 6.0 was used to measure the wound area. Data were collected from 3 independent experiments.
Dual-luciferase assay
Dual-luciferase assay was performed to determine the binding between miR-133b and 3ʹ-UTR of SKA3. The putative miR-133b binding sequences on SKA3 3ʹUTR were cloned into pmirGLO luciferase reporter carriers (Promega, USA) to establish SKA3-WT. Besides, mutant SKA3 3ʹUTR sequences were cloned to build SKA3-MUT. For luciferase activity examination, LUAD cell line A549/H1299 was inoculated in 96-well plates (3 × 105), and then miR-mimics/miR-NC and SKA3-WT/SKA3-MUT plasmids were co-transfected onto cells. After 48 h of culture, luciferase activity was determined through the dual-luciferase reporter system (Beyotime, Wuhan, China). The experiment was repeated 3 times.
Statistical analysis
All data were shown as Mean±SD and represented three independent experiments. Student’s t-test was performed to statistically analyze the data in two groups using GraphPad Prism 6.0 (La Jolla, CA). P < .05 was considered statistically significant.
Results
MiR-133b expression is significantly low in LUAD
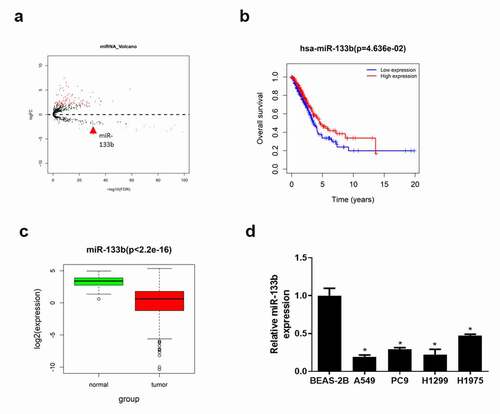
Differential expression analysis was applied to analyze TCGA-LUAD dataset. Altogether, 114 miRNAs which were abnormally expressed in LUAD tissue were acquired, wherein miR-133b expression was downregulated () (Detailed information see Supplementary ). Studies confirmed that miR-133b is downregulated in various cancers and indicates unfavorable prognosis.Citation17,Citation18 In this study, survival analysis showed that the overall survival rate of patients with low miR-133b expression was significantly lower than that of patients with high miR-133b expression (). In TCGA-LUAD dataset, miR-133b was also observed to be significantly lowly expressed in LUAD tissue (). Besides, qRT-PCR detected miR-133b expression in LUAD cells A549, PC9, H1299 and H1975 and human pulmonary bronchial epithelial cells BEAS-2B. It was found that the expression of miR-133b was remarkably downregulated in LUAD cell lines (). The above results indicated that miR-133b was lowly expressed in LUAD tissue and cells.
Figure 1. MiR-133b is lowly expressed in LUAD
Overexpressing miR-133b suppresses the proliferation, migration and invasion of LUAD cells
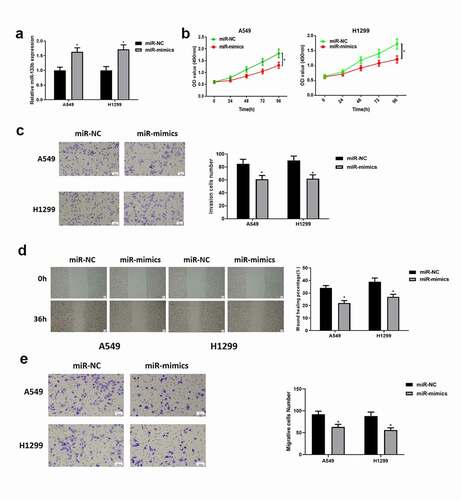
Above experiments presented that the expression of miR-133b in LUAD cell lines A549 and H1299 was markedly different from that in normal cell line. Therefore, LUAD cell lines A549 and H1299 were chosen for the following experiments. To explore the function of miR-133b in LUAD progression, miR-133b mimics were transfected with LUAD cells A549 and H1299. It was testified by qRT-PCR that the expression of miR-133b was significantly upregulated after overexpression (). Therefore, the cell lines with successfully overexpressed miR-133b could be used in the following experiments. The effect of miR-133b on cell viability was detected by CCK-8 assay. It was found that after overexpressing miR-133b, the proliferative ability of cells was significantly inhibited (). Similarly, it was indicated by Transwell assay (, ) and wound healing assay () that overexpressing miR-133b could significantly inhibit the invasive and migratory abilities of LUAD cells. The above results demonstrated that overexpressing miR-133b could inhibit the proliferation, migration and invasion of LUAD cells.
