ABSTRACT
Exosome DNA (exoDNA) can be used for liquid biopsy. This study was the first to use droplet digital PCR (ddPCR) to detect tumor-specific mutations in exoDNA and to evaluate the prognosis of hepatocellular carcinoma (HCC) patients. 60 HCC patients were enrolled in the study. We used ddPCR to detect c.747 G > T mutation in TP53 gene. We analyzed the correlation between detectable mutation in exoDNA and clinicopathologic characteristics using Multivariate logistics regression analysis. We performed Cox regression to assess the correlation between mutation frequency (mutant droplets/total droplets, MD/TD) and prognostic. We found that 48 of 60 patients had c.747 G > T mutation in TP53 gene in exoDNA (80.0%). We found that detectable mutation in exoDNA and age were associated with microvascular invasion (MVI) (P < .01). The ROC curve analysis revealed that the best cutoff value of mutation frequency to predict MVI was 67% (sensitivity 48.15%, specificity 93.94%,), the corresponding AUC was 0.761 (95%CI, 0.640–0.866; P < .01). Furthermore, we found that patients suffered high-frequency mutation (>67%) had shorted median recurrence-free survival (RFS) with 63 days (range, 53–202 days), compared with 368 days (range, 51–576 days) for patients with low-frequency mutation (<67%) (HR:4.61; 95% CI, 1.70–12.48; P = 0 .003). We also found that high-frequency mutation was associated with poor prognosis though patients had better pathological characteristics, such as AFP (<400 ng/mL), Liver cirrhosis (Negative), Tumor thrombus (Negative), Tumor numbers (Single) and Post-operation TACE (Executed). We provided evidence that the mutations in exoDNA might be used to predict patients with poor RFS.
Abbreviations: TP53: Tumor protein p53; ExoDNA: Exosomal DNA; HCC: Hepatocellular carcinoma; ddPCR: Droplet digital Polymerase Chain Reaction (PCR); MD/TD: The ratio of mutant droplets/total droplets; AFP: Alpha-fetoprotein; MVI: Microvascular invasion; RFS: Recurrence-free survival.
Background
Hepatocellular carcinoma (HCC) is currently the fifth most common malignancy with the third highest mortality rate in the world.Citation1 Effective treatment and diagnostic strategies for HCC remain difficult problems. Although there are many treatments for HCC, including surgery, transcatheter arterial chemoembolization, and targeted drugs, the recurrence rate of HCC is high.Citation1,Citation2 Therefore, timely diagnosis and real-time intervention are required for effective diagnostic and therapeutic measures for HCC. Unfortunately, the diagnostic efficacy of traditional markers, such as AFP, was not satisfactory.Citation3 In recent years, multiple studies have used circulating free DNA (cfDNA) from the blood of pancreatic and colorectal cancer patients to identify tumor-specific mutations as cancer markers.Citation4,Citation5 However, cfDNA originates from dead cells in damaged tissue or accumulates as a result of physiological cell turnover.Citation6,Citation7 This may lead to reduced sensitivity of cfDNA bio-markers and make the identification of cancer-specific mutations more challenging.
Exosomes are extracellular vesicles 40–150 nm in size that are rich in DNA, RNA, and proteins.Citation8 Although exoDNA shows biological stability and has the potential for clinical application, few studies have examined exoDNA compared with the multiple studies already performed on exosomal RNA. Exosomes are secreted by living and dead cells, as well as cancer cells, which produce high numbers of exosomes.Citation8–10 Recent studies have shown that circulating exoDNA can be used to identify cancer-specific mutations,Citation9,Citation11,Citation12 which is of great significance for the diagnosis and treatment of HCC patients as well as prognosis assessment. Genetic mutations have been traditionally identified by genome sequencing of biopsy tumor fragments or cfDNA, but this represents only a small fraction of tumor heterogeneity because cfDNA is released by dead tumor cells.Citation6 ExoDNA is more stable than cfDNA because of the lipid bilayer structure of exosomes,Citation13,Citation14 and exosomes contain larger DNA fragments, which is conducive to the detection of mutations. Additionally, because exosomes can be released from various cells in tumors, exoDNA in plasma may reflect tumor heterogeneity. Based on these advantages, increasing studies have been reported on tumor-specific mutations in circulating exoDNA, such as EGFR mutations in lung adenocarcinoma patients and KRAS mutations in pancreatic cancer patients.Citation9,Citation11,Citation15 However, no studies have been reported on the detection of tumor-specific mutations in circulating exoDNA in HCC patients.
