ABSTRACT
Mounting evidence has demonstrated that fatty acid binding protein 5 (FABP5) is commonly upregulated in many human malignancies. However, the mechanisms explaining the involvement of FABP5 in hepatocellular carcinoma (HCC) remain unclear. In this study, we demonstrated the involvement of FABP5 and its downstream signaling molecules in HCC progression. We first confirmed that FABP5 expression was upregulated in HCC. Additionally, FABP5 promoted HCC cells proliferation, migration, and invasion. Mechanistic investigation showed that FABP5 could improve cAMP-response element binding protein (CREB) phosphorylation. Meanwhile, CREB, as a transcription factor, upregulated the miR-889-5p expression by binding to the miR-889-5p promoter region. Consequently, miR-889-5p led to downregulation of Krüppel-like factor 9 (KLF9) by binding to the 3ʹ-UTR of the KLF9 mRNA, potentiating the PI3K/AKT signaling pathway and promoting the proliferation, migration, and invasion of HCC cells. Our findings have identified a FABP5/CREB/miR-889-5p/KLF9 axis for HCC progression, and we postulate that blocking this key signaling pathway may represent a promising strategy for HCC treatment.
Introduction
The occurrence and development of liver cancer is a complex process involving multi-factor, multi-stage, and multi-gene mutation accumulation. It is of utmost importance in the prevention and treatment of liver cancer to identify certain key genes in liver cancer. Currently, a critical challenge is distinguishing key molecules that confer a selective advantage to tumorigenesis from the random alterations due to the inherent genomic instability, gene linkage, and spontaneous mutagenesis.Citation1,Citation2 In our previous study, we found that some genes in the differentially expressed gene profile of human HCC were shared by HCC tissues from other animals with liver cancer.Citation3 Therefore, we speculate that it is possible to identify key genes that affect the development of liver cancer by exploring common gene changes between human HCC and other species. Hence, we carried out a strategy of cross-species comparative oncogenomics by collecting the differentially expressed genes in the liver cancer tissues of tree shrews (Tupaia belangeri) and the differentially expressed proteins in the liver cancer tissues of rats, combined with the Gene Expression Omnibus (GEO) database (including humans and mice).Citation4,Citation5 Among the different potential targets, the epidermal fatty acid binding protein 5 (FABP5), also known as E-FABP, attracted our attention.
FABP5, a member of the fatty acid binding protein family, is a highly conserved small molecule protein derived from epidermal cells and is located in the cytoplasm.Citation6 As a carrier of hydrophobic ligands (including retinoic acids and various long-chain fatty acids), it not only participates in the binding, transport, storage, and metabolism of long-chain fatty acids, which provide energy and raw materials for cell growth but also affects signal transduction in cell growth.Citation7,Citation8 Thus far, FABP5 has been confirmed as an oncogene in prostate cancer, gliomas, breast cancer, colorectal cancer, and so on.Citation9–12
Mounting evidence has recently revealed the molecular mechanism by which FABP5 promotes tumor development. Researches have shown that FABP5 may be involved in tumorigenesis by influencing lipid metabolism, including promoting lipid droplet decomposition, enhancing de novo fatty acid synthesis pathways, and upregulating the expression of key enzymes involved in lipid metabolism and lipolysis.Citation13,Citation14 FABP5 enhances lipid reprogramming by promoting hypoxia-inducible factor (HIF-1) synthesis, preventing the interaction of factor-inhibiting hypoxia-inducible factor (FIH)/HIF-1, and improving HIF-1 activity in liver cancer cells.Citation15 Additionally, FABP5 can regulate tumor progression by activating key signaling pathways. In prostate cancer, FABP5 activates the NF-κB signaling pathway by increasing reactive oxygen species (ROS) and protein kinase C production and induces inflammation and the production of IL-6 and IL-8.Citation16 FABP5 can promote angiogenesis in HCC by activating the IL6/STAT3/VEGFA pathway.Citation17 However, the specific molecular mechanism by which FABP5 regulates the proliferation and migration of hepatoma cells has not been clearly defined, and it needs to be further elucidated. Therefore, this study aimed to investigate the role and potential molecular mechanisms of FABP5 in HCC.
Materials and methods
Patients and tissue samples
48 HCC tissues and corresponding adjacent normal tissues from Guangxi Medical University Cancer Hospital between January 2018 and February 2019 were collected. All HCC patients, who received no pre-operative radiotherapy, chemotherapy and surgical treatment, excluding other tumors and serious psychosomatic diseases, were initially diagnosed as hepatocellular carcinoma by pathology after operation. Tumor staging was classified into stages A, B, C, and D following the Barcelona Clinic Liver Cancer classification criteria.Citation18 This study was approved by the Ethical Committee of Guangxi Medical University Cancer Hospital (No. LW2021086). All participants signed a written informed consent.
Cell culture
In the previous experiment, the expression of FABP5 mRNA was verified in four kinds of HCC cells, including HepG2, Huh7, SK-Hep-1, Hep3B cells. It was found that the mRNA expression level of FABP5 was higher in HepG2 and SK-Hep-1, which was used for this study. The HCC cell lines HepG2 and SK-Hep-1 were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) medium containing 10% fetal bovine serum (FBS) and 1% antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin) and incubated at 37°C with 5% CO2。
Lentiviral transfection
FABP5-knockdown, FABP5-overexpressed, and Krüppel-like factor 9 (KLF9)-knockdown and their corresponding control lentiviruses were constructed by Hanyin Biotechnology Limited Company (Shanghai, China). The shRNA target sequences are listed in Supplementary Table 1. HCC cells were seeded into 6-well plates, and 10 μL of the lentivirus with polybrene (with a final concentration of 5 g/ml) were added per well. The infected cells were selected by Puromycin (2 μg/ml) after 48 h of transduction.
SiRNA and vector infection
miR-889-5p mimic, siRNAs of NF-κb, c-Myc, cAMP-response element binding protein (CREB), and the corresponding negative controls were synthesized by RiboBio (Guangzhou, China). These were transfected into HCC cells using Lipofectamine 3000 (Invitrogen, Cat. no. L3000015) according to the manufacturer’s protocol. miR-889-5p mimic and the siRNAs sequences are listed in Supplementary Table 2.
RT-qPCR
Total RNA extracted by TRIzol reagent (Invitrogen, Cat. no. 15596026) was reversely transcribed into cDNA using a PrimeScript™ RT Reagent kit (Takara, Cat. no. RR037A) according to the manufacturer’s guidelines. qPCR was performed using the SYBR® Premix Ex Taq kit (Takara, Cat. no. RR820A). GAPDH and U6 were used as the reference genes for mRNA and miRNA respectively. The results were analyzed using the 2-ΔΔCT method.Citation19 Primer sequences used in this study are listed in Supplementary Table 3.