Figure 2. Overexpressing miR-133b suppresses the proliferation, migration and invasion of LUAD cells
SKA3 is upregulated and correlated with poor prognosis in LUAD
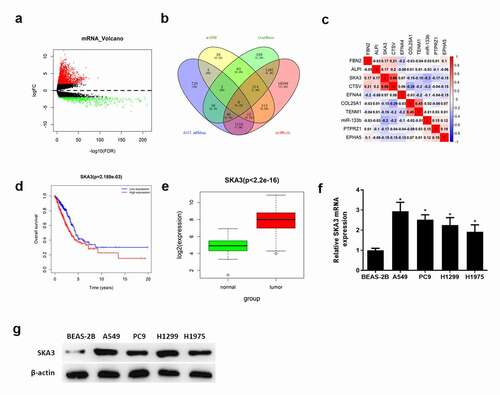
To explore the downstream mechanism of miR-133b in LUAD cells, edgeR was applied to analyze the expression of differential mRNAs in TCGA-LUAD dataset. Totally, 2,503 differential mRNAs were selected at last (). Target genes of miR-133b were predicted by miRDB, starBase and miRWalk databases. The predicted genes were then intersected with 1,975 upregulated mRNAs, and 9 target genes were obtained (). Pearson correlation analysis showed that miR-133b was negatively correlated with SKA3 and the correlation coefficient was the highest among the 9 mRNAs (). Survival analysis indicated that the survival rate of patients with highly expressed SKA3 was lower than that of patients with lowly expressed SKA3 (). Moreover, the expression of SKA3 was significantly high in LUAD tissue (). The expression of SKA3 mRNA and protein in LUAD cell lines and normal cell line were detected by qRT-PCR () and western blot (). It was found that the expression of SKA3 mRNA and protein were significantly upregulated in LUAD cell lines. Hence, it was assumed that SKA3 may be a target gene of miR-133b and associated with poor survival of LUAD.
Figure 3. SKA3 is upregulated and correlated with poor prognosis in LUAD
MiR-133b can target SKA3 in LUAD
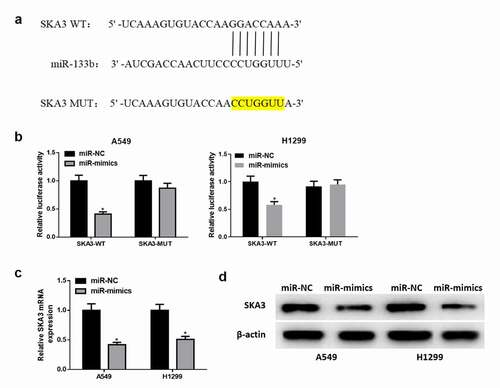
The purpose of this part is to explore the targeting relationship between miR-133b and SKA3 in LUAD cells. As shown in , it was found by bioinformatics analysis that miR-133b had binding sites on SKA3 3ʹUTR. Next, luciferase reporter gene assay revealed that miR-133b directly targeted SKA3 3ʹ-UTR (). The expressions of SKA3 mRNA () and protein () in cells transfected with miR-133b mimics were significantly decreased. The above results illustrated that miR-133b downregulated the expression of SKA3 protein by directly binding to SKA3 3ʹ-UTR.