In this study, here we report the first investigation of the value and significance of detection of tumor-specific mutations in circulating exoDNA in HCC patients using droplet digital PCR (ddPCR). And we found that patients with high-frequency mutation are more likely to microvascular invasion and associated with poor prognosis.
Results
Patient characteristics
This study included 60 patients with primary HCC who were enrolled between October 2018 and August 2020 at the Second Affiliated Hospital of Nanchang University. The median age in this study was 56 years, with 40 (66.7%) males and 20 (33.3%) females. The proportion of HBV infection and cirrhosis was 58 (96.7%) and 33 (55.0%) respectively, 27 patients (45.0%) had Microvascular invasion, 8 patients (13.3%) had Tumor thrombus and 40 patients (33.3%) with preoperative AFP ≥400 ng/ml. The median of tumor size was 6.2 cm, with the liver function of all patients was Child A. In these patients, 48 (80.0%) had been detected TP53 mutation in circulating exoDNA. The clinical characteristics of these 60 patients are listed in . Additionally, we presented the detailed preoperative information of these patients in Supplementary Table S1.
Table 1. Clinical characteristics of patients in this study.
TP53 mutation in exoDNA and clinicopathologic characteristics correlation
The c.747 G > T mutation in TP53 had been identified as one of the hottest mutants in HCC patients.Citation16–18 And our previous studies had confirmed the presence of TP53 mutations in circulating tumor DNA in HCC patients.Citation16 Thus, these mutations might be detectable in exoDNA, which also contained abundant tumor cell-derived genetic information. We detected TP53 gene mutation status (c.747 G > T) in circulating exoDNA and presented the copy number of wild-type and mutant TP53 gene in all patients. We found that 48 of 60 patients had c.747 G > T mutations in TP53 gene in exoDNA (sensitivity, 80.0%). The high sensitivity may be due to the wide source and stability of exosomes. Multivariate logistics regression analysis revealed that TP53 mutation and age was associated with microvascular invasion (MVI) (P < .001), but were not significantly associated with other clinicopathological features such as Satellite nodules, tumor number, and AFP () .
Table 2. Results of correlation multivariate regression analysis between patient characteristics and MVI.
In order to explore the clinical application of exoDNA, we calculated and analyzed the ratio of mutant droplets/total droplets (MD/TD). We executed the ROC curve analysis and found that the best cutoff value of MD/TD to predict MVI was ≥67% (sensitivity 48.15%, specificity 93.94%), the corresponding AUC was 0.761 (95%CI [0.640–0.866], P < .01). The detailed results are shown in Supplementary Figure S1.
TP53 mutation in exoDNA and prognosis correlation
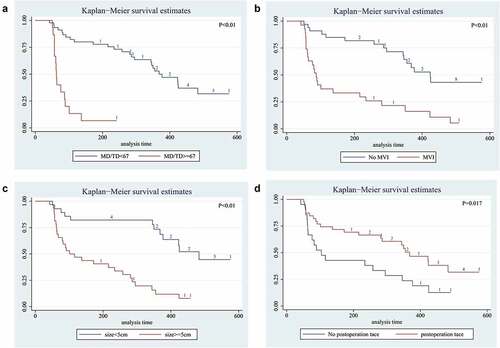
We also analyzed whether TP53 mutation status in exoDNA could predict the prognosis of HCC patients who had undergone surgical treatment. We developed a regular and rigorous follow-up strategy of these 60 HCC patients up to 576 days. The results shown that patients suffered high-frequency mutations (MD/TD ≥67%) had shorted median RFS with 68 days (range, 53–202 days), compared with 368 days (range, 51–576 days) for patients with low-frequency mutations (MD/TD <67%). This result had statistically different (P < .01, log-rank test, ). This result shown that patients in the group with high-frequency mutations were more likely to relapse than those in the group with low-frequency mutations. Similarly, the patients with pathological characteristics (such asMVI (+), tumor size >5 cm or postoperative tace (-)) had shorted median RFS (log-rank test, ).