Western blot analysis
The total protein was extracted from the prepared cells and tissues by radioimmunoprecipitation assay (RIPA) lysis buffer containing 1% phenylmethylsulfonyl fluoride (1 mM), followed by the quantification. 50 μg of the assigned protein was separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and loaded on polyvinylidene fluoride membranes. Nonspecific antigens were blocked in 5% skimmed milk for 1 h. Subsequently, the membranes were incubated overnight at 4°C with the different primary antibodies as follows: anti-FABP5 rabbit polyclonal antibody (Abcam, Cat. no. ab84028), anti-KLF9 rabbit polyclonal antibody (Abcepta, Cat. no. Ap16249a), anti-PI3K rabbit polyclonal antibody (Cell Signaling Technology, Cat. no. 3811), anti-Phospho-PI3K rabbit polyclonal antibody (Cell Signaling Technology, Cat. no. 13857), anti-AKT rabbit monoclonal antibody (Cell Signaling Technology, Cat. no. 4685), anti-Phospho-AKT rabbit monoclonal antibody (Abways, Cat. no. cy6569), anti-mTOR rabbit monoclonal antibody (Cell Signaling Technology, Cat. no. 2986), anti-Phospho-mTOR rabbit monoclonal antibody (Cell Signaling Technology, Cat. no. 5536), anti-CREB rabbit monoclonal antibody (Cell Signaling Technology, Cat. no. 9197), anti-Phospho-CREB rabbit monoclonal antibody (Cell Signaling Technology, Cat. no. 9198), anti-β-actin mouse monoclonal antibody (Santa Cruz, Dallas, Cat. no. sc-8432), and anti-GAPDH mouse monoclonal antibody (Abcam, Cat. no. ab59164).
Cell viability
The transfected cells were harvested after transfection for 48 hours and seeded into a 96-well plate. At 0, 24, 48, 72, and 96 h after cell attachment, 10% Cell Counting Kit (CCK)-8 solution (Dojindo, Cat. no. CK04) was added. Cells were incubated at 37°C with 5% CO2 for 1 h. Absorbance was measured at 450 nm using a microplate reader (Berthold).
Transwell assay
Transwell assay was performed to detect cell migration and invasive ability. For the invasion assay, the transwell chamber was coated with Matrigel® (BD Biosciences, Cat. no. 354480). 105 cells were added to the upper chamber, followed by incubation for 24 h. The migratory cells on the lower surface of the insert were fixed in 4% paraformaldehyde and methanol, and stained with crystal violet.
Cell cycle assay
106 cells for each group were harvested and washed twice with cold phosphate-buffered saline (PBS). Then, the cells were fixed with 70% ethanol overnight and stained with propidium iodide (PI; Multi Science, Cat. no. 70-AP105-60) solution containing RNase A (10 μg/ml) for 30 min. The cells were analyzed on a FACSCalibur™ flow cytometer (BD Biosciences).
Apoptosis assay
The collected cells were stained with 20 μl of PI and 20 μl of fluorescein isothiocyanate (FITC; Multi Science, Cat. no. 70-CCS012) for 20 min at room temperature. Apoptotic cells were analyzed on a FACSCalibur™ flow cytometer (BD Biosciences).
Dual-luciferase assay
The full-length KLF9 UTR (wild-type or mutant sequence) and miR-889-5p promoter (wild-type or mutant sequence) were cloned into the psiCHECK-2 vector (GenePharma). 293 T cells were then co-transfected with the miR-889-5p mimic and wild-type KLF9 or mutant KLF9 luciferase reporter plasmids to study the interaction between miR-889-5p and KLF9. They were also co-transfected with the CREB overexpression plasmid and wild-type miR-889-5p promoter or mutant miR-889-5p promoter to study the interaction between CREB and miR-889-5p. After 48 h of incubation, the collected cells were lysed. Luciferase activity was detected using a dual-luciferase assay kit (Promega, Cat. no. E1910) according to the manufacturer’s protocol.
Xenograft tumor growth model
The Ethical Committee of Guangxi Medical University Cancer Hospital approved the animal experiments (No. LW2021087). All experimental procedures and animal care were conducted in accordance with institutional ethics guidelines. A total of 12 BABL/c nude mice, 5 weeks old, weighing 18–20 g, were randomly divided into sh-negative control (NC) group (transfected cells with negative control empty lentiviral particles), and shFABP5 group (transfected cells with lentiviruses carrying shRNA to inhibit FABP5). Approximately 1 × 106 SK-Hep-1 cells were injected into the right axilla (0.5 cm) of the mice. The longest and shortest diameter of the tumor were measured and the tumor volume was calculated every week. Four weeks after injection, the nude mice were euthanized, and the xenograft tumors were harvested and weighed.
Immunohistochemistry (IHC) assay
Xenograft tumors were fixed in 4% paraformaldehyde for 48 h. The fixed tumors were then dehydrated, embedded in paraffin, and cut into 5 μm sections. The sections were incubated at 4°C overnight with rabbit anti-human FABP5 primary antibody (Proteintech, cat. no. 12348-1-AP) and then with goat anti-rabbit secondary antibody (Proteintech, cat. no. PR30009) for 50 min at room temperature. Ten random fields of positive cells were examined at 200X magnification.
Statistical analysis
Statistical analyses were performed using the SPSS software (v19.0, IBM Inc., Armonk, NY, USA). Continuous variables were presented as mean ± standard deviation. The differences between two groups were verified using the Student’s t-test. The Kruskal-Wallis test and Mann-Whitney U test were performed for group comparisons. Categorical variables were compared using the χ2 test or Fisher’s exact test. Univariate survival analysis was performed using the Kaplan-Meier method. Correlational analysis was performed using Pearson’s correlation coefficient. In this study, p < .05 was defined as a significant difference.
Results
FABP5 was upregulated in HCC samples
FABP5 expression in HCC tissues was significantly higher than that in adjacent normal tissues from the TCGA data set using GEPIA database (http://gepia.cancer-pku.cn) (). We examined FABP5 expression in 48 paired HCC tissues and adjacent normal tissues using qPCR and consistently found that it was significantly upregulated in the tumor tissues (). The results were further confirmed by western blotting (). The median level of FABP5 protein in the 48 HCC tissues was calculated and divided into high (n = 24) and low (n = 24) FABP5 expression groups. By analyzing the clinicopathological characteristics and clinical outcomes of the 48 HCC patients, we discovered that the expression of FABP5 was closely related to tumor staging and postoperative metastasis (, p < .05). In addition, Kaplan-Meier curves demonstrated a poor prognosis in HCC patients in the high FABP5 expression group (from TCGA data set) (). Taken together, FABP5 is a critical gene involved in the occurrence and development and of HCC.
Figure 1. FABP5 expression was upregulated in HCC. a, expression of FABP5 in HCC samples (N = 369) and normal samples (N = 160) from the TCGA data set. b, relative expression of FABP5 mRNA in HCC tissues compared with adjacent normal tissues were detected by RT-qPCR. c, relative expression of FABP5 protein in HCC tissues and adjacent normal tissues were detected by western blotting. Representative images of FABP5 expression in 48 paired HCC tissues(t) and adjacent normal tissues (n). d, the expression of FABP5 was negatively correlated with overall survival time in HCC patients from the TCGA data set. *P < .05 and ***P < .001 as compared with the vehicle control.