Figure 4. MiR-133b downregulates the expression of SKA3
MiR-133b suppresses the proliferation, migration and invasion of LUAD cells via targeting SKA3
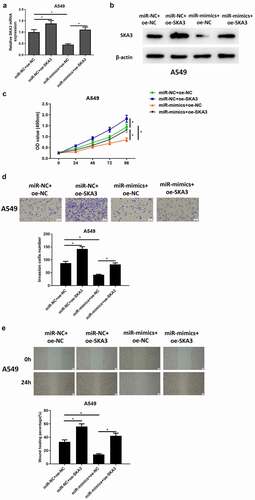
Afterward, we focused on studying whether overexpressing miR-133b inhibits LUAD cell progression by downregulating SKA3. A549 cell lines with overexpressed SKA3 (miR-NC+oe-SKA3), overexpressing miR-133b (miR-mimics+oe-NC) and simultaneously overexpressed miR-133b and SKA3 (miR-mimics+oe-SKA3) were transfected to research the biological function of cells. Compared to miR-NC+oe-NC group, the expression of SKA3 mRNA and protein in miR-NC+oe-SKA3 group were significantly upregulated as detected by qRT-PCR and western blot assays; The expression of SKA3 was significantly downregulated in the miR-mimics+oe-NC group, while the expression of SKA3 was recovered after simultaneously overexpressing miR-133b and SKA3 (). This finding further explained that the expression of SKA3 could be downregulated by miR-133b. CCK-8 manifested that, transfecting SKA3-overexpressed plasmids could markedly strengthen the proliferative ability of A549 cells, while transfecting miR-133b mimics could suppress such ability; Overexpression of miR-133b and SKA3 at the same time weakened the inhibitory effect of miR-133b overexpression on cell proliferation (). Later, Transwell assay and wound healing assay found that compared with miR-NC+ oe-NC group, invasion and migration of SKA3 cells were significantly stimulated after SKA3 overexpression. After overexpression of miR-133b, the invasive and migratory abilities of cancer cells were significantly hampered. However, when miR-133b and SKA3 were overexpressed at the same time, these abilities were significantly stimulated compared with that when miR-133b was overexpressed alone (). These data all verified that miR-133b inhibited LUAD cell proliferation, migration and invasion by downregulating SKA3 expression.
Figure 5. MiR-133b suppresses the proliferation, migration and invasion of LUAD cells through targeting SKA3
Discussion
miRNAs are involved in many crucial physiological and pathological processes, such as cell proliferation, development, differentiation, viral infection and tumorigenesis.Citation19,Citation20 Currently, studies indicated that many miRNAs play a vital role in the progression of LUAD. For example, miR-204-3p,Citation21 miR-423-5p,Citation22 miR-135-5p,Citation23 miR-511-3p,Citation2 and miR-22-3p suppress the malignant progression of LUAD, while miR-96-5pCitation24 promotes the metastasis and proliferation of LUAD cells. Generally, miR-133b is considered as a cancer suppressor and is downregulated in breast cancer,Citation18 colorectal cancer,Citation18 etc. Jiyong Pan and other authors pointed out that miR-133b is involved in apoptosis, proliferation, migration, invasion, angiogenesis, and drug resistance of lung cancer cells.Citation11 MiR-133b upregulates FBN1 expression to suppress gastric cancer cell proliferation and invasion.Citation25 MiR-133b targets PKM2 to influence the proliferation and drug sensitivity of A549 lung cancer stem cells.Citation26 MiR-133b restrains SOX9/β-catenin signaling pathway to inhibit lung cancer progression.Citation27 In this study, it was identified that the expression of miR-133b was significantly downregulated in LUAD, using bioinformatics analysis and cell experiments in vitro. MiR-133b overexpression could suppress the proliferation, migration and invasion of LUAD cells.
Furthermore, it is reported that miRNAs can play a regulatory role by decreasing the expression of its target mRNA.Citation28,Citation29 On this basis, we studied the downstream regulatory mechanism of miR-133b in LUAD. In this study, bioinformatics analysis predicted that SKA3 could be a target gene of miR-133b. A previous study reported that biological behaviors of various tumors can be regulated by SKA3.Citation30 Hu DD et al.Citation16 found that SKA3 was overexpressed in LUAD, which can promote cell proliferation, induce tumor cell angiogenesis and enhance the migration and invasion of cells. Although studies reported the effect of SKA3 in LUAD, the upstream regulatory mechanism of SKA3 in LUAD is not clear. Through cell biological assays, it was found in this study that SKA3 was upregulated in LUAD cells and overexpressing SKA3 could strengthen the proliferative, migratory and invasive abilities of LUAD cells. It demonstrated that highly expressed SKA3 could promote the malignant progression of LUAD, which is consistent with previous research results. Moreover, the binding relationship between miR-133b and SKA3 was further testified by dual-luciferase assay. Results of the rescue assay clarified that miR-133b affected the biological behaviors of LUAD cells through regulating the expression of SKA3. These conclusions indicated that miR-133b and SKA3 may be prognostic indicators for LUAD.
All in all, this study revealed that the expression of miR-133b was downregulated in LUAD cells and was relevant to the poor prognosis of patients. MiR-133b inhibited cell proliferation, migration and invasion. It was also found that SKA3, negatively regulated by miR-133b, was abnormally highly expressed in LUAD cells. The regulatory mechanism by which miR-133b affects biological behaviors of LUAD via regulating SKA3 was revealed. Both miR-133b and SKA3 can be new therapeutic targets for LUAD treatment.
Ethics approval and consent to participate
Not applicable.