Figure 1. Long-term survival outcomes in HCC using Kaplan-Meier’s analysis:A. Survival time of mutation frequency in HCC. The results shown that the patients with MD/TD≥67 had shorter median survival time; B. MVI positive shown worse survival time; C. Tumor size >5 cm had worse result shorter survival time; D. Patient had not been postoperative tace had worse median survival than patients without satellite nodules. P value was assessed using the log-rank test.
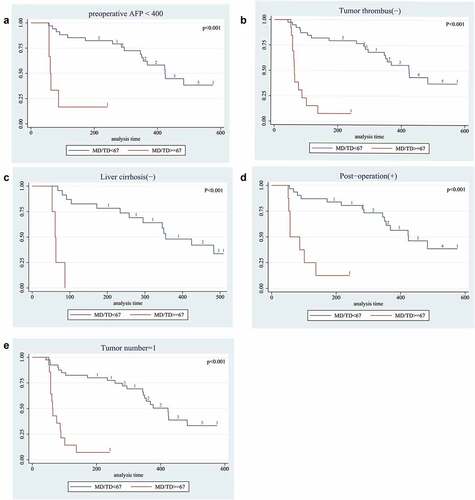
In addition, we also wonder whether there had correlation between mutation frequency and RFS in different pathological status. The results shown that the patients with high-frequency mutations (MD/TD ≥67%) had shorted RFS when preoperative AFP <400 ng/ml had been detected (median RFS:61 VS. 368 days, P < .01). In patients suffered single tumor, high-frequency mutations also predicted shorter survival times (median RFS:63 VS. 423 days, P < .01). Similar results could be seen in patients without tumor thrombus and liver cirrhosis (median RFS:63 VS. 424 days, P < .01; median RFS:61 VS. 355 days, P < .01). When patients executed TACE after operation, the status of high-frequency mutations also predicted shorter survival times (median RFS:57 VS. 424 days, P < .01). These results had been shown in .
Figure 2. Long-term survival outcomes in HCC with mutation frequency using Kaplan-Meier’s analysis:A. Survival curve of mutation frequency in patients with AFP<400. The results shown that the patients with MD/TD≥67 had worse survival time; B. Survival curve of mutation frequency in patients with Tumor thrombus. The results shown that the patients with MD/TD≥67 had worse median survival time; C. Survival curve of mutation frequency in patients without liver cirrhosis. The results shown that patients with MD/TD≥67 had worse median survival time; D. Survival curve of mutation frequency in patients who treated by Post-operation TACE. The results shown that patients with high MD/TD had worse survival time; E. Survival curve of mutation frequency in patients with Single lesion. The results shown that MD/TD≥67 result had worse median survival time. P value was assessed using the log-rank test.
Based on multivariable Cox regression analysis, we found that high-frequency mutations in TP53 gene detected in exoDNA (MD/TD ≥67%) was associated with poor prognosis (hazard ratio [HR] = 4.61; 95% CI: 1.71–12.48; P = .003). In addition, we also found that MVI (Positive), postoperative tace (negative), and tumor size (>5 cm) were all independently associated with poor prognoses (). Furthermore, we also found when the patients with different status, such as AFP (<400 ng/mL), Liver cirrhosis (Negative), Tumor thrombus (Negative), Tumor numbers (Single) and TACE (Post-operation), high-frequency mutations was associated with poor prognosis ().
Table 3. Multivariate Cox’s Proportional Hazards Model Assessing Factors Associated with RFS.