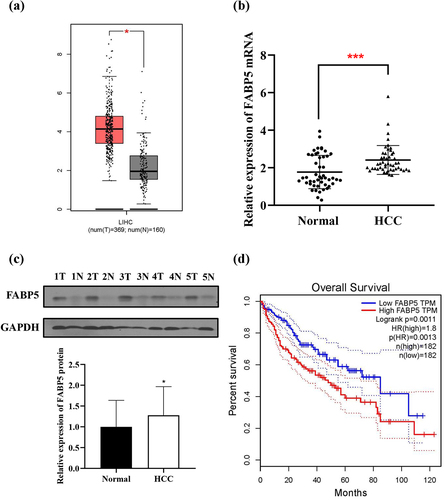
Table 1. Correlation between FABP5 expression and clinicopathological characteristics
Knockdown of FABP5 suppresses proliferation, migration, and invasion of HCC
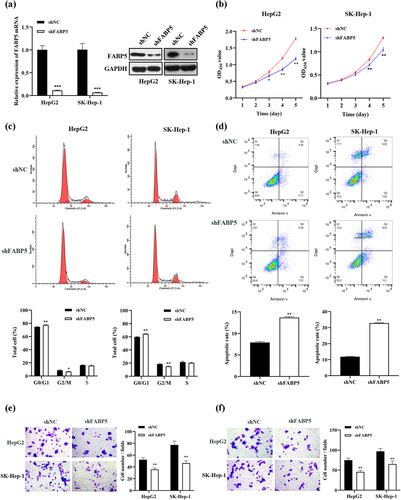
To investigate the role of FABP5 in HCC progression, HepG2 and SK-Hep-1 cells were transfected with FABP5 shRNA, and FABP5 expression was verified. These cells were successfully transfected with FABP5 shRNA, as indicated by the significantly lower levels of FABP5 mRNA in the shFABP5 group compared with their respective negative controls (P < .001). Western blot analysis revealed that FABP5 protein expression dramatically decreased in HepG2 and SK-Hep-1 cells after transfection (). As demonstrated in , the viability of these cell lines was significantly reduced in the shFABP5 group when compared with the shNC group on days 4 and 5 following transfection (HepG2 cells at days 3, 4, and 5 following transfection). To further explore the mechanisms underlying the effects of FABP5 on HCC cell proliferation, flow cytometry was used to analyze cell cycle distribution and apoptosis in FABP5 knockdown and NC groups. As shown in , the cell population in the G0/G1 phase increased compared to that in the NC group after transfection with FABP5 shRNA. Additionally, the G2/M phase ratio of the shFABP5 cells was lower than that in the shNC group. This indicated that FABP5 silencing led to cell cycle arrest at the G0/G1 phase in HepG2 and SK-Hep-1 cells. Next, we explored the effects of FABP5 on apoptosis. Compared with the control cells, the FABP5 knockdown group showed a significantly increased apoptotic proportion, indicating that FABP5 knockdown can induce apoptosis in HCC cells (). These data indicated that FABP5 increased the proliferation of tumor cells by promoting cell division and inhibiting apoptosis. Transwell assays were used to investigate the effects of FABP5 on the migration and invasion of HCC cells. As indicated in ), our results showed that the migration and invasion of HepG2 and SK-Hep-1 cells with FABP5 knockdown was significantly lower than that of the vector-transfected control group. FABP5 silencing significantly inhibited the migration and invasive abilities of HCC cells. These data confirmed that the FABP5 knockdown significantly inhibited the proliferation, migration, and invasion of HepG2 and SK-Hep-1 cells.
Figure 2. FABP5 knockdown inhibited cell growth, migration and invasion in HCC cells. a, the mRNA and protein expression levels of FABP5 were analyzed in FABP5-knockdown HepG2 and SK-Hep-1 cells by RT-qPCR and western blotting. b, the growth curve of HCC cells was determined by CCK-8 assay. c, PI fluorescence pattern was applied for cell-cycle distribution in HCC cells transfected with shFABP5 or shNC. d, HCC cells transfected with shFABP5 or shNC were stained with Annexin-V FITC and PI to be measured apoptosis. Apoptotic cells are presented in the right-lower (Q3, early apoptosis) and right-upper (Q2, late apoptosis) quadrants of the plots. e, transwell assay revealed the migration abilities of HCC cells transfected with shFABP5 or shNC. f, transwell assay revealed the invasive abilities of HCC cells transfected with shFABP5 or shNC. Results are presented as mean ± S.E.M. (N = 3). Results were averaged from three independent experiments and presented as percentage of control levels. *p < .05, **p < .01 and ***p < .001 as compared with the vehicle control. FITC, fluorescein isothiocyanate; PI, propidium iodide.
Suppression of FABP5 inhibits tumor growth in vivo
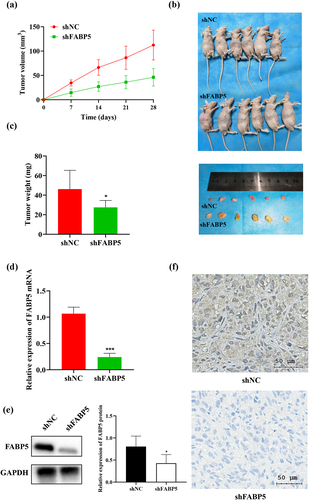
SK-Hep-1 cells successfully transfected with stable shFABP5 RNA were injected into the BALB/c mice to create a xenograft tumor model. Mice were assigned to the shFABP5 and shNC groups. We found that tumors grew more rapidly in the shNC groups, as shown in . The mice were sacrificed on the 28th day after injection. Tumors from the shNC group were larger and heavier than in the FABP5 shRNA group, as shown in ). We performed RT-qPCR, IHC staining, and western blot analysis to evaluate FABP5 expression at the transcriptional and translational levels. Compared to the shNC groups, FABP5 mRNA and protein expression in the FABP5 shRNA group was significantly decreased (P < .001) . Representative images of the immunohistochemical staining in shNC and shFABP5 groups were shown in .
Figure 3. Decreased expression of FABP5 inhibited the growth of HCC cells in vivo. a, HepG2 cells and HepG2 cells stably transfected with shFABP5 RNA or shNC were implanted into nude mice by subcutaneous injection. Tumor volume was measured each 7 days by a calliper until the end of the experiment. The tumor growth curve was drawn. b, the mice and the tumors of different groups on the 28th day after injection. c, the weight of each xenograft tumor was measured at the end of the experiment. d, the differences in FABP5 mRNA levels between the shFABP5 group and the control groups were detected by RT-qPCR. e, the protein levels of FABP5 in the shFABP5 group and the control groups were measured by western blotting. f, the tumors were analyzed by immunohistochemical staining with anti-FABP5. Original magnification: 400 × . ***p < .001 as compared with the vehicle control.
The KLF9 gene was the downstream target of FABP5
In order to better understand the molecular mechanisms by which FABP5 regulates the progression of HCC, we analyzed potential downstream target genes of FABP5 using transcriptional profiling of FABP5 knockdown SK-Hep-1 cells compared with the control group. A total of 832 genes were identified as having significant changes (FC > |2|; P < .01). Among these, 432 genes were upregulated, and 400 genes were downregulated in the SK-Hep-1 cells following FABP5 knockdown. An interactive heat map shows the abundance of these differentially expressed (DE) genes (). According to Gene Ontology (GO) analysis, the DE genes were mainly enriched in epidermal growth factor receptor binding, neurotrophin binding, and other related molecular functions, as well as in the regulation of platelet activation, monocyte differentiation, and other biological processes, as shown in . To validate the microarray results, we selected ten DE genes that were previously reported to be related to tumors, as shown in , and verified their expression using RT-qPCR in SK-Hep-1 and HepG2 cells before and after FABP5 knockdown. We found that four genes in both methods (microarray analysis and PCR) showed similar patterns for the selected genes ( and Supplementary Figure 2). Of the genes analyzed, KLF9 attracted our attention. It has been reported that KLF9 regulates diverse biological processes, including cell proliferation, differentiation, migration, and apoptosis. It is also closely associated with the occurrence and development of tumors.Citation20 In this study, KLF9 was significantly upregulated in the SK-Hep-1 and HepG2 cells upon FABP5 knockdown. We examined KLF9 expression in 48 HCC tissues using RT-qPCR and western blot analysis. KLF9 was downregulated in tumor tissues compared with matched adjacent normal tissues () and Supplementary Figure 1B). HCC patients with low levels of KLF9 mRNA expression showed low overall survival in TCGA data set (). FABP5 protein expression was negatively correlated with KLF9 expression (r = −0.310, p < .05, ). These results suggest that FABP5 may promote HCC development by regulating KLF9 expression.