Availability of data and materials
The data used to support the findings of this study are included within the article. The data and materials in the current study are available from the corresponding author on reasonable request.
Supplemental Material
Download MS Word (114 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Morgensztern D, Campo MJ, Dahlberg SE, Doebele RC, Garon E, Gerber DE, Goldberg SB, Hammerman PS, Heist RS, Hensing T, et al. 2015. Molecularly targeted therapies in non-small-cell lung cancer annual update 2014. J Thorac Oncol. 10:S1–63. https://doi.org/10.1097/JTO.0000000000000405.
- Li J, Ge J, Yang Y, Liu B, Zheng M, Shi R. 2020. Long noncoding RNA ZFPM2-AS1 is involved in lung adenocarcinoma via miR-511-3p/AFF4 pathway. J Cell Biochem. 121(3):2534–2542. https://doi.org/10.1002/jcb.29476.
- Tan J, Wang W, Song B, Song Y, Meng Z 2020. Integrative Analysis of Three Novel Competing Endogenous RNA Biomarkers with a Prognostic Value in Lung Adenocarcinoma. Biomed Res Int. 2020:2837906. https://doi.org/10.1155/2020/2837906.
- Wu L, Fan J, Belasco JG. 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 103(11):4034–4039. https://doi.org/10.1073/pnas.0510928103.
- Schickel R, Boyerinas B, Park SM, Peter ME. 2008. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 27(45):5959–5974. https://doi.org/10.1038/onc.2008.274.
- Wu Y, Shi W, Tang T, Wang Y, Yin X, Chen Y, Zhang Y, Xing Y, Shen Y, Xia T, et al. 2019. miR-29a contributes to breast cancer cells epithelial-mesenchymal transition, migration, and invasion via down-regulating histone H4K20 trimethylation through directly targeting SUV420H2. Cell Death Dis. 10(3):176. DOI:https://doi.org/10.1038/s41419-019-1437-0
- Mitchelson KR, Qin WY. 2015. Roles of the canonical myomiRs miR-1, −133 and −206 in cell development and disease. World J Biol Chem. 6(3):162–208. https://doi.org/10.4331/wjbc.v6.i3.162.
- Guo Y, Lu G, Mao H, Zhou S, Tong X, Wu J, Sun Q, Xu H, Fang F 2020. miR-133b Suppresses Invasion and Migration of Gastric Cancer Cells via the COL1A1/TGF-beta Axis. Onco Targets Ther. 13:7985–7995. https://doi.org/10.2147/OTT.S249667.
- Wang Q-Y, Zhou C-X, Zhan M-N, Tang J, Wang C-L, Ma C-N, He M, Chen G-Q, He J-R, Zhao Q. 2018. MiR-133b targets Sox9 to control pathogenesis and metastasis of breast cancer. Cell Death Dis. 9(7):752. https://doi.org/10.1038/s41419-018-0715-6.
- Yang L, Hou J, Cui XH, Suo LN, Lv YW 2017. MiR-133b regulates the expression of CTGF in epithelial-mesenchymal transition of ovarian cancer. Eur Rev Med Pharmacol Sci. 21:5602–5609. https://doi.org/10.26355/eurrev_201712_14001.
- Pan JY, Sun CC, Bi ZY, Chen ZL, Li SJ, Li QQ, Wang YX, Bi YY, Li DJ 2017. miR-206/133b Cluster: a Weapon against Lung Cancer? Mol Ther Nucleic Acids. 8:442–449. https://doi.org/10.1016/j.omtn.2017.06.002.
- Jeyaprakash AA, Santamaria A, Jayachandran U, Chan YW, Benda C, Nigg EA, Conti E. 2012. Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol Cell. 46(3):274–286. https://doi.org/10.1016/j.molcel.2012.03.005.
- Zhang QH, Qi ST, Wang ZB, Yang CR, Wei YC, Chen L, Ouyang YC, Hou Y, Schatten H, Sun QY. 2012. Localization and function of the Ska complex during mouse oocyte meiotic maturation. Cell Cycle. 11(5):909–916. https://doi.org/10.4161/cc.11.5.19384.
- Jiao X, Hooper SD, Djureinovic T, Larsson C, Warnberg F, Tellgren-Roth C, Botling J, Sjoblom T. 2013. Gene rearrangements in hormone receptor negative breast cancers revealed by mate pair sequencing. BMC Genomics. 14(1):165. https://doi.org/10.1186/1471-2164-14-165.