Table 4. Multivariate Cox’s Proportional Hazards Model Assessing Mutation frequency Associated with RFS in different Clinical characteristics.
Discussion
ExoDNA is considered as stable cargo in exosomes and contained the mutations derived from cancers.Citation19 These mutations that detected in random exoDNA fragments spanning multiple chromosomes and could be mimicked as “peripheral reservoir”.Citation20 Recently, exosomal DNA had been considered as one of the most prospective liquid biopsy in oncology research.Citation21 Many studies had found that the correlation between exoDNA and clinical outcomes in several cancers.Citation22 However, no studies thus far tried to explore its application value in HCC patients. Therefore, we used ddPCR to detect TP53 mutation in circulating exoDNA of HCC patients and explored the possibility of circulating exoDNA as a new noninvasive liquid biopsy method in prognosis of HCC.
In our study,80.0% patients had been detected the TP53 mutations in exoDNA. The detection efficiency was immensely higher than that was used in cfDNA.Citation16This result should be attributed to longer DNA fragments which were protected by exosomes in peripheral circulation. The total number of circulating DNA fragments <1.0%,Citation23,Citation24 and this made traditional technology difficult to detect. This was reason why cfDNA was considered as poor robustness using for HCC diagnosis.Citation25 Targeted sequencing of circulating exoDNA had been applied to a variety of tumors. For example, Allenson et al. detected KRAS mutations in 66.7% (22/33), 80% (12/15) and 85% (17/20) of patients with localized, locally advanced and metastatic pancreatic cancer, respectively.Citation9 It was similar to our sensitivity detection results.
In addition, our results shown that TP53 mutation in exoDNA was associated with MVI. It is easy to understand because the exosome secretion could be increased when microvascular invasion was presented. That means we can use the detectable tumor-associated mutations in exoDNA to predict vascular invasion status in patients indirectly. Wang et al. reported that circulating tumor DNA correlated with MVI and could use to predict tumor recurrence of HCC,Citation17 this results were similar to ours. Futhermore, our results shown that there had no significantly correlation between mutations in exoDNA and other clinicopathological features such as Satellite nodules, tumor number, and AFP. It confirmed that the relevance between detectable tumor-associated mutations in exoDNA and MVI was uniqueness. This indicated that exoDNA had wide application prospect.
The greatest contribution of our study was that we found the correlation between exoDNA and poor prognosis in HCC patients. In our study, we found that the patients with high-frequency mutations in exoDNA had poor prognosis compared with patients with low-frequency mutations (P < .01), even with good clinicopathologic characteristics. Furthermore, although patients suffered single tumor, or without tumor thrombus and liver cirrhosis, the prognosis was poor if high-frequency mutations in exoDNA was existed. That means, the frequency mutations detected in exoDNA might be an independent risk factor for prognosis in HCC. This phenomenon might be the result for the burden of tumor. Normally, exoDNA was DNA fragments originating from either normal physiologic tissues or unnormal pathological tissues as a result of DNA accumulation.Citation10 The equilibrium would be broken if the tumor-derived exosomes secretion was increased, and this would be reflected in the detection of high-frequency mutations in exoDNA. This is the reason why patients with high-frequency mutations in exoDNA would suffer poor prognosis.
In order to improve the detection efficiency of mutations in exoDNA, we used ddPCR described by Huggett JF.Citation26 This method could provide higher sensitivity and precision for discrimination of rare mutant variants in exoDNA, and was extensive used.Citation27
We anticipated that exoDNA would provide beneficial information for the prognosis of personalized HCC therapy. The detection of circulating exoDNA might enable further development of precision medical technologies to realize prognosis evaluation of HCC and to customize personalized treatment strategies.
Methods
Sample collection
A total of 60 patients with primary HCC at the Second Affiliated Hospital of Nanchang University were enrolled between October 2018 and August 2020. All patients agreed to the analysis of exoDNA mutations in collected blood before operation. Pathology was used to diagnose HCC. HCC patients were eligible if they agreed to have their blood collected for detection of exoDNA before surgery. None of the HCC patients received preoperative cancer-related therapies. Written informed consent was obtained from all patients enrolled in the study. Blood samples were collected in EDTA tubes, and within 1 h, samples were centrifuged at 3000 × g for 20 min. Serum samples were stored at −80°C. This study was approved by the Ethics Review committee of the Second Affiliated Hospital of Nanchang University.