Figure 4. The expression of KLF9 was negatively correlated with FABP5. a, gene microarray analysis were applied to examine the differential expression genes in SK-Hep-1 cells before and after FABP5 knockdown (FC > |2|; P < .01). b, gene ontology (GO) enrichment analysis of differential expression genes in accordance with the biological processes, cellular component, and molecular function. c, KLF9 mRNA expression in HepG2 and SK-Hep-1 cells with FABP5 knockdown determined by RT-qPCR. d, the KLF9 expression in HCC tissue and adjacent tissues were detected by RT-qPCR. e, relative expression of KLF9 protein in HCC tissues and adjacent normal tissues were detected by Western blotting. Representative images of KLF9 expression in 48 paired HCC tissues (t) and adjacent normal tissues (n). e, the expression of KLF9 was positively correlated with overall survival time in HCC patients from the TCGA data set. F, A negative correlation was found between the protein expression level of FABP5 and KLF9 in HCC samples. *p < .05, **p < .01, and ***p < .001 as compared with the vehicle control.
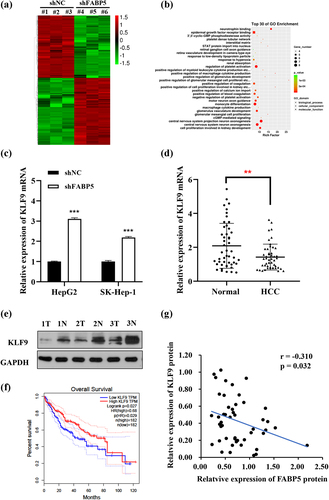
Table 2. Ten genes differentially expressed in the microarray selected and verified their expression by RT-qPCR.
FABP5 promoted HCC cell proliferation, migration, and invasion by inhibiting KLF9 in vitro
We hypothesized that KLF9 may mediate the function of FABP5 in suppressing HCC cell proliferation and metastasis. To test this hypothesis, rescue assays were conducted by co-transfection with shFABP5 with shKLF9 in SK-Hep-1 and HepG2 cells. We selected three KLF9 targets for the knockout experiment. Compared with the shFABP5 group, SK-Hep-1 and HepG2 cells co-transfected with shFABP5 and KLF9-sh1 showed significantly reduced KLF9 expression levels ). Thus, shFABP5 with KLF9-sh1 was selected for subsequent experiments. As shown in , suppression of KLF9 rescued the inhibition of cell proliferation induced by FABP5 knockdown. Transwell assays showed that attenuation of KLF9 reversed the effects of FABP5 silencing on the migration of HCC cells (). The invasion assay revealed that shKLF9 significantly rescued the inhibition of invasion induced by FABP5 silencing in HCC cells (). In addition, inhibition of KLF9 expression reversed cell cycle arrest and apoptosis induced by FABP5 knockdown. ). These results demonstrated that FABP5 promotes HCC progression by suppressing the expression of KLF9 in vitro.
Figure 5. KLF9 knockdown reversed the phenotypes caused by FABP5 knockdown in HCC cells. a-b, HepG2 and SK-Hep-1 cells upon co-transfection of shFABP5 with shKLF9 showed significantly reduced KLF9 expression at the mRNA and protein levels. c, CCK-8 assay showed that shKLF9 significantly reversed the inhibition of proliferation induced by FABP5 knockdown in HCC cells. d, transwell migration assay revealed that shKLF9 significantly rescued the migratory inhibition induced FABP5 knockdown in HCC cells. e, transwell invasion assay revealed that shKLF9 significantly rescued the inhibition of invasion induced by FABP5 knockdown in HCC cells. F, KLF9 knockdown reversed the cell cycle arrest caused by FABP5 knockdown in HCC cells. PI fluorescence pattern was applied for cell-cycle distribution. G, KLF9 knockdown reversed the apoptosis caused by FABP5 knockdown in HCC cells. HepG2 and SK-Hep-1 cells were stained with Annexin-V FITC and PI to be measured apoptosis. Apoptotic cells are presented in the right-lower (Q3, early apoptosis) and right-upper (Q2, late apoptosis) quadrants of the plots. Results are presented as mean ± S.E.M. (N = 3). * p < .05, ** p < .01, and ***p < .001 as compared with the vehicle control. Results were averaged from three independent experiments and presented as percentage of control levels. FITC, fluorescein isothiocyanate; PI, propidium iodide.
FABP5 suppresses KLF9 to potentiate PI3K/AKT signaling
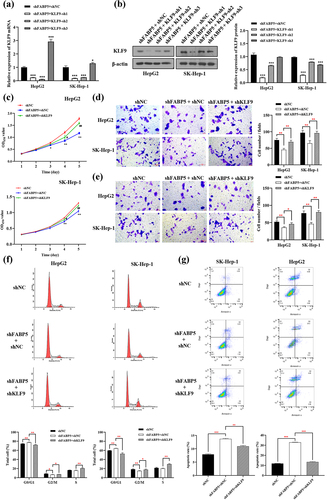
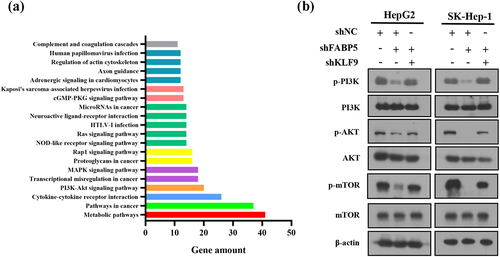
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DE genes revealed that PI3K/AKT was one of the top twenty differentially expressed signaling pathways (), suggesting that the PI3K/AKT signaling pathway may be involved in FABP5-induced cell proliferation and migration. To examine the potential involvement of PI3K/AKT signaling in FABP5-induced KLF9 downregulation, the phosphorylation levels of PI3K, AKT, and mTOR were measured by western blot analysis in stable HepG2 shFABP5 and SK-Hep1 shFABP5 clones. We found that phosphorylation of PI3K, AKT, and mTOR was decreased in FABP5-knockdown HCC cells and could be reversed by KLF9 gene silencing ( and Supplementary Figure 3). Taken together, these data suggest that reduced FABP5 expression inhibits KLF9 expression and the PI3K/AKT signaling pathway.