- Lee M, Williams KA, Hu Y, Andreas J, Patel SJ, Zhang S, Crawford NP. 2015. GNL3 and SKA3 are novel prostate cancer metastasis susceptibility genes. Clin Exp Metastasis. 32(8):769–782. https://doi.org/10.1007/s10585-015-9745-y.
- Hu DD, Chen HL, Lou LM, Zhang H, Yang GL SKA3 promotes lung adenocarcinoma metastasis through the EGFR-PI3K-Akt axis. Biosci Rep. 2020;40. doi:https://doi.org/10.1042/BSR20194335.
- Huang S, Wa Q, Pan J, Peng X, Ren D, Li Q, Dai Y, Yang Q, Huang Y, Zhang X, et al. 2018. Transcriptional downregulation of miR-133b by REST promotes prostate cancer metastasis to bone via activating TGF-beta signaling. Cell Death Dis. 9(7):779. DOI:https://doi.org/10.1038/s41419-018-0807-3
- Li X, Deng S, Pang X, Song Y, Luo S, Jin L, LncRNA PY NEAT1 Silenced miR-133b Promotes Migration and Invasion of Breast Cancer Cells. Int J Mol Sci. 2019;20. doi:https://doi.org/10.3390/ijms20153616.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2): 281-97. doi:https://doi.org/10.1016/S0092-8674(04)00045-5.
- Carlsson J, Davidsson S, Helenius G, Karlsson M, Lubovac Z, Andren O, Olsson B, Klinga-Levan K. 2011. A miRNA expression signature that separates between normal and malignant prostate tissues. Cancer Cell Int. 11(1):14. https://doi.org/10.1186/1475-2867-11-14.
- Yang S, Liu T, Sun Y, Liang X. 2019. The long noncoding RNA LINC00483 promotes lung adenocarcinoma progression by sponging miR-204-3p. Cell Mol Biol Lett. 24(1):70. https://doi.org/10.1186/s11658-019-0192-7.
- Li W, Zhang B, Jia Y, Shi H, Wang H, Guo Q, LncRNA LH. 2020. LOXL1-AS1 regulates the tumorigenesis and development of lung adenocarcinoma through sponging miR-423-5p and targeting MYBL2. Cancer Med. 9(2):689–699. https://doi.org/10.1002/cam4.2641.
- Zheng C, Li X, Ren Y, Yin Z, Zhou B 2019. Long Noncoding RNA RAET1K Enhances CCNE1 Expression and Cell Cycle Arrest of Lung Adenocarcinoma Cell by Sponging miRNA-135a-5p. Front Genet. 10:1348. https://doi.org/10.3389/fgene.2019.01348.
- Zhao M, Xin XF, Zhang JY, Dai W, Lv TF, LncRNA SY. 2020. GMDS-AS1 inhibits lung adenocarcinoma development by regulating miR-96-5p/CYLD signaling. Cancer Med. 9(3):1196–1208. https://doi.org/10.1002/cam4.2776.
- Yang D, Zhao D, Chen X. 2017. MiR-133b inhibits proliferation and invasion of gastric cancer cells by up-regulating FBN1 expression. Cancer Biomark. 19(4):425–436. https://doi.org/10.3233/CBM-160421.
- Mi Y, He M, Liu B 2017. [MiR-133b Affect the Proliferation and Drug Sensitivity in A549 Lung Cancer Stem Cells by Targeting PKM2]. Zhongguo Fei Ai Za Zhi. 20:376–381. https://doi.org/10.3779/j.1009-3419.2017.06.02.
- Liu S, Li S, Yu X, Wang Q, Sun H microRNA-133b represses the progression of lung cancer through inhibiting SOX9/beta-catenin signaling pathway. Int J Clin Exp Pathol. 2020;13:2270–2279.
- Lin C, Wang Y, Wang Y, Zhang S, Yu L, Guo C, Xu H. 2017. Transcriptional and posttranscriptional regulation of HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in esophageal squamous carcinoma cells. Oncogene. 36(38):5392–5406. https://doi.org/10.1038/onc.2017.133.
- Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. 2015. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 6(1):e1583. https://doi.org/10.1038/cddis.2014.541.
- Chuang TP, Wang JY, Jao SW, Wu CC, Chen JH, Hsiao KH, Lin CY, Chen SH, Su SY, Chen YJ, et al. 2016. Over-expression of AURKA, SKA3 and DSN1 contributes to colorectal adenoma to carcinoma progression. Oncotarget. 7(29):45803–45818. DOI:https://doi.org/10.18632/oncotarget.9960