Exosome extraction and exoDNA extraction
Exosomes were extracted from serum samples using the ExoQuick™ kit (System Bioscience, CA, USA) following the manufacturer’s instructions. We examined the exosomes by electron microscopy and detection of the exosome-specific protein CD63. Protein expression of CD63 was assessed by western blotting and three experiments were repeated to verify the presence of CD63. ExoDNA was extracted from exosomes using the ExoDNA™ Extraction kit (BioVision Inc, SF, USA) following the manufacturer’s instructions. We used this method to extract exosome DNA and the presence of DNA was confirmed in all samples.
Study design and droplet digital PCR (ddPCR)
DdPCR (also known as single-molecule PCR) generally consists of two steps: PCR amplification and fluorescence signal analysis. Our experimental procedures followed the guidelines proposed for reporting digital PCR data.Citation26 The PCR primers and probe sequences were designed using Primer Premier 5.0 software. Each PCR reaction contained 10 μl ddPCR Supermix for Probes (Bio-Rad, USA), 3.6 μl of primer (Sangon Company, China), 1 μl of probe (Sangon Company), 2 μl of template DNA from exoDNA and 3.4 μl of ddH2O to give a total volume of 20 μl. The PCR conditions were 96°C for 10 min; 40 cycles of 94°C for 30 s and 60°C for 60s, with a final incubation at 98°C for 10 min.
Preparation and analysis of droplets
We added a new DG8 Cartridge (BioAssay Systems, CA, USA) into the holder and added 20 μL of the samples into eight holes in the middle row. Next, 70 μL drop-generated Oil (DG Oil) was added into the bottom row of eight cartridges in the same chamber. The gasket was covered and the holes on both sides were firmly hooked. The holder was placed in the QX200 droplet generator (Bio-Rad, CA, USA) for generation of droplets, which generally took about 2 min to complete. Droplets were produced in the top row of cartridges and subsequently transferred into a 96-well plate. The plate was placed on a heat-sealing instrument and covered with a heat-sealing membrane for sealing. The QX200 Droplet analyzer was used for droplet analysis and detection, and the data were uploaded to the computer for final analysis. After the reactions were completed, the threshold line was adjusted to the appropriate position according to the specific reaction conditions for result interpretation. The 5’ primer ends of mutant and wild-type genes were labeled with the fluorescent dyes FAM (blue) and VIC (green), respectively, to assess the mutation status of the genotyped PCR product with the QX200 Droplet analyzer. Red fluorescence indicated that FAM and VIC were simultaneously detected.
Statistical analysis
We used Stata software (version 16.0; Stata Corporation LP; College Station, TX, USA) to perform statistical analyses. Logistics regression analysis was used to assess the correlation between mutation status and the clinical and pathologic characteristics. Recurrence-free survival (calculated as the time from operation to tumor recurrence) curves were plotted according to Kaplan–Meier method and assessed using the log-rank test. Independent predictors of RFS were determined by Cox proportional hazard regression. Results were considered statistically significant if the P-value was less than 0.05.
Conclusions
Our results provided evidence that the mutations in exoDNA might be used to predict patients with shorter recurrence-free survival.
Authors’ contributions
Research design and project supervision: WJ L
Data collection, statistical analysis and writing: YL, JJ W, EL L
Quality control of data and algorithms: ZQ X, J L, XB Y
Data analysis and interpretation: DD H, YL M, F Z
Literature review and manuscript proofreading: LQ W
Ethics approval and consent to participate
This study was approved by the Ethics Review committee of The Second Affiliated Hospital of Nanchang University.