Figure 6. PI3K/AKT signaling pathway was involved in FABP5-induced KLF9 downregulation. a, KEGG pathway enrichment analysis of top 20 differentially expressed signaling pathways in FABP5 knockdown group compared with control group in SK-Hep-1 cells. b, PI3K/AKT signaling pathway was analyzed by western blotting in the indicated groups.
miR-889-5p influences the regulatory effect of FABP5 on KLF9
To investigate the specific mechanism of FABP5-regulated KLF9 function, we analyzed the potential upstream target miRNA of KLF9 using targetscan v7.1 and miRDB databases. We focused on ten candidate miRNAs (as shown in ) related to HCC prognosis in the TCGA database. miR-889-5p, miR-26a-5p, miR-636, let-7d-3p, and miR-106b-5p were significantly decreased in the FABP5-knockdown HepG2 and SK-Hep-1 cells ( and Supplementary Figure 4). Compared with other potential miRNAs, the expression level of miR-889-5p was the lowest in the FABP5-knockdown HCC cells. Therefore, miR-889-5p was selected for further study. To investigate the relationship between miR-889-5p and FABP5, a dual luciferase assay was conducted in the 293 T cells. miR-889-5p was predicted to have a binding site at the 3-UTR of KLF9. The KLF9 mutation site is illustrated in . Luciferase activity was significantly decreased by treatment with pGL3-KLF9-wt and miR-889-5p mimics. However, the luciferase activity in cells transfected with pGL3-KLF9-mut showed no significant difference following treatment with the miR-899-5p mimic (). These results indicate that miR‑889-5p targets the 3’‑UTR of KLF9 mRNA directly. Next, we compared the miR-889-5p expression in HCC tissues to that in adjacent normal tissues. RT-qPCR analysis showed that miR-889-5p was significantly upregulated in tumor tissues (), while miR-889-5p expression was negatively associated with KLF9 expression in HCC tissues and positively associated with FABP5 expression in HCC tissues (). Next, the effect of miR-889-5p on FABP5-regulated KLF9 expression was further examined. The expression of KLF9 was increased by treatment with shFABP5; however, after combined treatment with the miR-889-5p mimic and shFABP5, the expression of KLF9 was significantly lower compared with the shFABP5 treatment group ). Overall, miR-889-5p may be associated with FABP5-regulated KLF9 expression.
Figure 7. miR-889-5p suppressed KLF9 expression by directly targeted KLF9. a, the expression of miR-889-5p was significantly decreased by FABP5 knockdown in HCC cells. b, bioinformatics analysis predicted that the 3ʹUTR sequence of KLF9 is complementary to the seed sequence of miR-889-5p.The core binding sequences of KLF9 were mutated (in red text). c, dual-luciferase reporter assay was performed to verify that miR-889-5p directly bound to the 3’-UTR sequences of KLF9 in the cells co-transfected with miR-889 mimics or NC with pGL3-KLF9-WT or pGL3-KLF9-Mut. d, relative expression of miR-889-5p in 48 HCC tissues compared with adjacent normal tissues was detected by RT-qPCR. E, miR-889-5p expression was negatively correlated with KLF9 expression and positively correlated with FABP5 expression in HCC tissues. f-g, increased expression of miR-889-5p attenuated KLF9 mRNA and protein expression of in HCC cells with shFABP5 treatment. *p < .05, **p < .01, and ***p < .001 as compared with the vehicle control.
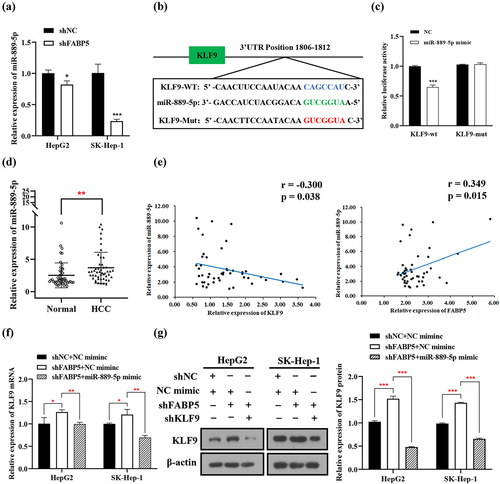
Table 3. Ten candidate miRNAs target KLF9 through targetscan v7.1 and miRDB databases.
FABP5 inhibits the expression of miR-889-5p by regulating CREB protein phosphorylation
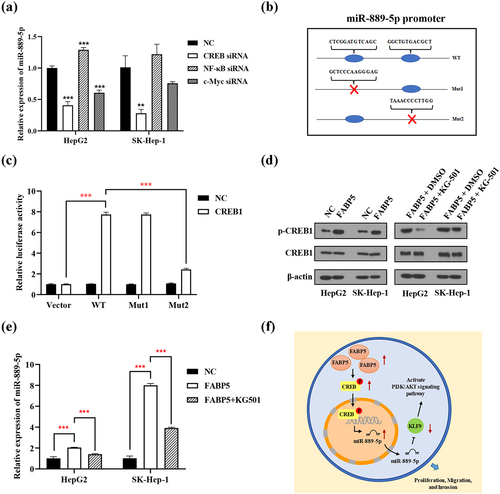
Both PROMO and JASPAR database were used to predict the transcription factors of miR-889-5p. Based on bioinformatics analysis and previous literatures, the transcription factors NF-κB, c-Myc, and CREB were selected candidates for transcriptional regulation of miR-889-5p promoter. NF-κB, c-Myc, and CREB siRNA were transiently transfected into HepG2 and SK-Hep-1 cells. The expression of miR-889-5p was detected using RT-qPCR. CREB knockdown reduced the expression of miR-889-5p in SK-Hep-1 and HepG2 cells. CREB, but not NF-κB and c-Myc, positively regulated the expression of miR-889-5p (). A luciferase reporter promoter system was used to confirm that CREB was binding to the miR-889-5p promoter. Wild-type (WT) or mutated miR-889-5p promoters (Mut-1 and Mut-2) were cloned into a luciferase reporter vector. The miR-889-5p mutation sites are illustrated in . Ectopic CREB expression markedly improved the luciferase activity of the WT and Mut-1 miR-889-5p promoters but not the Mut-2 promoter, implying that Mut-2 may be the primary binding positions of the CREB and miR-889-5p promoters (). Moreover, overexpression of FABP5 significantly increased the expression of phosphorylated CREB ( and Supplementary Figure 5). To investigate whether FABP5 upregulates miR-889-5p via CREB, the CREB inhibitor KG-501 was used. In the HCC cells overexpressing FABP5 treated with the CREB inhibitor KG-501, CREB phosphorylation were significantly decreased compared with the untreated FABP5 overexpression group (). The CREB inhibitor KG-501 reversed the effect of FABP5 overexpression on miR-889-5p expression (). Based on this evidence, we suggest that FABP5 promotes CREB phosphorylation and that this is a result of improved miR-889-5p transcriptional activity.
Figure 8. FABP5 inhibits the expression of miR-889-5p by regulating CREB protein phosphorylation. a, the siRNA of NF-κB, c-Myc and CREB were transiently transferred into HepG2 and SK-Hep-1 cells. The expression of miR-889-5p was detected by RT-qPCR. b, the designed mutative model of miR-889-5p promoter. c, a luciferase reporter promoter system was employed to confirm CREB bind to miR-889-5p promoter. d, the expression of total and phosphorylated CREB protein were detected by western blotting in the indicated groups. e, CREB inhibitor KG-501 reverse the effect of FABP5 overexpression on miR-889-5p expression in HCC cells. **p < .01 and ***p < .001 as compared with the vehicle control. e, the scheme of the mechanism by which FABP5 affects HCC tumorigenesis. FABP5 improves CREB protein phosphorylation to upregulate the miR-889-5p expression by CREB binding to the miR-889-5p promoter region, whereby leading to downregulation of KLF9 by miR-889-5p binding to the 3ʹ-UTR of the KLF9 mRNA, potentiating the PI3K/AKT signaling pathway and promoting the proliferation, migration, and invasion of HCC cells.