Supplemental Material
Download Zip (1.2 MB)Acknowledgments
We thank Sangon Biotech Company (Shanghai, China) for technical assistance with ddPCR technology. We thank Gabrielle White Wolf, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384047.2022.2094666
Additional information
Funding
Notes on contributors
Yong Li
Yong Li: Graduate from Nanchang University.
Junjun Wu
Junjun Wu: Graduate from Nanchang University.
Enliang Li
Enliang Li: MD, Graduate from Zhejiang University.
Zhouqing Xiao
Zhouqing Xiao: Graduate from Nanchang University.
Jun Lei
Jun Lei: MD, Graduate from HUST.
Fan Zhou
Fan zhou: MD, Graduate from Sun Yat-Sen University.
Xiangbao Yin
Xiangbao Yin: MD, Graduate from Sun Yat-Sen University.
Dandan Hu
Dandan Hu: MD, Graduate from PUMC.
Yilei Mao
Yilei Mao: Professor, Graduate from harvard university.
Linquan Wu
Linquan Wu: Professor, Graduate from Zhejiang University.
Liao Wenjun
Wenjun Liao: MD, Graduate from PUMC.
References
- Altekruse SF, Mcglynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27(9):1485–1491. doi:10.1200/JCO.2008.20.7753.
- Molla MD, Dessie G, Akalu Y, Ayelign B. Hepatocellular expression of SIRT1 and its effect on hepatocellular carcinoma progression: a future therapeutic perspective. Int J Hepatol. 2020;2020:1–10. doi:10.1155/2020/2374615.
- Benowitz S. Liver cancer biomarkers struggling to succeed. J Natl Cancer Inst. 2007;99:590–591. doi:10.1093/jnci/djk174.
- Zvereva M, Roberti G, Durand G, Voegele C, Calvez-Kelm FL, Delhomme TM, Chopard P, Fabianova E, Adamcakova Z, Holcatova I. Circulating tumour-derived KRAS mutations in pancreatic cancer cases are predominantly carried by very short fragments of cell-free DNA. EBioMedicine. 2020:102462. doi:10.1016/j.ebiom.2019.09.042
- Lee J, Ji HK, Sun HK, Yoo HM. 2020. Improvement of digital PCR conditions for direct detection of KRAS mutations. J Clin Lab Anal. 34. DOI:10.1002/jcla.23344
- Schwarzenbach H, Hoon D, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer Clin Oncol. 2011;1:426.
- Jahr S, Hentze H, Englisch S, Hardt D, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665.
- Sharma A, Johnson A, Chen S, Li Y, Hu H, Cai Y, Hou L. Exosome DNA: critical regulator of tumor immunity and a diagnostic biomarker. J Cell Physiol. 2020:235. doi:10.1002/jcp.29837
- Allenson K, Castillo J, Lucas FS, Scelo G, Kim DU, Bernard V, Davis G, Kumar T, Katz M, Overman MJ, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017;28:741–747. doi:10.1093/annonc/mdx004.
- Bakhoum SF, Ngo B, Laughney AM, Cavallo J-A, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553(7689):467–472. doi:10.1038/nature25432.
- Yang S, Che S, Kurywchak P, Tavormina JL, Gansmo LB, Correa de Sampaio P, Tachezy M, Bockhorn M, Gebauer F, Haltom AR, et al. Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biology & Therapy. 2017;18(3):158–165. doi:10.1080/15384047.2017.1281499.
- Bernard V, Dong UK, Lucas F, Castillo J, Alvarez HA, Mulu Feven C, Stephens Bret M, Huang Jonathan, Semaan Alexander, Guerrero Paola A, Kamyabi Nabiollah, Zhao Jun, Hurd Mark W, Koay Eugene J, Taniguchi Cullen M, Herman Joseph M, Javle Milind, Wolff Robert, Katz Matthew, Varadhachary Gauri, Maitra Anirban et al. Circulating nucleic acids associate with outcomes of patients with pancreatic cancer. Gastroenterology. 2018;156(1):108–118.e4.
- Liu Y, Wang Y, Lv Q, Li X. Exosomes: from garbage bins to translational medicine. Int J Pharm. 2020;583:119333. doi:10.1016/j.ijpharm.2020.119333.