Discussion
In this study, we confirmed that FABP5 plays a role in HCC. We demonstrated that FABP5 is highly expressed in human HCC tissues, which confirms the findings of previous studies.Citation21 Moreover, we found that FABP5 was involved in HCC cell proliferation, migration, and invasion, which is in agreement with a previous study.Citation22 The specific mechanisms underlying the upregulation of FABP5 in various human tumors have also been discussed for other cancers. Overexpression of FABP5 in prostate cancer cells can be attributed to hypomethylation of the CpG island in its promoter region, along with upregulation of the direct trans-acting factors SP1 and c-MYC.Citation9 Hypoxia downregulated the expression of miR-144-3p, which subsequently increased the expression of FABP5 in cervical cancer.Citation13 The different regulatory networks and molecular mechanisms of FABP5 in numerous other cancers still require further study. In this study, we focused on the mechanism by which FABP5 influences HCC progression. Mechanically and most importantly, we found that the effects of FABP5 appear to work through the upregulation of phosphorylated CREB binding to the miR-889-5p promoter, which in turn leads to upregulation of miR-889-5p. Increased expression of miR-889-5p seems to contribute to the downregulation of KLF9, which in turn inhibits the PI3K-AKT signaling pathway. Notably, we made the novel discovery that FABP5 influences the progression of HCC via the CREB/miR-889-5p/KLF9/PI3K-AKT signaling pathway.
CREB belongs to the leucine zipper class of transcription factors and participates in numerous cellular processes, including proliferation, apoptosis, and autophagy.Citation23 Transcriptional regulation depends on CREB phosphorylation status.Citation24 Phosphorylated CREB enters the nucleus and binds to specific sites of cAMP response element (CRE) as a transcription factor, thus regulating the expression of target genes.Citation25 Accumulating evidence has established the proto-oncogenic role of CREB in tumorigenesis in many different types of malignancies. Previous studies have indicated that CREB was ubiquitously overexpressed in almost all malignant tumors and the overexpression of CREB could dramatically promote tumor proliferation and tumorigenesis in breast cancer, lung cancer, and liver cancer.Citation26–28 Moreover, the expression levels of CREB in HCC have been shown to be negatively correlated with prognosis.Citation29 In recent years, it has been found that CREB in human tumors regulates the expression of multiple miRNAs, such as miR-9 in glioma and miR-373 in pancreatic cancer.Citation30,Citation31 Interestingly, we found strong evidence that FABP5 regulates CREB phosphorylation. Further in vitro experiments proved that the upregulation of FABP5 in HCC could increase the expression of miR-889-5p via CREB, which enhances the promoter activity of miR-889-5p. But whether FABP5 is the direct kinase of CREB and how FABP5 regulates CREB phosphorylation have not been determined, which needs further research.
Previous studies have demonstrated that miR-889 is dysregulated in various cancers, such as colon cancer, osteosarcoma, and esophageal cancer. miR-889 promotes colon cancer cell proliferation by inhibiting DAB2IP transcription.Citation32 miR-889 can inhibit the expression of myeloid cell nuclear differentiation antigen (MNDA) targets cell cycle regulation in order to influence the progression of osteosarcoma.Citation33 However, miR-889 has been shown to have the opposite effect on non-small cell lung cancers (NSCLCs). miR-889 expression levels were found to be significantly reduced in NSCLC, and miR-889 overexpression suppressed the proliferation and invasiveness of NSCLC cells.Citation34 These observations suggest that miR-889 may play different roles in different cancers. However, its expression in liver cancer is not well understood. In our study, we found that miR-889-5p expression was significantly upregulated in HCC tissues.
Based on gene chip analyses, we identified KLF9 as a downstream gene target of FABP5. Krüppel-like factors (KLF) is a family of transcription factors with three highly conserved C2H2 zinc finger structures at the carboxyl end, which is widely found in various tissues and has been shown to control essential cellular processes such as proliferation, apoptosis, migration, and differentiation.Citation35,Citation36 KLF9, one of the most important members of the KLF family, regulates gene expression by activating or inhibiting promoters with rich GC boxes.Citation37 Aberrant expression of KLF9 leads to cellular disorders and influences the occurrence and development of various diseases, including cancer. Emerging studies have demonstrated that KLF9 is commonly downregulated and serves as a tumor suppressor in multiple malignancies, which is related to the poor prognosis of cancer patients with downregulated KLF9 expression.Citation38–40 Fu et al.Citation41 reported that KLF9 mRNA and protein levels were decreased in HCC tissues compared with normal liver tissues and that the upregulation of KLF9 has anti-proliferative and pro-apoptotic properties in HepG2 cells. Sun et al.Citation42 obtained similar results and found that by binding to the p53 promoter, KLF9 upregulates p53 levels, and KLF9 overexpression significantly promotes tumor regression in xenograft models. These results suggest an antitumor role for KLF9 in HCC. In the present study, we demonstrated that FABP5 promoted the proliferation and migration of HCC cells by inhibiting KLF9 expression. KLF9 was significantly downregulated in HCC tissues and was negatively associated with FABP5 expression.
KLF9 is regulated by various miRNAs. Huang et al.Citation43 reported that miR-140-5p significantly suppressed the expression of KLF9 by binding to the 3ʹ-UTR of KLF9 mRNA, which promoted the progression of renal cell carcinoma. He et al.Citation44 reported that miR-636 promotes the proliferation of bladder cancer cells by decreasing the expression of KLF9 upon binding to the 3’-UTR of its mRNA. miR-135a-5p and miR-135b-5p are involved in FABP5-regulated KLF9 expression in colorectal cancer.Citation45 Interestingly, we found that miR-889-5p can target the 3ʹ-UTR of KLF9 mRNA. miR-889-5p expression was positively correlated with FABP5 expression and negatively correlated with KLF9 expression. In addition, treatment with the miR-889-5p mimic attenuated the increase in KLF9 expression induced by FABP5 knockout. These results suggest that FABP5 inhibits the expression of KLF9 by miR-889-5p, which promotes the proliferation and migration of HCC cells.
The results of transcriptional profiling demonstrated that FABP5 knockdown obviously inhibited the PI3K/AKT signaling pathway. PI3K/AKT pathway activation by extracellular signals has been observed in HCC and contributes to tumorigenesis.Citation46 Intriguingly, LV et al. found that FABP5 regulated the proliferation of clear cell renal cell carcinoma via the PI3K/AKT signaling pathway.Citation47 In the present study, FABP5 silencing significantly decreased the expression of PI3K, p-AKT, and p-mTOR. We hypothesized that FABP5 may regulate the PI3K/AKT signaling pathway via KLF9. To test this hypothesis, KLF9 was silenced in FABP5 knockdown HCC cells. The results demonstrated that KLF9 silencing reversed the p-PI3K, p-AKT, and p-mTOR expression levels in HCC cells, suggesting that FABP5 regulates the PI3K/AKT signaling pathway by inhibiting KLF9 expression.
Conclusion
In summary, our study demonstrated that FABP5 promotes the proliferation and migration of HCC through the CREB/miR-889-5p/KLF9 axis and provided novel insights into the pathogenesis of HCC. The schematic representation was as shown in . The discovery of this signaling pathway and its critical involvement in the progression of HCC serves as the first step in improving our understanding of HCC and aiding treatment. There are some limitations of the current study. First, the number of clinical samples was relatively small, the results still need to verify by larger number of cohorts. Second, we confirmed that FABP5 affect CREB protein phosphorylation, but the mechanism of regulation has not been studied in this study. Although we revealed the involvement of FABP5, CREB, miR-889-5p and KLF9 in HCC progression, we cannot prove entirely that they constitute a single signaling pathway. Therefore, further experiments could identify and confirm the complete upstream-downstream relationship of these signaling molecules.
Notes on contributions
TY performed the experiments, analyzed data, and wrote the first draft of the manuscript. LK helped design the study and reviewed the manuscript. CZ and TH performed the experiments. HB supervised the experiments. LJ collected and performed the statistical analysis. CJ contributed to conception and design of the study. All authors have read and approved the manuscript in its current state.
Ethical statement
This study which reports in vivo experiments on humans or animals was approved by the Ethical Committee of Guangxi Medical University Cancer Hospital.
Supplemental Material
Download Zip (8.7 MB)Acknowledgments
We thank all members participated in this work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384047.2022.2094670
Additional information
Funding
Notes on contributors
Yanping Tang
Yanping Tang, Medicine M.D, currently works at Guangxi Medical University Cancer Hospital (China), mainly study the pathogenesis and chemical prevention of liver cancer.
References
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature. 463(7283):899–905. DOI:10.1038/nature08822
- Mattison J, Kool J, Uren AG, de Ridder J, Wessels L, Jonkers J, Bignell GR, Butler A, Rust AG, Brosch M, et al. Novel candidate cancer genes identified by a large-scale cross-species comparative oncogenomics approach. Cancer Res. 2010;70(3):883–895. DOI:10.1158/0008-5472. CAN-09-1737.
- Lu X, Sun W, Tang Y, Zhu L, Li Y, Ou C, Yang C, Su J, Luo C, Hu Y, et al. Identification of key genes in hepatocellular carcinoma and validation of the candidate gene, cdc25a, using gene set enrichment analysis, meta-analysis and cross-species comparison. Mol Med Rep. 2016;13(2):1172–1178. DOI:10.3892/mmr.2015.4646
- LiY, WanDF, SuJJ,CaoJ, OuC, QiuXK, BanKC, YangC,QinLL, LuoD, etal. Differential expression of genes during aflatoxin B (1)-induced hepatocarcinogenesis in tree shrews. World JGastroenterol. 2004;10(4):497–504.DOI:10.3748/wjg.v10.i4.497
- Liang HJ, Wei W, Kang XN, Guo K, Cao J, Su JJ, Yang C, Yang C, Li Y, Liu YK, et al. Differentially expressed proteins in the precancerous stage of rat hepatocarcinogenesis induced by diethylnitrosamine. Chin J Hepato. 2009;17(9):669–674. DOI:10.3760/cma.j.1007-3418.2009.09.008.
- Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr. 2008;28(1):73–95. doi:10.1146/annurev.nutr. 27.061406.093710.
- Zimmerman AW, Veerkamp JH. 2002. New insights into the structure and function of fatty acid-binding proteins. Cell Mol Life Sci. 59(7):1096–1116. DOI:10.1007/s00018-002-8490-y.
- Hotamisligil GS. 2006. Inflammation and metabolic disorders. Nature. 444(7121):860–867. doi:10.1038/nature05485.
- Kawaguchi K, Kinameri A, Suzuki S, Senga S, Ke Y, Fujii H. 2016. The cancer-promoting gene fatty acid-binding protein 5 (FABP5) is epigenetically regulated during human prostate carcinogenesis. Biochem J. 473(4):449–461. doi:10.1042/BJ20150926.
- Wang Y, Wahafu A, Wu W, Xiang J, Huo L, Ma X, Wang N, Liu H, Bai X, Xu D, et al. 2021. FABP5 enhances malignancies of lower-grade gliomas via canonical activation of NF-κB signaling. J Cell Mol Med. 25(9):4487–4500. DOI:10.1111/jcmm.16536
- Apaya MK, Hsiao PW, Yang YC, Shyur LF. 2020. Deregulating the CYP2C19/Epoxy-Eicosatrienoic acid-associated FABP4/FABP5 signaling network as a therapeutic approach for metastatic triple-negative breast cancer. Cancers (Basel). 12(1):199. doi:10.3390/cancers12010199.
- Kawaguchi K, Senga S, Kubota C, Kawamura Y, Ke Y, Fujii H. 2016. High expression of fatty acid-binding protein 5 promotes cell growth and metastatic potential of colorectal cancer cells. FEBS Open Bio. 6(3):190–199. doi:10.1002/2211-5463.12031.
- Zhang C, Liao Y, Liu P, Du Q, Liang Y, Ooi S, Qin S, He S, Yao S, Wang W. FABP5 promotes lymph node metastasis in cervical cancer by reprogramming fatty acid metabolism. Theranostics. 2020;10(15):6561–6580. doi:10.7150/thno. 44868.
- Carbonetti G, Wilpshaar T, Kroonen J, Studholme K, Converso C, d’Oelsnitz S, Kaczocha M. 2019. FABP5 coordinates lipid signaling that promotes prostate cancer metastasis. Sci Rep. 9(1):18944. DOI:10.1038/s41598-019-55418-x.
- Seo J, Jeong DW, Park JW, Lee KW, Fukuda J, Chun YS. 2020. Fatty-acid-induced FABP5/HIF-1 reprograms lipid metabolism and enhances the proliferation of liver cancer cells. Commun Biol. 3(1):638. DOI:10.1038/s42003-020-01367-5.
- Senga S, Kobayashi N, Kawaguchi K, Ando A, Fujii H. 2018. Fatty acid-binding protein 5 (FABP5) promotes lipolysis of lipid droplets, de novo fatty acid (FA) synthesis and activation of nuclear factor-kappa B (NF-κB) signaling in cancer cells. Biochim Biophys Acta Mol Cell Biol Lipids. 1863(9):1057–1067. DOI:10.1016/j.bbalip.2018.06.010.
- Pan L, Xiao H, Liao R, Chen Q, Peng C, Zhang Y, Mu T, Wu Z. 2018. Fatty acid binding protein 5 promotes tumor angiogenesis and activates the IL6/ STAT3/ VEGFA pathway in hepatocellular carcinoma. Biomed Pharmacother. 106:68–76. DOI:10.1016/j.biopha.2018.06.040.
- Llovet J, Bru C, Bruix J. 1999. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 19(3):329–338. DOI:10.1055/s-2007-1007122.
- Winer J, Jung CKS, Shackel I, Williams PM. 1999. Development and validation of real - time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Analytical Biochem. 270(1):41–49. DOI:10.1006/abio.1999.4085.
- Simmen RC, Pabona JM, Velarde MC, Simmons C, Rahal O, Simmen FA. The emerging role of Krüppel-like factors in endocrine-responsive cancers of female reproductive tissues. J Endocrinol. 2010;204(3):223–231. DOI:10.1677/JOE-09-0329
- Ohata T, Yokoo H, Kamiyama T, Fukai M, Aiyama T, Hatanaka Y, Hatanaka K, Wakayama K, Orimo T, Kakisaka T, et al. 2017. Fatty acid-binding protein 5 function in hepatocellular carcinoma through induction of epithelial-mesenchymal transition. Cancer Med. 6(5):1049–1061. DOI:10.1002/cam4.1020
- Ohira M, Yokoo H, Ogawa K, Fukai M, Kamiyama T, Sakamoto N, Taketomi A. 2021. Serum fatty acid-binding protein 5 is a significant factor in hepatocellular carcinoma progression independent of tissue expression level. Carcinogenesis. 42(6):794–803. DOI:10.1093/carcin/bgab025.