- Jin Y, Chen K, Wang Z, Wang Y, Liu J, Lin L, Shao Y, Gao L, Yin H, Cui C, et al. DNA in serum extracellular vesicles is stable under different storage conditions. BMC Cancer. 2016;16(1):753. doi:10.1186/s12885-016-2783-2.
- Qu X, Li Q, Yang J, Zhao H, Xiao W, Zhang F, Zhang S, Zhang H, Wang R, Wang Q. Double-stranded DNA in exosomes of malignant pleural effusions as a novel DNA source for EGFR mutation detection in lung adenocarcinoma. Front Oncol. 2019;9:931. doi:10.3389/fonc.2019.00931.
- Liao W, Yang H, Xu H, Wang Y, Ge P, Ren J, Xu W, Lu X, Sang X, Zhong S. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget. 2016;7(26):40481–40490. doi:10.18632/oncotarget.9629.
- Wang J, Huang A, Wang YP, Yin Y, Zhou J, Zhang X, Zhou J. Circulating tumor DNA correlates with microvascular invasion and predicts tumor recurrence of hepatocellular carcinoma. Ann Transl Med. 2020;8:237. doi:10.21037/atm.2019.12.154.
- Shen T, Li SF, Wang JL, Zhang T, Zhang S, Chen H-T, Xiao Q-Y, Ren W-H, Liu C, Peng B, et al. TP53 R249S mutation detected in circulating tumour DNA is associated with prognosis of hepatocellular carcinoma patients with or without hepatectomy. Liver Int. 2020;40:2834–2847. doi:10.1111/liv.14581.
- Chennakrishnaiah S, Meehan B, D’Asti E, Montermini L, Rak J, Karatzas N, Buchanan M, Tawil N, Choi D, Divangahi M. Leukocytes as a reservoir of circulating oncogenic DNA and regulatory targets of tumorヾerived extracellular vesicles. J Thrombosis Haemostasis. 2018;16(9):1800–1813. doi:10.1111/jth.14222.
- Wang L, Li Y, Guan X, Zhao J, Shen L, Liu J. Exosomal double-stranded DNA as a biomarker for the diagnosis and preoperative assessment of pheochromocytoma and paraganglioma. Mol Cancer. 2018;17(1):128. doi:10.1186/s12943-018-0876-z.
- Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. doi:10.1038/nature14581.
- Lino M, Huang, Helen HJ, Hong DS, Tsimberidou AM, Fu S, Piha-Paul SA, Subbiah V, Karp DD, Naing A, et al. Liquid biopsies using plasma exosomal nucleic acids and plasma cell-free DNA compared with clinical outcomes of patients with advanced cancers. Clin Cancer Res. 2017;24(1):181–188. doi:10.1158/1078-0432.CCR-17-2007.
- Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. doi:10.1038/nm.1789.
- Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–586. doi:10.1200/JCO.2012.45.2011.
- Liao W, Mao Y, Ge P, Yang H, Xu H, Lu X, Sang X, Zhong S. Value of quantitative and qualitative analyses of circulating cell-free DNA as diagnostic tools for hepatocellular carcinoma: a meta-analysis. Medicine. 2015;94(14):e722. doi:10.1097/MD.0000000000000722.
- Huggett JF, Foy CA, Benes V, Emslie K, Garson Jeremy A, Haynes Ross, Hellemans Jan, Kubista Mikael, Mueller Reinhold D, Nolan Tania, Pfaffl Michael W, Shipley Gregory L, Vandesompele Jo, Wittwer Carl T, Bustin Stephen A et al. The digital miqe guidelines: minimum information for publication of quantitative digital PCR experiments. Clin Chem. 2013;59(6):892–902.
- Wang ZY, Wang RX, Ding XQ, Zhang X, Tong JH. A protocol for cancer-related mutation detection on exosomal DNA in clinical application. Front Oncol. 2020;10:558106. doi:10.3389/fonc.2020.558106.