- Sapio L, Salzillo A, Ragone A, Illiano M, Spina A, Naviglio S. 2020. Targeting CREB in cancer therapy: a key candidate or one of many? An update. Cancers (Basel). 12(11):3166. DOI:10.3390/cancers12113166.
- Yamamoto KK, Gonzalez GA, Biggs WH, Montminy MR. 1988. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 334(6182):494–498. DOI:10.1038/334494a0.
- Han J, Li E, Chen L, Zhang Y, Wei F, Liu J, Deng H, Wang Y. 2015. The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature. 524(7564):243–246. DOI:10.1038/nature14557.
- Hu PC, Li K, Tian YH, Pan WT, Wang Y, Xu XL, He YQ, Gao Y, Wei L, Zhang JW, et al. CREB1/Lin28/miR-638/VASP interactive network drives the development of breast cancer. Int J Biol Sci. 2019;15(12):2733–2749. DOI:10.7150/ijbs.36854.eCollection2019
- Chen Z, Li JL, Lin S, Cao C, Gimbrone NT, Yang R, Fu DA, Carper MB, Haura EB, Schabath MB, et al. 2016. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest. 126(6):2267–2279. DOI:10.1172/JCI85250
- Wang J, Ma L, Weng W, Qiao Y, Zhang Y, He J, Wang H, Xiao W, Li L, Chu Q, et al. 2013. Mutual interaction between YAP and CREB promotes tumorigenesis in liver cancer. Hepatology. 58(3):1011–1020. DOI:10.1002/hep.26420
- Li CJ, Lin HY, Ko CJ, Lai JC, Chu PY. 2020. A novel biomarker driving poor-prognosis liver cancer: overexpression of the mitochondrial calcium gatekeepers. Biomedicines. 8(11):451. DOI:10.3390/biomedicines8110451.
- Tan X, Wang S, Yang B, Zhu L, Yin B, Chao T, Zhao J, Yuan J, Qiang B, Peng X, et al. The CREB-miR-9 negative feedback minicircuitry coordinates the migration and proliferation of glioma cells. PLoS One. 2012;7(11):e49570. DOI:10.1371/journal.pone.0049570
- Zhang Y, Yang J, Cui X, Chen Y, Zhu VF, Hagan JP, Wang H, Yu X, Hodges SE, Fang J, et al. A novel epigenetic CREB-miR-373 axis mediates ZIP4-induced pancreatic cancer growth. EMBO Mol Med. 2013;5(9):1322–1334. DOI:10.1002/emmm.201302507
- Xiao Y, Li ZH, Bi YH. 2019. MicroRNA-889 promotes cell proliferation in colorectal cancer by targeting DAB2IP. Eur Rev Med Pharmacol Sci. 23(8):3326–3334. DOI:10.26355/eurrev_201904_17695.
- Ge D, Chen H, Zheng S, Zhang B, Ge Y, Yang L, Cao X. Hsa-miR-889-3p promotes the proliferation of osteosarcoma through inhibiting myeloid cell nuclear differentiation antigen expression. Hsa-miR-889-3p promotes the proliferation of osteosarcoma through inhibiting myeloid cell nuclear differentiation antigen expression. Biomed Pharmacother. 2019;114:108819. doi:10.1016/jbiopha.2019.108819
- Dong Z, Li BC, Wang XJ. 2019. ΜicroRNA-889 plays a suppressive role in cell proliferation and invasion by directly targeting TAB1 in non-small cell lung cancer. Mol Med Rep. 20(1):261–269. DOI:10.3892/mmr.2019.10245.
- McConnell BB, Yang VW. 2010. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 90(4):1337–1381. DOI:10.1152/physrev.00058.2009.
- Kaczynski J, Cook T, Urrutia R. 2003. Sp1- and Krüppel-like transcription factors. Genome Biol. 4(2):206. DOI:10.1186/gb-2003-4-2-206.
- Imataka H, Sogawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11(10):3663–3671. doi:10.1002/j.1460-2075.1992.tb05451.x.
- Li Y, Sun Q, Jiang M, Li S, Zhang J, Xu Z, Guo D, Gu T, Wang B, Xiao L, et al. KLF9 suppresses gastric cancer cell invasion and metastasis through transcriptional inhibition of MMP28. FASEB J. 2019;33(7):7915–7928. DOI:10.1096/fj.201802531R.Epub2019Mar26
- Brown AR, Simmen RC, Raj VR, Van TT, MacLeod SL, Simmen FA. 2015. Krüppel -like factor 9 (KLF9) prevents colorectal cancer through inhibition of interferon-related signaling. Carcinogenesis. 36(9):946–955. DOI:10.1093/carcin/bgv104.
- Bai XY, Li S, Wang M, Li X, Yang Y, Xu Z, Li B, Li Y, Xia K, Chen H, et al. 2018. Krüppel-like factor 9 down-regulates matrix metalloproteinase 9 transcription and suppresses human breast cancer invasion. Cancer Lett. 412:224–235. DOI:10.1016/j.canlet.2017.10.027.
- Fu DZ, Cheng Y, He H, Liu HY, Liu YF. 2014. The fate of Krüppel-like factor 9-positive hepatic carcinoma cells may be determined by the programmed cell death protein 5. Int J Oncol. 44(1):153–160. DOI:10.3892/ijo.2013.2147.
- Sun J, Wang B, Liu Y, Zhang L, Ma A, Yang Z, Ji Y, Liu Y. Transcription factor KLF9 suppresses the growth of hepatocellular carcinoma cells in vivo and positively regulates p53 expression. Cancer Lett. 2014;355(1):25–33. DOI:10.1016/j.canlet.2014.09.022
- Huang C, Li J, Zhang X, Xiong T, Ye J, Yu J, Gui Y. 2020. The miR-140-5p/KLF9/ KCNQ1 axis promotes the progression of renal cell carcinoma. FASEB J. 34(8):10623–10639. DOI:10.1096/fj.202000088RR.
- He Q, Huang L, Yan D, Bi J, Yang M, Huang J, Lin T. 2019. CircPTPRA acts as a tumor suppressor in bladder cancer by sponging miR-636 and upregulating KLF9. Aging (Albany NY). 11(23):11314–11328. DOI:10.18632/aging.102530.
- Zhang Y, Zhang Z, Yi Y, Wang Y, Fu J. 2020. CircNOL10 acts as a sponge of miR-135a/b-5p in suppressing colorectal cancer progression via regulating KLF9. Onco Targets Ther. 13:5165–5176. DOI:10.2147/OTT.S242001.
- Khemlina G, Ikeda S, Kurzrock R. The biology of hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer. 2017;16(1):149. DOI:10.1186/s12943-017-0712-x.
- Lv Q, Wang G, Zhang Y, Han X, Li H, Le W, Zhang M, Ma C, Wang P, Ding Q. FABP5 regulates the proliferation of clear cell renal cell carcinoma cells via the PI3K/AKT signaling pathway. Int J Oncol. 2019;54(4):1221–1232. DOI:10.3892/ijo.2019.4721
